Abstract
Ensemble learning is an important element in machine learning. However, two essential tasks, including training base classifiers and finding a suitable ensemble balance for the diversity and accuracy of these base classifiers, are need to be achieved. In this paper, a novel ensemble method, which utilizes a multimodal multiobjective differential evolution (MMODE) algorithm to select feature subsets and optimize base classifiers parameters, is proposed. Moreover, three methods including minimum error ensemble, all Pareto sets ensemble, and error reduction ensemble are employed to construct ensemble classifiers for executing classification tasks. Experimental results on several benchmark classification databases evidence that the proposed algorithm is valid.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The advent of the information age prompts us to mine valuable knowledge from big data and complete diverse classification tasks, forming an extensive research field, i.e., machine learning that includes a variety of methods. Thereinto, ensemble learning receives widespread attention from researchers owing to its more dependable accuracy and generalization performance than an individual classifier. Hence, numerous ensemble learning algorithms have been employed in a variety of areas, such as texture image classification [1], medical information analysis [2] and synthetic aperture radar image classification [3].
Ordinarily, ensemble learning consists of two steps, training a set of base learners and integrating predictions of these learners. As for training base classifiers, the most prevailing strategies are Bagging [4], Adaboost [5], random forest [6], rotation forest [7]. Recently, many studies focus on employing feature selection to train different classifiers. For example, in [8], an optimal feature and instance subsets were obtained by embedding both parameters searching in a multiobjective evolutionary algorithm with a wrapper approach. And in [9], the Pareto sets of image features obtained from a multiobjective evolutionary trajectory transformation algorithm was utilized for generating base classifiers. Meanwhile, an increasing number of researches have coped with feature selection (FS), which can be generally classified into three sorts: filter [10, 11], wrapper [12,13,14] and embedded methods [15]. As for filter, feature selection is independent of the generalization performance of the learning algorithms by scoring and ranking features. Thus, the selected feature subsets may not enhance the performance of the classification algorithms. In contrast, wrapper methods utilize search strategies to quest the optimal feature subsets and evaluate these subsets by learning algorithms. Obviously, wrapper methods have a larger amount of computation than filter approaches but more credible. As for embedded approaches, feature selection and learner are incorporated in a single model, such as decision tree learner. In feature selection, studies always focus on two aspects: classification accuracies of learners and the size of selected feature subsets. Actually, multiple feature subsets of the same number of features can be able to achieve the same accuracy. If unimodal multiobjective evolutionary methods are utilized to deal with these problems, only one of them may be retained, which may cause some excellent feature subsets to be lost. These studies do not consider the multimodal [16] of Pareto sets in multiobjective optimization problems. Specifically, different solutions could have the same objective results.
Motivated to solve this problem, we utilize the evolutionary algorithms (EAs), which are highly popular on multimodal multiobjective issues. Usually, they are called multimodal multiobjective EAs (MMOEAs). Recently, there are numerous multi-objective evolutionary algorithms [17, 18]. Meanwhile, several MMOEAs [19,20,21] are proposed to solve multimodal multiobjective optimization (MMO) issues that may exist multiple Pareto sets which corresponds to the same Pareto front (PF) point. In [19], a multiobjective PSO by means of ring topology was proposed, which could produce stable niches and employ a special crowding distance. Here, we concentrate on utilizing an MMOEA for generating the base classifiers ELMs by optimizing feature subsets and the number of ELMs hidden nodes simultaneously then constructing an ensemble model in different ways.
In this paper, we present an ensemble method via multimodal multiobjective differential evolution (EMMODE), a novel approach that performs MMOEA to optimize the size of feature sets and the performance of ELM, by way of feature selection and selecting the number of hidden nodes. Due to the characteristics of MMODE, we are able to get a series of non-dominated solutions from it. As for the strategies of combining the base classifiers, EMMODE fulfills an operation on the Pareto sets for constructing them into an ensemble. This intent is accomplished by three strategies that are: (1) minimum error ensemble; (2) all Pareto sets ensemble, and; (3) error reduction ensemble approach. The experiments are conducted for classification problems. The experiments results of benchmark datasets from the UCI Machine Repository [22] show the effectiveness of our proposal, being capable to obtain multiple solutions with the same number of features and similar classification accuracy. Meanwhile, the EMMODE is also able to achieve solutions with an excellent tradeoff between the reduction rate in the number of features and accuracy.
