Abstract
Soil and plant root are known as the microbial reservoir, and these microbes are found broadly in the plant rhizosphere and tissues. Phytobiome generally exists as epiphytic, endophytic, and rhizospheric that undertakes a critical role in plant development. These microbiomes may shape networks, to stabilize the function among different kinds of plant-associated factors to propagate or transmit in a different part of the plant. Microbial networks linked with plant health give crucial beneficial insights to look upon. The present section covers the features of such microbial networks that build the phytobiome. The chapter highlights their ability to better uptake nutrients or plant growth regulators in a stressed environment and further extends an evolution of studies depicting the supporting components that shape the phylogenetic and plant-related networks. The chapter advocates the possibility to understand the techniques by which plants select and connect with their microbiomes and affect plant improvement and well-being, thereby laying the foundation of novel microbiome-driven systems to the advancement of sustainable agriculture. The microbiome is unpredictably engaged with plant well-being providing extra qualities to the plant. To understand the guideline of plant characteristic articulation, henceforth plant execution, and how this impacts the biological systemic network, it is required to get well versed with phytobiome and its usefulness. In the present section, the significance of the phytobiome to plant genomics is tended to describe the phytobiome in assembly to the environment of the outline with attention on natural surroundings happening subterranean at the plant-soil between face, where the center is around the job of exudates as currency in this framework.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction
Nature allows the coexistence of healthy and asymptomatic plants with diverse microbes such as archaea, bacteria, fungi, and protists where a complex microbial consortium is formed to impact plant growth and productivity (Vorholt 2012; Kumari et al. 2019; Solanki et al. 2019). Phytobiome has either neutral or helpful roles in the plants’ fitness (Mendes et al. 2013). The useful impacts on plants include disease suppression (Ritpitakphong et al. 2016; Solanki et al. 2012), plant immunization (Van der Ent et al. 2009), induction of systemic resistance (Zamioudis et al. 2015), increased nutrient acquisition (Van der Heijden et al. 2016), increased tolerance to abiotic stresses (Rolli et al. 2015; Wang et al. 2018), adaptation to environmental variations (Haney et al. 2015), or enhancement of the mycorrhizal colonization (Garbaye 1994). Microorganisms can also target agricultural productivity by providing nutrient availability/acquisition (Kavamura et al. 2013). Lack of precise methodologies has led to limited access to nonculturable microbial groups, and thus, most of the work relies on single microbial groups associated with plants (Andreote et al. 2009). Mycorrhizal association with a plant (Chagnon et al. 2013) and microbial diazotrophs (Raymond et al. 2004) are the few examples that need to be explored in-depth. Nevertheless, an inclusive map of this system laid stress on the interactions happening between diverse groups of microbes, permitting the term “microbiome.” Joshua Lederberg coined the term “microbiome” for the first time and described it to be the “ecological community of commensal microorganisms, symbionts or pathogens that occupy a space in our body” (Lederberg and McCray 2001). New terminology for “microbiome” was suggested by Boon et al. (2014) that relates to host-associated genes in a defined surrounding, thereby bypassing the abundance of the microbial community of low significance. Plant-associated microbial groups work in multidimensional ways as host plant delivers unique metabolic adeptness to attract beneficial microbial niches that can have a positive (mutualistic), neutral (communalistic), or deleterious (pathogenic) effect on plant health (Thrall et al. 2007). Microbes are the main component of plant functional traits such as soil formation, organic matter decomposition, nutrient mobilization, and improvement in plant productivity (deBello et al. 2010). Rhizospheric prokaryotes are known as plant helpers due to their beneficial activities such as nitrogen (N2) fixation (Martinez-Romero 2006), solubilization of insoluble minerals, and stimulation of phytohormones (Hardoim et al. 2008). Genome duplication (polyploidization) is defined as macroevolutionary events of host that can change microbiome structure. The phytobiome exerts influences on plant trait expression through upstream and downstream regulation of nutritional uptake, thus supervising plant’s performance. To unlock the subtleties inside the ecosystem, and the regulation of plant trait expression, impacts of the microbiome are needed to be observed. Bernedsen et al. (2012) reported that plant microbiome interface aligned as “microbe-soil-microbe-plant-microbe interface” rather than the “soil-microbe-plant interface.” Plant genome is itself a complex system, and microbial interaction is coined as the plant’s “second genome” because it extends the plants’ genetic compendium extensively.
This chapter also contains a detailed description of beneficial phytobiome interactions. Three microbial groups (bacteria, fungi, and protists) that abundantly originate on plant tissues are deliberated, and diverse mechanisms used to cooperate and compete in planta are defined. Nevertheless, the activity of microbiomes is a new systematic approach that is required to understand the multidimensional actions of microbial communities (Bashiardes et al. 2018). To some degree, microbiome applications would include an emphasis on enlightening basic components that can improve crop production such as management of plant nutrients, soil health, and environmental safety (Syed Ab Rahman et al. 2018).
3.2 Soil Microbiome Characterization
Phytobiomes are discrete that comprise unfavorable pathogens, potential endophytes, and helpful symbionts (Rosenblueth and Martínez-Romero 2006; Wang et al. 2017; Malviya et al. 2019). Be that as it may, traditionally, the microbial assorted variety was assessed by segregating and refined on various supplement media and development conditions. Microbial metabolism fulfills the nutritional and regulatory prerequisites of plants (Lugtenberg and Kamilova 2009). These healthful necessities, for the most part, incorporate nitrogen, phosphorous, and iron. Moreover, these elements also control plant growth by stimulating the production of plant growth regulators. Screening of the most suitable bacteria would require culture-based methods (Taulé et al. 2012). On a routine basis, the procedures normally contain an agar plate assay or a broth medium to multiply the microbes. These assays also help in locating genetic components of microbes. However, these protocols failed to explore the microbial diversity of nonculturable microbiota.
For investigating the entire microbiome, the very first effort is initiated with sequencing of a conserved gene region such as the 16S rRNA gene that is widely applied for microbial identification (Mullis et al. 1987). To contemplate and comprehend the microbiome in a brief span, thorough upgrades have been accomplished by this technique, thereby yielding metagenomics. These techniques incorporate beginning with the entire metagenome examining, trailed by refinement, partition, and sequencing and lastly information investigation and elucidation. Particularly, the sequencing innovation is experiencing fast improvement, as it gives wide and top to bottom perspectives on metagenomics, and now it is extensively named as high-throughput sequencing (HTS) or cutting-edge sequencing technology. HTS methods incorporate the utilization of the AB SOLiD System (Life Technologies), the HiSeq 2000 (Illumina), and the 454 Genome Sequencer (Roche Diagnostics) (Yergeau et al. 2014). Besides, other propelled methods, for example, DNA/RNA-SIP and DNA arrays (PhyloChip and practical quality exhibits), likewise have prospective highlights in the examination of microbiomes, mostly their useful parts (Uhlik et al. 2013). At present, there is a change from metagenomics to metatranscriptomics, as the latter helps in understanding the numerous microbial functions and structure (Turner et al. 2013).