The rest of this paper is organized as follows. Section 2 describes the related works. Section 3 details the EMMODE methods. The experimental settings and results are introduced in Sect. 4. Finally, Sect. 5 is the conclusion.
2 Related Works
This section describes related works on multimodal multiobjective optimization and feature selection for ensemble learning.
2.1 Multimodal Multiobjective Optimization
MMO problems are those which have multiple Pareto sets corresponding to the same PF [23]. Evidently, it is significant to find all Pareto solutions which are equivalent to PF. Give an example, decision-makers can use more Pareto sets to solve the real-world tasks. The MMO problem is vividly demonstrated in Fig. 1, where three Pareto solutions with similar objective values.
In the real world, many applications belong to MMO problems [24], which conclude optimization of truss-structures, metabolic network modeling, femtosecond laser pulse shaping problem, automatic determination of point, and so on. To solve these problems, many MMOEAs have been proposed. In [25], a multimodal multiobjective differential evolution (MMODE) algorithm which formulated a decision variable preselection strategy was proposed. The niching mechanism was employed in Niching-CMA [26] and MO-Ring-PSO-SCD [19]. In this paper, we use the MMODE, due to its good performance on MMO problems [27].
MMODE is an enhanced version of differential evolution (DE). The process of MMODE is as follows. Firstly, users define a possible solution search space \(\varOmega _{d} \subseteq \varOmega \). The boundary can be just defined as the endpoint of the actual range of values for the decision variables. In particular, the bounds of each element \(X^{(e)}\) in the decision variables \(x=\left( X^{(1)}, X^{(2)}, \ldots , X^{(m)}\right) ^\mathrm{T} \in R^{m}\) are indicated as \(X^{(e), \mathrm{L}} \le X^{(e)} \le X^{(e), \mathrm{U}}\), in which the \(X^{(e), \mathrm{L}}\) and \(X^{(e), \mathrm{U}}\) are applied to express the lower and upper values of the solution space for the element in the decision vectors respectively, and the variable m is defined as problem dimension [25]. Secondly, initialize the population P which consists of N individuals. There is a common method for initialization.
where the sub index 1 in \(X_{i, 1}^{(e)}\) is utilized to express the element of an initial decision solution, \(i=1,2, \dots , N\), and \(e=1,2, \ldots , m\). Meanwhile, the rand(0, 1) produces random real numbers between 0 and 1.
The next step is the preselection scheme, which applies both an objective space crowding distance (SCD) and a decision SCD indicators, to select population Q of size N/2 for producing offspring. In addition, let the notation \(x_{i, G}=\left( X_{i, G}^{(1)}, X_{i, G}^{(2)}, \dots , X_{i, G}^{(m)}\right) ^\mathrm{T}\) denotes the selected individual of the G-th generation whose elements \(X_{i, G}^{(e)}\), \(e=1,2, \ldots , m\) are subjected to MMODE mutation. Then generate a mutation vector \(v_{i, G}=\left( V_{i, G}^{(1)}, V_{i, G}^{(2)}, \ldots , V_{i, G}^{(m)}\right) ^\mathrm{T}\). One possible way for obtaining the elements of the mutation vector is the DE/rand/2 technique [25] that is as follows.
where \(V_{i, G}^{(e)}\) is the e-th element of the mutation vector, \(X_{r_{s}^{i}, G}^{(e)}\) is the e-th element of \(x_{r_{r}^{i}, G}\), and the indices \(r_{k}^{i}\), \(k=1,2, \ldots , 5\), are randomly selected integers in the [1, N/2]. The factors \(F_{1} \in (0,1)\) and \(F_{2} \in (0,1)\) are scaling factors of difference terms, and set \(F_{1}=F_{2}=F\) in MMODE. If a left-hand side value of Eq. (2) was outside the decision space boundary, the MMODE would implement an alternative mutation bound scheme
Then, use a common method to implement crossover process:
where the cross probability \(Cr \in (0,1)\) is set by users. And a vector \(u_{i}=\left( U_{i}^{(1)}, U_{i}^{(2)}, \ldots , U_{i}^{(m)}\right) ^\mathrm{T} \in R^{m}\) stores the crossover results. For the case of MMO problems, MMODE applies the following selection offspring generation method:
In addition, when neither branch is true in i-th individual, vector \(u_{i}\) would be added to the \(c_{i, G}\).
Finally, splice \(c_{i, G}\) and \(x_{i, G}\), and use a nondominated sorting scheme on the spliced vector to generate the \((G + 1)\) – st generation. A complete algorithm for conducting MMODE is shown in [25].
2.2 Feature Selection for Ensemble Learning
In this subsection, we classify feature selection methods in two kinds: ordinary feature selection algorithms and feature selection by evolutionary algorithms.
On the one hand, ordinary feature selection algorithms exist several defects, such as difficult to set the value of important parameters, nesting effect, falling into local optima. For instance, common feature selection methods, Sequential Forward Selection (SFS) [28] and Sequential Backward Selection (SBS) [29], affect by the nesting effect [30].
On the other hand, evolutionary algorithms [31, 32] supply a valid strategy for coping with feature selection owing to the three reasons: (1) We can acquire quite acceptable feature subsets without searching the entire decision space. (2) They are capable to search the decision space comprehensively. (3) They get over falling into local optima and nesting effect for they set no restriction on selecting features. Recently, an increasing number of studies apply evolutionary approaches for feature selection. For example, classic EAs such as PSO [33], DE [34], GA [35], and ACO [36] were used. The above-mentioned methods utilized a single objective or multiobjective EAs to select feature subsets. In addition, there also exist some studies optimizing both the learners parameters and feature selection. For example, [35] encoded the parameters of support vector machine (SVM) and the feature subsets into GA chromosomes.
Moreover, to evaluate feature subsets, we employ Extreme Learning Machine (ELM) as base classifier of an ensemble. ELM [37] is an efficient method for single-hidden layer feedforward neural networks (SLFNs). ELM randomly generates the parameters of hidden nodes and input weights, while the output weights are determined analytically. In [38], the universal approximation and effective generalization performance of ELM are proved. Compared with conventional learning methods, such as the back-propagation algorithm (BP) and SVM, ELM can learn extremely fast because it need not adjust network parameters iteratively. For ELM, it is vital that hidden layer parameters, especially the number of hidden nodes for the generalization performance. Thus, Huang et al. proposed an Incremental Extreme Learning Machine (I-ELM) method by adding the hidden nodes one by one [38]. Another method called Error Minimized ELM (EM-ELM) is proposed in [39]. The difference from I-ELM is that EM-ELM adjusts all output weights iteratively when it adds one or more new hidden nodes. In [40], an improved method of EM-ELM called Incremental Regularized Extreme Learning Machine (IR-ELM) is proposed by utilizing the RELM. However, the expected termination accuracy may be difficult to set in the real world, which can cause the overfitting or fail to achieve the desired testing accuracy.
Based on the above, the purposes of feature selection and the classifiers model selection are obtaining the representation of datasets and appropriate model parameters that are adequate for classification tasks. In addition, there exists more than one such solution in the same objectives. Thus, MMODE is utilized to overcome these drawbacks in this paper.
3 The EMMODE Method
In this section, we present the EMMODE approach formulating the feature and model parameters selection as a multimodal multiobjective one. The flow chart of the EMMODE method is depicted in Fig. 2. Like DE, this process begins with the initial population generation whose each individual encodes a possible solution. For each individual, calculate its error rate and feature selection rate on the datasets. After that, new individuals are generated by means of differential evolution operations over the existing ones. Then, repeated this process iteratively until a termination condition is satisfied. The detail of the EMMODE approach is introduced as follows.
3.1 Encoding
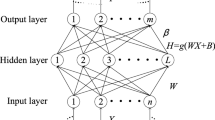
MMODE works by chromosomes whose each chromosome encodes a potentially feasible solution for the optimization task, i.e., the number of hidden nodes for an ELM and the selected feature subsets. The first process is to encode a potentially feasible solution for the task. In this paper, the feature is encoded in a binary variable demonstrating if the corresponding feature is selected. As for the parameter of ELM, the number of selected hidden nodes is encoded with an integer variable. The encoding of the chromosome is shown in Fig. 3.
3.2 Evolutionary Operators and Fitness Functions
This subsection presents the evolutionary operators which are different from MMODE, namely the mutation process and mutation-bound process. Meanwhile, the fitness functions for determining the quality of a solution are explained. For the model and feature selection task, the range of decision variables is relatively small, thus this paper uses fewer perturbation vectors and a mutation vector is generated by the DE/rand/1 method, which is shown in the mutation equation.
where \(V_{i, G}^{(e)}\), \(X_{r_{s}^{i}, G}^{(e)}\), \(r_{\ell }^{i}\), \(\ell =1,2,3\), and \(F \in (0,1)\) represent the same meaning as mentioned in the Eq. (2) formula.