In the metatranscriptomics approach, complementary DNA analysis aligned with quantitative reverse transcription-PCR and RNA-SIP explored the microbial functionality associated with the soil and rhizosphere (Uhlik et al. 2013). RNA-SIP significantly is used to crack the complexity in interactions especially between root-derived carbon and microbiome so as to provide sequence as first and second utilizers of carbon within the microbiome. This method is dissimilar to DNA-SIP because it provides higher amounts of labeling and does not rely on cell multiplication. Challenges coming with these cutting-edge innovations include choosing either mRNA or rRNA alone and accomplishing more extensive inclusion of environmental RNA pool that gives naturally vital information through the sequencing. Peiffer et al. (2013) demonstrated noteworthy community contrasts among 27 maize innate lines (a genetic variant of a single species) with a normal enhanced population in the maize rhizosphere. Metaproteomics, on the other hand, has a different approach as it focuses on the dynamic function of the phytobiome and extracts samples of metaproteome and performs peptide fingerprinting by mass spectrometry (Kolmeder and de Vos 2014; Lakshmanan et al. 2014). Using metagenomic and metaproteomic (existing and future) information is an essential process-driven methodology and should be supplemented by different strategies to decide the diversity and functional relatedness of the rhizospheric microbiome (Keiblinger et al. 2012).
Molecular methods (molecular fingerprinting) and plate count anomaly (culture-dependent methods) demonstrate the entire bacteria community structure (Amann et al. 1995). Therefore, both approaches are utilized, for thoughtful knowledge of separate classification and communication with host plants. Dini-Andreote and van Elsas (2013) have, however, stressed the present need for a change in outlook from HTS (or comprehensive endeavors) to investigations of basic studies.
3.3 Structural and Compositional Factors in Plant-Associated Microbial Network
3.3.1 Plant-Associated Bacterial and Archaeal Microbiomes
Plant-associated bacterial population detected on plants does not look arbitrary; relatively numerous components participate in controlling the structure of microbiomes such as soil type (Lundberg et al. 2012), plant compartment (Leff et al. 2015), host genotype/species (Tkacz et al. 2015), plant invulnerable framework (Horton et al. 2014), plant attribute variety/developmental stage (Donn et al. 2015), and residence time/season (Shi et al. 2015). Hacquard et al. (2015) described that Proteobacteria, Actinobacteria, and Bacteroidetes are the major bacterial phyla that exist in both substrata such as above- and belowground plant tissues, and they influence the plant metabolism. Broad cover among root- and leaf-related network individuals has been portrayed at OTU (operational taxonomic unit) level determination in different plants such as Arabidopsis thaliana, wild mustard, grapevine, and agave (Wagner et al. 2016), and microbiota reconstitution experiments with germ-free A. thaliana approved that root- and leaf-related bacterial networks have reciprocal relocation. Regardless of the conspicuous elementary uniformities saw between A. thaliana leaf- and root-related bacterial networks, it is observed that related microbiota individuals are particular and adjusted to their separate related plant organs (Bai et al. 2015). Among all phytobiomes, the nonpathogenic segregation of archaea has been depicted. The plant endophytic archaeal taxa of the phyla Thaumarchaeota, Crenarchaeota, and Euryarchaeota have plant-associated functional significance (Müller et al. 2015).
3.3.2 The Fungal Microbiota of Plants
Ascomycota and Basidiomycota are two noteworthy phyla that colonize both above- and belowground plant tissues (Hardoim et al. 2015). In roots, even though arbuscular (Glomeromycota phylum) and ectomycorrhizal growths have been for the most part contemplated, ongoing network profiling information demonstrates that other endophytic organisms too make up for root microbiota (Toju et al. 2013). The structure of fungal communities on plants relies upon different kinds of soil, plant parts, plant genotypes, or seasons (Coince et al. 2014) and is subjected to stochastic variations (Wang et al. 2013) and reacts distinctively to ecological elements (Thomson et al. 2015). Thus, mostly dispersal restriction and atmosphere clarify the worldwide biogeographic conveyance of growths and have been recommended to compel contagious dispersal, supporting high endemism in parasitic populaces (Talbot et al. 2014). Steady with that, the synchronous examination of both contagious and bacterial networks related to plants recommended a more prominent significance of biogeography for organizing parasitic networks contrasted with bacterial networks (Hacquard. 2016). Regardless of using molecular markers such as 16S rRNA and ITS, their loci need to be elucidated (Peay et al. 2016).
Recently, Yunshi et al. (2018) quantified the prokaryotic and fungal groups within the phyllosphere and rhizosphere of six spruce (Picea spp.) tree species through illumine amplicon sequencing. In brief, this microbial quantification experiment is performed in a common garden, and linkages among phenotypic characters of their plant hosts and bacterial/archaeal and fungal community are analyzed. Correlation results among plant microbiome and different phenotypic characters of host plants (such as leaf morphology, water content, water storage ability, dry biomass, nitrogen, etc.) which suggests that plant genotype played a significant role to shape its microbiota by improving plant phenotypes.
3.3.3 Plant-Associated Protists: The Outcasted Fraction of the Plant Microbiota
Protists are a vital constituent of the soil microbiome, and method progresses now extended to our thoughts of the real taxonomic and efficient diversity of soil protists. The Stramenopiles-Alveolata-Rhizaria (SAR) group is known as a large group of plant-associated protists (Ruggiero et al. 2015) and especially those having a place with the Oomycota (Stramenopiles) and Cercozoa (Rhizaria) lineage. Inside Oomycota, a couple of individuals having a place with the genera Peronospora, Phytophthora, Pythium (and other wool buildup genera), or Albugo frequently exist in the plant roots or leaves (Agler et al. 2016). Root colonization by oomycetes (i.e., Pythium oligandrum) provides positive benefits to the host (Van Buyten and Hofte 2013). Even though plant tissue-associated oomycete network profiling stays scanty, an exceptionally low decent variety is demonstrated with individuals from the Pythiaceae family being the most spoken about to be present on plant tissues (Sapp et al. 2018). Inside Cercozoa, one of the prevailing protistan bunches in biological systems, network profiling information uncovered a surprisingly high diversity in plant roots and leaves (Ploch et al. 2016), also giving a piece of strong evidence that indicates the plant stress tolerance and metabolic behavioral changes governed by special community structure. Thus, Oomycota and Cercozoa individuals are significantly important for holobiont wellness. Recent reports concluded that plant microbe linkages are outlined well under the evolutionary measure and it helps to unlock the complex interactions of plant and microbes in more depth, and plant-microbe or plant bacterial interaction is new as compared to the bacterial and other kingdom interactions (Lücking et al. 2009; Hassani et al. 2018).