As for the mutation-bound process, if a left-hand side value of Eq. (6) was outside the decision space boundary, the EMMODE would implement an alternative mutation bound scheme
Our aim is to attain optimal feature subsets and a corresponding number of the hidden nodes. We optimize the following functions: the feature selection rate (\(f_{1}\)) and the error rate (\(f_{2}\)).
where \(\mathcal {F}\) and \(\mathcal {S}\) represent the total number of features and number of selected features, respectively. \(T(\varvec{\theta })\) is the number of correctly classified samples for each corresponding selected feature subset, and N is the number of total samples.
The proposed approach aims to explore the space of parameter and FS techniques for attaining the solutions that suffice the best trade-off. Moreover, the result of a multimodal multiobjective optimization is not a single solution, but a series of them. The next subsection presents the methods of integrating a final classification model.
3.3 Ensembles Strategy
In this subsection, we focus on enhancing the prediction accuracy and generalization. All these Pareto solutions obtained from MMODE are equally appropriate for the task when no other preference information is used. Nevertheless, in the task we face, the purpose is to construct an ensemble with both a selected feature set and corresponding an ELM model, which is employed in the classification. Thus, it is significant to perform an ensemble step over the trade-off solutions so as to acquire a final classification model. In this case, we integrate the Pareto solutions of MMODE to reduce the risk of choosing an unstable solution and provide a better approximation to the optimal solutions.
Each solution in nondominated sets corresponds to an ELM-classifier trained with different parameter and different subsets of the original feature set. An ensemble of classifiers can combine the individual information acquired from each model and provide more information on the predicted label than a single classifier. In this regard, we study three different strategies of combining the ELMs which are described in the following.
-
(1)
All Pareto Sets Ensemble (APSE): The basic method here is to construct an ensemble applying all Pareto solutions of MMODE.
-
(2)
Minimum Error Ensemble (MEE): The opinion of this method does not use all Pareto solutions, but a subset of them. The ensemble consists of n solutions which have low error rate. As recommended in [41], it satisfies to combine 5 to 35 ELMs for most practical applications. In view of this, we set the n as 11. The ensemble \(\mathcal {E}\) is defined by the equation
$$\begin{aligned} \mathcal {E}=\underset{n}{{\text {argmin}}}\,\mathcal {PS}_{2} \end{aligned}$$(9)where the index n is the number of the selected ELMs, and \(\mathcal {PS}_{2}\) indicates the second target value of the Pareto solutions, namely error rate.
-
(3)
Error Reduction Ensemble (ERE): This approach is also not to employ all solutions. First, the solutions in the Pareto sets are sequenced in ascending order according to the corresponding error rate, and the solutions with the error greater than 0.5 are eliminated. Second, the misclassification samples numberings of each classifier constructed by the Pareto solutions in the dataset classification are stored in the matrix Misnum. Third, set the matrices \(Misnum_{1}, Misnum_{2}\) of the first two solutions in the sorted Pareto solution set as the reference matrices and decipher the two as part of the integration. And then, operate on each of the remaining solutions. For example, for the i-th \((i>2)\) solution, calculate the number of identical elements of its matrix Misnum and two reference matrices.
$$\begin{aligned} {\left\{ \begin{array}{ll} a_{i 1}= Misnum_{i} \cap Misnum_{1}\\ a_{i 2}= Misnum_{i} \cap Misnum_{2}\\ q_{i}=numel(a_{i 1})+numel(a_{i 2}) \end{array}\right. } \end{aligned}$$(10)where the \(a_{i 1}\) is the intersection matrix of matrix \(Misnum_{1}\) and matrix \(Misnum_{i}\), \(a_{i 2}\) denotes the intersection matrix of matrix \(Misnum_{2}\) and matrix \(Misnum_{i}\). And \(q_{i}\) represents the sum of the number of elements of the matrix \(a_{i 1}\) and the number of elements of the matrix \(a_{i 2}\).
Next, sort the quantity values of identical elements in ascending order and select the solutions corresponding to the first \(n-2\) values as part of the integration. At last, these n base classifiers form the final ensemble model. The main framework is demonstrated in Algorithm 1.