3.4 Microbial Currency: Exudates
Plants and microbes release certain chemicals called exudates, which help them to communicate with each other and to accelerate the disease tolerance against biotic and abiotic factors, stabilize the plant and microbial growth during nutrient scarcity, and remediate the toxic elements. Microbes utilized exudates as a food source, particularly carbon and other acids. This section discussed the two-way interaction of plant and microbial exudate that is influenced by plant and microbial metabolism. Huang et al. (2014) reported the significance of plant root exudates to regulate the microbial structure in the plant rhizosphere that is influenced by plant variety, growth stages, disease-suppressive soils, root exudate composition, and plant hormone signaling. Plant-microbe interaction is a complex system that is mediated by numerous compounds, and these compounds are released under specific conditions. These compounds play an indispensable role to shape the microbial community and unified the microbes and their functions up to species level. For example, legumes and rhizobia symbiosis is signaled by flavonoids, plant mycorrhizal association is stimulated by strigolactones, malic acid regulates the quorum sensing (QS) of plant microbial helpers and major chemoattractants of microbes such as sugars and amino acids attract the beneficial microbial niches toward the plant roots to protect the plant against the multiple stresses. However, various protein molecules are released from the root in the rhizosphere that are less explored to understand their mechanism in plant fitness. Besides, root exudates played intermediate role in several other interactions such as plant attract the nematodes, and these nematodes are the vectors of rhizobia that enhanced the nodulation of root to fix the nitrogen, plant nodulation efficiency enhanced by the interaction of rhizobia with PGPR and arbuscular mycorrhiza (Huang et al. 2014). A multitude of rhizospheric interactions is mediated by root exudates, which are depicted in Fig. 3.1.
3.4.1 Plant Uptake and Release
Plant exudate components and assembly are specific to plant species and incorporate high-molecular-weight particles (e.g., sugar molecules, proteins, and fatty acid) and low-molecular-weight signaling molecules (e.g., natural compounds, metabolites, and amino acids) (Badri and Vivanco 2009). Jaeger et al. (1999) reported that plant root exudate contains sugar molecules and amino acids that help bacteria and other microbes to attract toward the plant root. Exudates assist numerous jobs such as stimulate the antagonism, allelopathic particles, and pathogen/herbivore safeguards. A large number of these exudates likewise fill in as a vitality hotspot for the microbiome; prokaryotes can use plant exudates as nutrient sources. For instance, grass Sorghum halepense excretes the exudate sorgoleone from root hairs having allelopathic properties (Kagan et al. 2003) which can be used as microbial nutrients (Gimsing et al. 2009). The different elements of plant exudate repeat the significance it fills in as numerous monetary forms of the phytobiome.
Roots participate in taking up the nutrients and signaling molecules from the rhizosphere while at the same time saving these supplements and concoction signaling molecules into this equivalent space required for evoking defense reactions. Terpenoids, flavonoids, and isoflavonoids contain a large number of the plant’s antimicrobial barriers. Isoprenoids being the most diverse primary metabolite is required to control cellular processes such as photosynthesis (as phytopigments) and seed growth stimulation (as gibberellic and abscisic acids), and allelopathic molecules also protect the plants from the pathogens (Hardoim et al. 2008).
3.4.2 Microbial Uptake/Release
Nitrogen fixation requires a constant need for rhizobium-legume symbiosis inside the biosphere. The ability of this methodology to enhance agricultural yield has produced attention in knowledge to manipulate this process for better use. A number of the study approaches were procured inside the examiner of rhizobia, and the precise knowledge collected from these numerous tools is used to focus on the genome- and systems-level procedures (diCenzo et al. 2019). Exudates are essential methods for correspondence within the environment for the microbial network. The uptake of exudates such as sugars, organic, and amino acids has been a noteworthy focal point of many years of research in a microbial environment utilizing different estimations of respiration or carbon substrate use measures, for example, those utilizing ECO MicroPlates™ (Biolog®). Microbial community behaves according to expanded or diminished centralizations of promptly accessible supplements that require insignificant vitality to absorb. Moreno et al. (2009) reported that a PGPB Pseudomonas putida KT2440 utilized amino acids and sugar that is concluded by the identification of proteins that regulate the amino acid and sugar uptake. The microbiome of rhizosphere has a perplexing task in nutrient cycling and includes a horde of nutrient transformations in soils. Microorganisms act as a catalyst for chemical changes in the soil during biogeochemical cycling. These changes plant supplement (N and P) uptake, soluble metal (Ca2+, Mg2+, K+, and Na+) uptake, and micronutrient uptake (Zn2+, Fe2/3+, Cu+, and Mn2+) (Stevenson and Cole. 1999).
Nitrogen-fixing prokaryotes played a vital role to fix nitrogen gas (N2) into ammonia (NH3), and this method helps the plants. Howard and Rees (1996) reported that physiological and genetic drivers N2-fixing prokaryotes have a highly conserved protein complex that is nitrogenase, and it is used to assess the abundance of N2-fixers in diverse ecological zones (Zehr et al. 2003). Specific bacterial metabolites worked as important plant hormones, for example, indole-3-acidic corrosive (IAA) engaged with managing plant hormone flagging (e.g., 1-aminocyclopropane-1-carboxylate (ACC) deaminase). These hormone flagging particles can advance plant development.
When wheat is inoculated with rhizosphere bacteria expressing ACC deaminase activity, expanded root improvement, and consequently expanded nutrient uptake, has been recorded (Shaharoona et al. 2008; Honma and Shimomura. 1978). ACC deaminase-producing bacteria manage the ethylene production in the plant, in this manner limiting effects of different ecological anxieties, which typically trigger expanded ethylene generation (Hardoim et al. 2008). Microbial exudates involve a significant part of antimicrobial and antifungal compounds. Several prokaryotic and eukaryotic microorganisms are able to secrete or discharge antimicrobial substances, but among all very few are cultivable (Piel 2011); due to this to understand the complex functions of these substances, metatranscriptomics and metabolomics tools need to be applied. It is found that 35% of Escherichia coli strains produce the antimicrobial compound known as calcium. The discoveries bolster the theory that antimicrobial cooperations inside microbial networks serve to look after diversity; this thought was created utilizing recreation models (Czaran et al. 2002).