These schemes introduce different approaches to select the ELM classification model from Pareto sets. The next procedure is that integrating the results obtained by base learners to acquire a final prediction. We deal with this problem in the following. When we combine the models into an ensemble model, we take a majority voting to acquire the final prediction of the model.
4 Experiments and Results
In this section, we present the experiments implemented and the results acquired by the proposed approach by means of different classification datasets.
4.1 Experimental Settings
For our study, we used 6 datasets available in the UCI repository. Table 1 shows the characteristics of these datasets, such as the number of samples, the number of classes and the number of features. In our experiments, the results are the mean values by ten executes of ten-fold cross-validation. The process of EMMODE is a nested loop: as for inner loop, one-third of the training dataset is set as a validation set randomly to estimate each solution, while the rest is applied to train learners. In outer loop, these datasets are divided into ten subsets previously using the k-fold cross-validation method. In ten-fold cross-validation, a dataset is partitioned into ten subsets [42], and other processes are similar to the above.
We apply two standards to evaluate the performance of the EMMODE. One is the testing accuracy, and the other is the selection rate attained in the FS. In our experiments, the population size and maximum fitness evaluation are set to 100 and 5000. While the mutation rate and crossover rate are set to 0.9 and 0.6. For different datasets, the upper and lower bounds of the number of hidden layer nodes of the ELM are as shown in Table 2.
4.2 Experimental Results
This subsection introduces the experimental assessment of the EMMODE. First, we compare the performance of the three ensembles strategies, which aims at comparing among the different ensemble strategies to find one of them that performs best. Second, we compare the performance of EMMODE with traditional feature selection method and standard learning algorithms.
Tables 3 and 4 illustrate the obtained results by each of the ensembles. The displayed results are the average testing accuracy and the selection rate in feature set. These results are the mean and standard deviation values obtained by the algorithm running 10 times in the dataset. For each case, the best result is highlighted in boldface.
From the results in Tables 3 and 4, we can see that ERE is the excellent ensemble strategy among the three methods. It achieves the best performance when classifying test sets while reducing the feature set size. Hence, the ERE is used to compare with other methods, namely single ELM [37], wrapper feature selection method: PSO-SVM [43], whose SVM is the implementation of LibSVM [44]. and standard ensemble learning algorithms: random forest (RF) [6] and Adaboost [5]. For the random forest and Adaboost, their base classifiers are decision trees and the number of trees is set to 100.
In Table 5, we compare EMMODE-ERE with ELM, PSO-SVM, random forest (RF), Adaboost. The ELM is utilized as a baseline for comparing the performance of other methods. From the results indicated in the table, we can see the following. (1) EMMODE-ERE are capable of enhancing the performance of classification. (2) Traditional FS and ensemble approaches outperform the standard ELM.
Therefore, EMMODE is a competitive method for performing feature reduction and parameter selection for an ELM and can be adopted to far-going supervised learning problems. Meanwhile, EMMODE is an intensely efficient classification algorithm when compare it with traditional learning algorithms.
5 Conclusion
In this paper, we have presented EMMODE. The significance and importance of solving MMO problems of selecting features and the parameter are analyzed. EMMODE deals with the MMO problem by selecting features and the parameter of an ELM simultaneously. Moreover, it also presents three different strategies, including the APSE, MEE, and ERE, for combining the Pareto solutions into an ensemble. Experimental results prove the effectiveness of the proposed EMMODE approach.
The datasets used in this paper relatively small-scale. When the dimension of the dataset is higher, the result of ELM may be unstable. In the future, utilizing our method on unbalanced classification datasets and improving the performance of our method on large-scale datasets will be studied.