Notwithstanding oozing antimicrobials that encourage plant resistance, microorganisms additionally release low-molecular-weight compounds to the plants, and plant sensors identify the microbes as a pathogen or beneficial and then trigger the reaction (Boller and He 2009). In this way, microbial metabolites can act straightforwardly on different microorganisms inside the microbiome in suppressive components or can act specifically on the plant to revitalize secure reactions, regularly activating plant exudation. As modern biotechnological tools improve our knowledge to measure these microbial metabolites in the soil matrix, multiple functions of the same microbiological substances are discovered.
3.5 Ecological Considerations for Utilizing Plant’s Benefits in the Farmer’s Field
An effective microbial inoculant needs to attack the pathogens and survive in varying abiotic conditions, also to set up good cooperation with the host that incorporates molecular passivity with the plant resistant framework. All through the developing season, microbial network experiences progression in both over the ground and subterranean (Edwards et al. 2015; (Copeland et al. 2015) portions of the plant. Along these lines, regardless of whether PGP inoculants colonize the plant at first, their constancy after some time isn’t ensured. Estimating the determination of bacterial inoculants in the soil presents technical difficulties, as the inoculant should be distinguished from a complex network. Strategies such as culture-based count utilizing re-separation of antibiotic-resistant inoculants or culture-autonomous estimation of relative bounty of the inoculant’s 16S rRNA quality in the soil, by means of DGGE (Schreiter et al. 2014), amplicon sequencing (Haney et al. 2015), or metagenomic sequencing (Krober et al. 2014), are used to determine the persistence of microbes.
Ecological components impacting root exudate organization and amount include raised dimensions of CO2, dry season, and nutrient deprivation (especially nitrogen and phosphorus). Increased carbon allocated in roots is observed in CO2-fertilization experiments, resulting in shifts in exudate composition and concentration that differ with plant species (Cheng and Gershenson 2007). These species-explicit effects can result in increased yield, in no net profitability increment, or can be unfavorable to plant development and generation. For example, positive biomass reactions in rye and clover to CO2 preparation were observed, while maize demonstrated no net biomass advantage (Phillips et al. 2006). Be that as it may, maize showed expanded exudation of a few amino acids under CO2 treatment. These discoveries are not amazing thinking about that the C4 photosynthetic pathway encourages development under elevated amounts of CO2; in any case, the effects of expanded arrival of amino acids into the rhizosphere by the C4 grass (maize) may assume a job in a large number of criticisms between different plants and organisms (Klironomos 2002).
3.5.1 Impact on Plant Functions
Plants participate in nutrient exchange and exudate correspondence depending on the molecule and energy required for the plant (alone or through help from the microbiome) to acquire or release exudate currency. An active transport system using ATP-restricting tape transporter participates in root exudation creation and fixation (Badri et al. 2009). Low-molecular-weight particles such as amino acids can be discharged through membrane diffusion or through protein channels (Badri and Vivanco 2009).
Plants utilize those microbes which can communicate with increased levels of N-acyl-L-homoserine lactones. AHL-degrading enzymes in the presence of a pathogen subsequently suppress gene expression of pathogens (Reading and Sperandio 2006). Plants in the same manner also help in AHL degradation inside the microbiome (Teplitski et al. 2000). Fluorescent pseudomonads which are fundamental to the rhizosphere of the different clusters of the plant are used to deliver the antimicrobials 2,4-diacetyl phloroglucinol (2,4-DAPG) and phenazine (Phz) derivatives (Mavrodi et al. 2011). These antimicrobials are of wide range and act against a number of plant pathogens that are contagious leading to their suppression (Raaijmakers et al. 2009). 2,4-DAPG and Phz derivatives are evident in the rhizosphere and are associated with the suppression of disease in wheat called as take-all in wheat. These plant hormones administrate the plant performance and its marking abilities to adjust to exudate profiles and enhance the plant’s immunity (Doornbos et al. 2012).
Most of the microorganisms are capable of producing and controlling the majority of plant hormones (Friesen et al. 2011), thereby modifying plant physiological pathways. In plant rhizospheres, 80% of bacterial taxa accounts for IAA production (Loper and Schroth 1986). In plants, root development is accelerated with a low concentration of IAA (Glick 1999), while its high concentration represses plant’s development, hence making plants prone to pathogen’s attack (Sarwar and Kremer 1995). An example of this is seen in Sorghum halepense; microbes of the invasive grass secrete high concentrations of IAA in contrast to other prokaryotes that secrete lower levels of IAA (Rout et al. 2013). Environmental stress and plant phenology drive the changes in plants that need to increase or decrease the hormones. PGPB are competent cells having multiple genes required for plant-microbial association (Hardoim et al. 2008). Yield expansion, organic methodologies, intercropping, and other cultural practices are utilized for possible farming production. New strategies are formulated to modulate the plant microbiome in an ideal course (Fig. 3.1). Distinctive microbiota is induced by diverse agro-management in viticulture (natural, biodynamic, or biodynamic with green compost) (Longa et al. 2017). It is observed that in an integrated management system, soil has diminished bacterial species richness as compared to organic management, even though microbial composition was similar to organically and biodynamically managed soils (Hendgen et al. 2018).
3.5.2 Impact on Bacterial Functions
Microbiome present in the rhizosphere possesses varied phenotypic expressions due to root exudates. Inhabitant microflora of plants perform different functions such as chemotaxis, stress tolerance (Amador et al. 2010), polychlorinated biphenyl degradation (Toussaint et al. 2012), modulation of genes involved in competence and sporulation (Mader et al. 2002), and biofilm formation on plant roots (Rudrappa et al. 2008). Plant exudates such as terpenoids, flavonoids, and isoflavonoids protect and control the internal structure of plants and outward surface of roots from microbial inhabitant (Hardoim et al. 2008). Microbiome symbionts can be epiphytes or endophytes, which survive for a short growth period and may encompass not only the pathogens but plant growth-promoting microbes using a mechanism of hormone signaling. Organic farming impacts the community composition on soil and roots of winter wheat (Hartman et al. 2018). The structure of bacterial communities is taken care of by tillage. Root bacteria respond to management types, whereas fungal communities respond to both. Different agricultural practices are parameters in affecting the microbial structure with differences in soil, roots, bacteria, and fungi and hence bringing around 10% of the variation in microbial communities.
3.6 Formation of Biofilm
Microbial communities act as a unit and secrete polymeric substances to produce a network known as biofilms (Stoodley et al. 2002). Microbes, when present in a biofilm as consortia, are highly protected from their competitor, antimicrobial agents, enzyme degradation, and acquisition of new genes through horizontal gene transfer (Van Acker et al. 2014; Nadell et al. 2009; Zhang et al. 2014). Enterobacter spp., a root-occupying bacterial endophyte, when forming a biofilm, inhibits the entry of root-colonizing pathogen Fusarium graminearum (Mousa et al. 2016). Bacteria commonly produce biofilms on fungi, but it is rarely seen on the hyphae of ascomycete fungi. An example of this is seen in Pseudomonas fluorescens BBc6 which formulates biofilm on the hyphal region of the ectomycorrhiza Laccaría bicolor specifically at its root tip, thereby establishing ectomycorrhizal beneficial symbiosis and promoting bacterial biofilm on fungal host surfaces (Guennoc et al. 2017).
3.7 Molecular Communications
The mechanism of quorum sensing is used by microbes to sense their counterparts. Gram-negative microorganism secretes signaling molecule N-acyl-l-homoserine lactone (AHL) to screen out their populace densities (Eberl 1999). Regulation and secretion of signaling molecules are evident in Saccharomyces cerevisiae and Candida albicans (human fungal pathogens) that secrete farnesol to control filamentation (Oh et al. 2001), constrain biofilm formation, and activate oxidative stress responses or drug efflux (Sharma and Prasad 2011). Quorum sensing mechanisms are not defined thoroughly for plant-associated fungi. Signaling compounds such as volatile organic compounds (VOCs), oxalic acid, trehalose, glucose, or thiamine accelerate fungal bacterial associations (Schmidt et al. 2016).
3.8 Ecology of the Microbiome
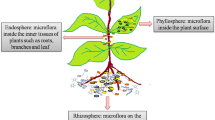
The plant microbiome is localized in three different regions, namely, rhizosphere, endosphere, and phyllosphere (Hirsch and Mauchline 2012). Rhizosphere presents a microbial community in requirement with plant metabolism; endosphere presents those microorganisms which interact with host closely and inhabit inner part of plant tissues asymptomatically (Hardoim et al. 2008); phyllosphere, on the contrary, is composed of those microbes which inhabit plant surfaces (Lambais et al. 2006). Irrespective of different plant habitats, specific microbes are present in all, also known as “keystone” species which interact with other microbes within the networks and affect the microbial structure (Bakker et al. (2014).
Rhizosphere and endosphere account for root microbiome that was key for the advancement of land plants and underlay crucial ecosystem processes. It is reported that nearly 30 angiosperm species affect root bacterial diversity and composition (Fitzpatrick et al. 2018). A competitive interaction gets affected when there is a similarity in root microbiomes between hosts among plant species. Climatic parameters such as drought affect the root microbiome composition, by elevating the Actinobacteria population. In the endosphere, Streptomyces are associated with host drought tolerance influencing drought response crosswise over host plant species bringing host-specific changes.
3.8.1 Rhizosphere and Rhizoplane
Plant health is influenced by the rhizosphere using next-generation and third-generation technologies (Hiltner 1904). Rhizospheric soil shows a significant difference in contrast to bulk soil due to abiotic and biotic stresses impacted by the atmosphere. Properties such as higher water holding capacity, expanded nutrient availability, and diverse microbial biomass mark its importance than bulk soil (Schade and Hobbie 2005). Spatiotemporal movements are observed in the rhizosphere microbiome (Kaplan et al. 2013); however, it is still to affirm how much abiotic stresses impact the microbiomes. Protection from a wide range of pathogens both aboveground and belowground is provided by microbiomes. For example, induction of systemic resistance (ISR) is initiated where jasmonic acid-inducible genes are secreted in leaves (Pineda et al. 2010).
3.8.2 Epiphytes and Endophytes
The epiphyte and endophyte microbial communities in root involve the acknowledgment and selection of those microbiomes that establish a homeostatic association with the plant. Technologies such as metabolomics and metatranscriptomics are used to observe microbial members that colonize in adherent (epiphytic) or internal (endophytic) parts of plants. Microbes colonize the outside root surfaces. For instance, secondary metabolite root exudates were released due to an ISR response in maize that appoints PGPB P. putida, based on chemotaxis inclinations (Neal et al. 2012). Plant chemical exudate is secreted, and valuable PGPB is selected in a plant-mediated reaction as observed in tomato, where natural acids are the major chemotactic operator (De Weert et al. 2002), while in rice, amino acids serve the purpose (Bacilio-Jimenez et al. 2003). Root microbes that laid distinctive qualities such as that code for the sort IV pilus and twitching motility (Bohm et al. 2007), isoflavonoid efflux siphon (Palumbo et al. 1998), and DNA improvements influence colony aggregation (Dekkers et al. 1998).
Endophytes are inhabitant to both wild and domesticated crops including intrusive species (Compant et al. 2008; Rout and Chrzanowski 2009). Biotechnology and agriculture ensure to utilize plant developing qualities of microbiomes as seen in phosphate-solubilizing Bacillus strains, where apart from secreting protein ACC deaminase, these also show plant development advancements (Baig et al. 2012). Thus, a microbiome serves a double attribute working together with mycorrhizal parasites to improve plant development advancement (Zaidi and Khan 2005).
Plant phenology correlates with endophyte microbiome composition shifts (van Overbeek and van Elsas 2008) which further depends upon colonization and similarity (Hardoim et al. 2008). This collaboration inclines more toward mutualism than parasitism. Most of known plant endophytes and epiphytes are horizontally transmitted (Friesen et al. 2011). This empowers host-to-host exchange of endosymbionts without the association of plant sexual reproduction. Endophytes also show vertical transmission depending upon host wellness and present more host benefits than horizontal transmission (Clay and Schardl 2002; Sachs et al. 2004). But the environmental hypothesis proposes that the presence of an accessible host allows for horizontally transmitted life forms. It is observed that horizontally transmitted endophytes were positively related to plant thickness reliance, while vertically transmitted endophytes did not demonstrate this pattern (Rudgers et al. 2009).