References
Song, Y., et al.: Gaussian derivative models and ensemble extreme learning machine for texture image classification. Neurocomputing 277, 53–64 (2018)
Piri, S., Delen, D., Liu, T., Zolbanin, H.M.: A data analytics approach to building a clinical decision support system for diabetic retinopathy: developing and deploying a model ensemble. Decis. Support Syst. 101, 12–27 (2017)
Zhao, Z., Jiao, L., Liu, F., Zhao, J., Chen, P.: Semisupervised discriminant feature learning for SAR image category via sparse ensemble. IEEE Trans. Geosci. Remote Sens. 54(6), 3532–3547 (2016)
Breiman, L.: Bagging predictors. Mach. Learn 24(2), 123–140 (1996)
Freund, Y., Schapire, R.E.: A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55(1), 119–139 (1997)
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001)
Rodriguez, J.J., Kuncheva, L.I., Alonso, C.J.: Rotation forest: a new classifier ensemble method. IEEE Trans. Pattern Anal. Mach. Intell. 28(10), 1619–1630 (2006)
Fernández, A., Carmona, C.J., Jose del Jesus, M., Herrera, F.: A Pareto-based ensemble with feature and instance selection for learning from multi-class imbalanced datasets. Int. J. Neural Syst. 27(06), 1750028 (2017)
Albukhanajer, W.A., Jin, Y., Briffa, J.A.: Classifier ensembles for image identification using multi-objective Pareto features. Neurocomputing 238, 316–327 (2017)
Lyu, H., Wan, M., Han, J., Liu, R., Wang, C.: A filter feature selection method based on the maximal information coefficient and Gram-Schmidt orthogonalization for biomedical data mining. Comput. Biol. Med. 89, 264–274 (2017)
Guyon, I., Elisseeff, A.: An introduction to variable and feature selection. J. Mach. Learn. Res. 3, 1157–1182 (2003)
Kohavi, R., John, G.H.: Wrappers for feature subset selection. Artif. Intell. 97(1–2), 273–324 (1997)
Xue, X., Yao, M., Wu, Z.: A novel ensemble-based wrapper method for feature selection using extreme learning machine and genetic algorithm. Knowl. Inf. Syst. 57(2), 389–412 (2017). https://doi.org/10.1007/s10115-017-1131-4
Zhang, Y., Gong, D., Cheng, J.: Multi-objective particle swarm optimization approach for cost-based feature selection in classification. IEEE/ACM Trans. Comput. Biol. Bioinf. (TCBB) 14(1), 64–75 (2017)
Quinlan, J.R.: Improved use of continuous attributes in C4.5. J. Artif. Intell. Res. 4, 77–90 (1996)
Kamyab, S., Eftekhari, M.: Feature selection using multimodal optimization techniques. Neurocomputing 171, 586–597 (2016)
Pan, L., Li, L., He, C., Tan, K.C.: A subregion division-based evolutionary algorithm with effective mating selection for many-objective optimization. IEEE Trans. Cybern. (2019). https://doi.org/10.1109/TCYB.2019.2906679
He, C., Tian, Y., Jin, Y., Zhang, X., Pan, L.: A radial space division based evolutionary algorithm for many-objective optimization. Appl. Soft Comput. 61, 603–621 (2017)
Yue, C., Qu, B., Liang, J.: A multiobjective particle swarm optimizer using ring topology for solving multimodal multiobjective problems. IEEE Trans. Evol. Comput. 22(5), 805–817 (2017)
Deb, K., Tiwari, S.: Omni-optimizer: a procedure for single and multi-objective optimization. In: Coello Coello, C.A., Hernández Aguirre, A., Zitzler, E. (eds.) EMO 2005. LNCS, vol. 3410, pp. 47–61. Springer, Heidelberg (2005). https://doi.org/10.1007/978-3-540-31880-4_4
Liang, J., Guo, Q., Yue, C., Qu, B., Yu, K.: A self-organizing multi-objective particle swarm optimization algorithm for multimodal multi-objective problems. In: Tan, Y., Shi, Y., Tang, Q. (eds.) ICSI 2018. LNCS, vol. 10941, pp. 550–560. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-93815-8_52
Dua, D., Graff, C.: UCI machine learning repository (2017). http://archive.ics.uci.edu/ml
Tanabe, R., Ishibuchi, H.: A review of evolutionary multimodal multiobjective optimization. IEEE Trans. Evol. Comput. 24(1), 193–200 (2020). ISSN 1941-0026
Li, X., Epitropakis, M.G., Deb, K., Engelbrecht, A.: Seeking multiple solutions: an updated survey on niching methods and their applications. IEEE Trans. Evol. Comput. 21(4), 518–538 (2017)
Liang, J., et al.: Multimodal multiobjective optimization with differential evolution. Swarm Evol. Comput. 44, 1028–1059 (2019)