3.8.3 Phyllosphere Region
Phyllosphere is regarded as a third segment of the plant microbiome that colonizes the outside region of the external area of plant tissues specifically when describing the leaf surface (Vorholt 2012). The microbiomes in the phyllosphere perform nitrogen fixation, securing plants against attacking pathogens and biosynthesizing phytohormones (Kishore et al. 2005). These can be beneficial in carbon sequestration (Bulgarelli et al. 2013), and they can also participate in sustainable agricultural practices. Fungi (filamentous and yeasts), bacteria, and algae make phyllosphere network, and at lower frequencies, protozoa and nematodes are seen (Lindow and Brandl 2003). The bacterial population is the most abundant group of microorganisms present in the phyllosphere at numbers ranging from 105 to 107 cells for each cm2 (Andrews and Harris 2000). These microbes can thrive in harsh environmental conditions such as limited availability of nutrients and variable conditions of humidity, UV radiation, pH, and temperature (Andrews and Harris 2000). The phyllosphere community is created with the help of various hotspots as air, soil, and water (Bulgarelli et al. 2013). Agricultural plants also show specificity to phyllosphere microbiomes as seen in beans, cucumber, grasses, lettuce, and maize (Rastogi et al. 2012). Plant genotype plays a significant effect on the composition of phyllosphere microbiomes (Bokulich et al. 2014). The microbial population shows intraspecific variations in its composition which are due to nutritional heterogeneity observed in regions on the leaf surface where heterogeneous carbon sources such as glucose, fructose, and sucrose are utilized near the stomata and surface appendages (Vorholt 2012). At times, this heterogeneity is observed when microbial cells aggregate to form a biofilm and hence defending themselves from unfavorable conditions (Lindow and Brandl 2003).
Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes are major phyla that account for the microbial community in the phyllosphere region (Vorholt 2012). Hence, this core is accepted to be made out of individuals exhibiting a co-developmental history with plant species, with the host physiology being complementary to the features found inside the microbial cells. Microbes such as protists largely act as predators on the bacterial community (Flues et al. 2017). Environmental conditions such as low nutrients, high UV, changing temperature, and humidity help in selecting consistent biological traits and low functional diversity at the community level for phyllosphere microbiomes as observed in next-generation sequencing (Lambais et al. 2017). The phyllosphere is dominated by oxygen-consuming organoheterotrophs, and metabolic diversity exists with regard to utilizable carbon compounds.
3.9 Competitive Interactions Among Plant Microbiota Members
Plant microbiomes show competitive behavior with closely or distantly related microbiota and affect microbial structure, its stability, and homeostasis.
3.9.1 Resource Competition
Microorganisms utilize limited resources and therefore compete indirectly with other microbes. For example, using advanced techniques, microorganisms sequestrate iron using the emission of siderophores, thereby affecting the growth of the opponent microbes present in their niche (Little et al. 2008). When advantageous Pseudomonas spp. secrete iron-chelating molecules, it suppresses the disease caused by fungal pathogens indicating that nutrient sequestration is a trait of biocontrol agents to outpower pathogens (Mercado-Blanco and Bakker 2007). In tomato plants, resource competition is said to be an essential factor connecting the bacterial network and pathogen attack on plants (Wei et al. 2015). In resource competition, individual microbes in a group share resources, but the ones who use the resources in an uncooperative can evade paying the price of cooperation while reaping the benefits of utilizing the resource, thereby increasing their fitness (Riehl and Frederickson 2016) and bringing the situation of distress to the commons. Nitrogen-deficient soil could harbor plants with rhizosphere having microbes that can capture nutrients for their usage. For instance, actively growing roots could signal for microorganisms that are capable of producing extracellular enzymes releasing nitrogen bound in soil organic matter (Lemanceau et al. 2017). In prokaryotes, mineralization processes are density-dependent and need a quorum of producers to sufficiently enter key nutrients in the soil. Such producers are taxonomically diverse microbiota that can biosynthetically produce the specific enzymes which are secreted into the soil. In this scenario, selection in the rhizosphere could favor microhabitats to promote coordinated group behaviors that enhance plant access to nitrogen or phosphorus upon cell turnover, while the microorganisms benefit from having an abundant supply of carbon and other nutrients from plant roots (Fig. 3.1). Phosphorous is also made available to plants using microbial taxa which could mobilize phosphorus in the soil via the production of extracellular compounds (Alori et al. 2017). Iron is an important plant nutrient which can be obtained through the production of siderophores (Radzki et al. 2013).
3.9.2 Contact-Dependent Competition
Plant microbiome participates in direct antagonistic cooperations interceded by the bacterial type VI secretion framework, a molecular weapon used by certain microscopic organisms (generally Proteobacteria) to convey effectors/toxins into both eukaryotic and prokaryotic cells (Records 2011). A few examples of contact-dependent competition are discussed as in the case of the plant pathogen Agrobacterium tumefaciens that utilizes a puncturing type VI secretion system to convey DNase effectors upon contact with a bacterial competitor in vitro and on the leaves of Nicotiana benthamiana. Moreover, the bacterial kind III secretion system can also be used in Burkholderia rhizoxinica, which uses this mechanism to control the productivity of its beneficial interaction with the contagious host, Rhizopus microspores. Physical parameters bring a change in plant-associated microbes. Soil condition (organic matter, nitrogen, and moisture content) identification helps in changing the macrophage activation potential of Echinacea purpurea and determining these changes in activity that relates to the shifts (Haron et al. 2019). Increasing soil organic matter in the root extracts of E. purpurea may increase the macrophage activation. Bacterial communities also differed significantly between root materials having varying levels of organic matter. The activity of E. purpurea roots is changed by the soil’s organic matter level. Use of bacterial preparation (e.g., probiotics) is reported to impact human health; similarly, Echinacea too shows therapeutic effects and is impacted by development conditions that change its related bacterial community (Haron et al. 2019).
3.9.3 Antimicrobial Compound Secretion
Various plant-related microorganisms appeared to emit chemical compounds that stifle the development of microbial rivals (Raaijmakers and Mazzola 2012). Filamentous eukaryotes are outstanding in delivering a large number of antifungal activity of secondary metabolites that have a low molecular weight spotted against phylogenetically distinct organisms (e.g., acetylgliotoxin and hyalodendrin) (Coleman et al. 2011). The secondary metabolites so obtained become activated in co-culture and remained inactive in pure culture. Netzker et al. (2015) indicate their specific role in competitive interactions. Antagonistic collaborations among microscopic organisms have been reported to be imperative in the organizing of soil-, coral-, or plant-related bacterial networks (Maida et al. 2016).