Shir, O.M., Preuss, M., Naujoks, B., Emmerich, M.: Enhancing decision space diversity in evolutionary multiobjective algorithms. In: Ehrgott, M., Fonseca, C.M., Gandibleux, X., Hao, J.-K., Sevaux, M. (eds.) EMO 2009. LNCS, vol. 5467, pp. 95–109. Springer, Heidelberg (2009). https://doi.org/10.1007/978-3-642-01020-0_12
Sikdar, U.K., Ekbal, A., Saha, S.: MODE: multiobjective differential evolution for feature selection and classifier ensemble. Soft Comput. 19(12), 3529–3549 (2015). https://doi.org/10.1007/s00500-014-1565-5
Whitney, A.W.: A direct method of nonparametric measurement selection. IEEE Trans. Comput. 100(9), 1100–1103 (1971)
Marill, T., Green, D.: On the effectiveness of receptors in recognition systems. IEEE Trans. Inf. Theory 9(1), 11–17 (1963)
Yusta, S.C.: Different metaheuristic strategies to solve the feature selection problem. Pattern Recogn. Lett. 30(5), 525–534 (2009)
Pan, L., He, C., Tian, Y., Wang, H., Zhang, X., Jin, Y.: A classification-based surrogate-assisted evolutionary algorithm for expensive many-objective optimization. IEEE Trans. Evol. Comput. 23(1), 74–88 (2018)
Pan, L., He, C., Tian, Y., Su, Y., Zhang, X.: A region division based diversity maintaining approach for many-objective optimization. Integr. Comput. Aided Eng. 24(3), 279–296 (2017)
Wang, X., Yang, J., Teng, X., Xia, W., Jensen, R.: Feature selection based on rough sets and particle swarm optimization. Pattern Recogn. Lett. 28(4), 459–471 (2007)
Yu, K., Qu, B., Yue, C., Ge, S., Chen, X., Liang, J.: A performance-guided jaya algorithm for parameters identification of photovoltaic cell and module. Appl. Energy 237, 241–257 (2019)
Huang, C.L., Wang, C.J.: A GA-based feature selection and parameters optimizationfor support vector machines. Expert Syst. Appl. 31(2), 231–240 (2006)
Wan, Y., Wang, M., Ye, Z., Lai, X.: A feature selection method based on modified binary coded ant colony optimization algorithm. Appl. Soft Comput. 49, 248–258 (2016)
Huang, G.B., Zhu, Q.Y., Siew, C.K.: Extreme learning machine: theory and applications. Neurocomputing 70(1–3), 489–501 (2006)
Huang, G.B., Chen, L., Siew, C.K., et al.: Universal approximation using incremental constructive feedforward networks with random hidden nodes. IEEE Trans. Neural Networks 17(4), 879–892 (2006)
Feng, G., Huang, G.B., Lin, Q., Gay, R.: Error minimized extreme learning machine with growth of hidden nodes and incremental learning. IEEE Trans. Neural Networks 20(8), 1352–1357 (2009)
Xu, Z., Yao, M., Wu, Z., Dai, W.: Incremental regularized extreme learning machine and it’s enhancement. Neurocomputing 174, 134–142 (2016)
Cao, J., Lin, Z., Huang, G.B., Liu, N.: Voting based extreme learning machine. Inf. Sci. 185(1), 66–77 (2012)
Rosales-Perez, A., Garcia, S., Gonzalez, J.A., Coello, C.A.C., Herrera, F.: An evolutionary multi-objective model and instance selection for support vector machines with Pareto-based ensembles. IEEE Trans. Evol. Comput. 21(6), 863–877 (2017)
García-Nieto, J., Alba, E., Jourdan, L., Talbi, E.: Sensitivity and specificity based multiobjective approach for feature selection: application to cancer diagnosis. Inf. Process. Lett. 109(16), 887–896 (2009)
Chang, C.C., Lin, C.J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2(3), 27 (2011)
Acknowledgments
This work is supported by the National Natural Science Foundation of China (61976237, 61922072, 61876169, 61673404).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Wang, J., Wang, B., Liang, J., Yu, K., Yue, C., Ren, X. (2020). Ensemble Learning via Multimodal Multiobjective Differential Evolution and Feature Selection. In: Pan, L., Liang, J., Qu, B. (eds) Bio-inspired Computing: Theories and Applications. BIC-TA 2019. Communications in Computer and Information Science, vol 1159. Springer, Singapore. https://doi.org/10.1007/978-981-15-3425-6_34
Download citation
DOI: https://doi.org/10.1007/978-981-15-3425-6_34
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-3424-9
Online ISBN: 978-981-15-3425-6
eBook Packages: Computer ScienceComputer Science (R0)