Strikingly, the investigation of adversarial collaborations among bacterial segregates from the rhizosphere, the roots, and the phyllosphere of the healing plant Echinacea purpurea proposes that plant-related microorganisms compete against one another through the discharge of antimicrobials (Maida et al. 2016). The microbiome related to plants has a robust influence on their strength and yield. The bacterial pathogen, Candidatus Liberibacter asiaticus (Las), causes Huanglongbing (HLB) disease and lives inside the phloem of citrus plants, including the root system. It has been proposed that Las negatively affects citrus microbiome. At the same time, the natural microbial flora of citrus also impacts the association between Las and citrus (Riera et al. 2017), i.e., two bacteria closely related to Las Agrobacterium tumefaciens and Sinorhizobium meliloti were found. Among them, Burkholderia metallica strain A53 and Burkholderia territorii strain A63 are within the β-proteobacteria class, whereas Pseudomonas granadensis strain 100 and Pseudomonas geniculata strain 95 are within the γ-proteobacteria class. It was observed that four bacterial strains Burkholderia territorii A63, Burkholderia metallica A53, Pseudomonas geniculate 95, and Bacillus pumilus 104 showed antagonistic action against the pathogen Phytophthora nicotianae (citrus root) on the basis of dual culture assays. Some of the antimicrobial-producing strains, Burkholderia metallica A53 and Burkholderia territorii stress A63, from a mandarin rhizosphere, belong to the Burkholderia cepacia complex (BCC) and its agricultural applications are restricted because of its high risk to human health (Depoorter et al. 2016). It remains to be determined whether Burkholderia metallica strain A53 and Burkholderia territorii strain A63 can cause human diseases. Both Burkholderia metallica A53 and Burkholderia territorii strain A63 can modulate citrus immune system beneath greenhouse situations while applied as a soil drench. Additionally, the Burkholderiaceae family changed into determined to be key taxa within the citrus microbiome of healthy trees in comparison to that of HLB-symptomatic trees in the discipline (Zhang et al. 2017).
3.9.4 Predation
Bacterial mycophagy comprises of microscopic organisms’ capacity to effectively develop at the expense of living contagious hyphae (De Boer et al. 2004). Mycophagous microbes colonize saprotrophic rhizosphere parasites and feed as auxiliary consumers on root-determined carbon (Rudnick et al. 2015). Some oomycetal species of family Trichoderma or Pythium can parasite or irritate other growths or oomycetes and can be utilized as biocontrol operators (Benitez et al. 2004). Root-related bacteria can prey on other microscopic organisms as described for Bdellovibrio spp. Protist predation on microscopic organisms is also well studied, and recent microbiota reconstitution tests in microcosm demonstrate a reasonable impact of Cercomonads (Rhizaria: Cercozoa) on the structure and the capacity of the leaf microbiota (Flues et al. 2017). Their results show that Alphaproteobacteria and Betaproteobacteria are less impervious to grazing and that predation rebuilds the bacterial system in leaves and impacts bacterial metabolic center capacities. Microbial assortment related to aphid inhabitants was characterized at species and intraspecies scales using a methodological structure (Guyomar et al. 2018). Utilizing this approach, on metagenomics read sets, high genomic diversity in different symbiont taxa can be uncovered in both between and within their hosts. The complete functional diversity related with host and microbiota was the first time it can be accessed using metatranscriptomics datasets which also helps in isolating the transcriptome of each member of the holobiont (Meng et al. 2018).
3.9.5 Genetic Management of Valuable Plant-Microbe Interactions
Host hereditary qualities add to plant microbiome assembly. Plants identify microorganisms through pattern recognition pattern that binds to the microbe-associated molecular pattern (MAMPs), setting off a basal barrier adequate to stop the development of most pathogenic organisms (Böhm et al. 2014). Plants can probably separate pathogens from nonpathogens and react by opposing microbial development, overlooking it, or effectively supporting it on or inside plant tissues. The transcriptional reaction of Arabidopsis leaves varies when vaccinated with various nonpathogenic individuals from its regular microbiota (Böhm et al. 2014). While Methylobacterium extorquens actuates no transcriptional reaction, Sphingomonas melonis initiates the defense-related genes that somewhat cover with those activated by the pathogen Pseudomonas syringae DC3000. This characterizes a mechanism of plant defense priming (Martinez-Medina et al. 2016) driven by the plant microbiome. The reaction example to nonpathogenic microorganisms can vary both crosswise over plant species (Ofek-Lalzar et al. 2014) and crosswise over promotions inside a single species (Haney et al. 2015). While some Arabidopsis accessions are colonized by and build up a valuable association with Pseudomonas fluorescens, different promotions effectively restrain the development of similar strains in their foundations. Given the basic capacity of defense phytohormones in the invulnerable plant framework, it is not astonishing that the plant microbiome organization is impacted by defense phytohormone flagging. Tests by a set of mutants with transformed protection phytohormone synthesis and notion stated that salicylic acid and salicylic acid-mediated events have an effect on the root microbiome composition at multiple taxonomic levels (Lebeis et al. 2015). The plant microbiome structure changes upon infection (Agler et al. 2016). Antifungal characteristics are enhanced in barley following infection with Fusarium graminearum, conceivably using changes in exudate arrangement (Dudenhöffer et al. 2016). An investigation of tomato plants tested with the pathogen Ralstonia solanacearum uncovered that the root exudation profile changed upon pathogen infection, expanding the discharge of phenolic mixes. Plant protection systems additionally sway different drivers of plant – organism cooperations, similar to plant sustenance (Hacquard et al. 2015). Present-day molecular methodologies are likewise being connected to understanding nitrogen-fixing symbioses in non-nodulating plants. Utilization of double host-organism transcriptomics depicted that the limit of a nitrogen-fixing Burkholderia strain to frame microaerobic biofilms on sugarcane roots is shown to have diminished motility and immunogenicity, trailed by metabolic adjustment to the sugar-rich plant condition. The plant does not enact an invulnerable reaction but, however, changes its root morphology and supplies the bacterium with photosynthates (Paungfoo-Lonhienne et al. 2016), a reaction pattern that is undifferentiated from the procedure of infection by BNF in legumes (Cao et al. 2017). These examples endorse that the coordination of defense and nutrition is crucial to driving microbiome characteristics.
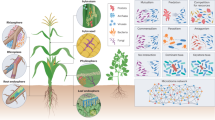
3.10 Implication of the Soil Microbiome on Sustainable Agriculture and Food Security
Conveying sustenance security, the way toward expanding nourishment creation, and improving nourishment quality to support populace development without trading off ecological well-being have been known as a worldwide green revolution (Gupta 2012). Sustainable agriculture improvement is expected to relieve these issues. A definitive objective of economical agribusiness, as per the US National Research Council, is to create cultivating frameworks that are gainful, beneficial, vitality saving, and environmentally solid, preserving natural materials, and that guarantee nourishment well-being and quality. This can be achieved by substituting risky agrochemicals (chemical fertilizers and pesticides) with environmentally friendly beneficial microorganisms, which could improve the sustenance of yields and animals and furthermore present protection from biotic (pathogens and pests) and abiotic (pollution and climatic change) stresses. The potential microbial segregates are detailed utilizing different natural and inorganic bearers through either solid or liquid fermentation technologies (outlined in Table 3.1).
Further improvement of microbial confines, and the plan procedure is required through broad research to present them in sustainable agricultural practices. Applications of microbial consortia are described in Table 3.2. Aside from the application of individual organisms, distinguishing sound and practically diverse microbiomes and their application for improving harvest yield poses another big challenge to meet.
3.11 Conclusions
Several illustrations depict the significance of understanding the multitude of plant microbiome relations that pay to plant versatility in a specified environment. Acknowledgment of the plant microbiome as a coordinated part of the plant genome develops the environmental idea of “feedback.” Disproportional accumulation of microbiome parasites (communicated as pathogenic impacts) prompts negative feedback, whereas the disproportional combination of microbiome mutualists stimulates positive feedback. An enhanced information about these interactions and how changes in biodiversity affect ecosystem functions (plant yield, biogeochemical pools, and fluxes) may be a vital feature for explaining plant microbiome boom and gene expression forms. Fast microbial generation time and the prevalence of horizontal gene transfer give probable systems to the improvement of localized genetic differences, or ecotypes, to emerge because of the impacts of local plant species and networks. As the plant-microbiome interaction unfolds, a new emerging methodology incorporates the study of microbial biology, microbiomes, and transcriptomes into plant genetics. The vast diversity documented in the rhizosphere microbiome is linked with the useful genes responsible for important nutrient changes, similar to those involved in N2-fixation. The age of expansive confine accumulations and the investigation of engineered microbial networks in the mix with plant genetic properties will enable us to connect this hole and to direct reductionist, theory-driven tests in progressively complex environmental settings up to handle field tests. These developments can convert our expertise of plant-microbe interactions in nature and agriculture and could make contributions extensively to the next green revolution. The key player(s) regarding microbiome structure have not been recognized. As needs are, there is a major break in the identification of the molecular segments associated with the collaboration among the host plant and the microbial populace. Also, these ongoing microbiome examinations attempted only to distinguish its structure and multifaceted nature instead of to decide how these microbial gatherings are adjusting the plant phenome, which is basic to investigate its usage. Likewise, there would be a cross talk using signal transduction among aboveground and belowground plant tissues that can be modified by an outer biotic or abiotic stress impacting the rhizospheric microbiome.
References
Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM (2016) Microbial hub taxa link host and abiotic factors to phytobiome variation. PLoS Biol 14(1):e1002352
Alori ET, Glick BR, Babalola OO (2017) Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Microbiol 8:971
Amador CI, Canosa I, Govantes F, Santero G (2010) Lack of CbrB in Pseudomonas putida affects not only amino acids metabolism but also different stress responses and bio-film development. Environ Microbiol 12:1748–1761
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59(1):143–169
Andreote FD, Azevedo JL, Araújo WL (2009) Assessing the diversity of bacterial communities associated with plants. Braz J Microbiol 40:417–432
Andrews JH, Harris RF (2000) The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol 38:145–180
Bacilio-Jimenez M, Aguilar-Flores S, Ventura-Zapata E, Perez-Campos E, Bouquelet S, Zenteno E (2003) Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 249:271–277
Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32:666–681
Badri DV, Quintana N, El Kassis EG, Kim K, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM (2009) An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 151:2006–2017
Bai Y, Muller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, Hüttel B, McHardy AC, Vorholt JA, Schulze-Lefert P (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528:364–369
Baig KS, Arshad M, Shaharoona B, Khalid A, Ahmed I (2012) Comparative effectiveness of Bacillus spp. possessing either dual or single growth-promoting traits for improving phosphorus uptake, growth and yield of wheat (Triticum aestivum L.). Ann Microbiol 62:1109–1119
Bakker MG, Schlatter DC, Otto-Hanson L, Kinkel LL (2014) Diffuse symbioses: roles of plant-plant, plant-microbe and microbe-microbe interactions in structuring the soil microbiome. Mol Ecol 23:1571–1583
Bashiardes S, Godneva A, Elinav E, Segal E (2018) Towards utilization of the human genome and microbiome for personalized nutrition. Curr Opin Biotechnol 51:57–63
Benitez T, Rincon AM, Limon MC, Codon AC (2004) Biocontrol mechanisms of Trichoderma strains. Int Microbiol 7:249–260
Berendsen RL, Vismans G, Yu K, Song Y, de Jonge R, Burgman WP, Burmølle M, Herschend J, Bakker AHM, Pieterse CMJ (2018) Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J 12:1496–1507
Berg M, Koskella B (2018) Nutrient- and dose-dependent microbiome-mediated protection against a plant pathogen. Curr Biol 28:2487–2492
Bernedsen RL, Pieterse CMJ, Bakker AHM (2012) The rhizosphere micro-biome and plant health. Trends Plant Sci 17:478–486
Bohm M, Hurek T, Reinhold-Hurek B (2007) Twitching motility is essential for endophytic rice colonization by the N2-fixing endophyte Azoarcus sp. strain BH72. Mol Plant Microbe Interact 20:526–533
Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T (2014) Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol 20:47–54
Bokulich NA, Thorngate JH, Richardson PM, Mills DA (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci 111:E139–E148
Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324:742–744
Boon E, Meehan CJ, Whidden C, Wong DHJ, Langille MGI, Beiko RG (2014) Interactions in the microbiome: communities of organisms and communities of genes. FEMS Microbiol Rev 38:90–118
Bulgarelli D, Spaepen SS, Themaat EVL, Shulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Cao Y, Halane MK, Gassmann W, Stacey G (2017) The role of plant innate immunity in the legume-rhizobium symbiosis. Annu Rev Plant Biol 68:535–561
Chagnon PL, Bradley RL, Maherali H, Klironomos JN (2013) A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant Sci 18:484–491
Cheng W, Gershenson A (2007) Carbon fluxes in the rhizosphere. In: Cardon ZG, Whitbeck JL (eds) The rhizosphere – an ecological perspective. Academic, San Diego, pp 31–56
Clay K, Schardl C (2002) Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat 160:99–127
Coince A, Cordier T, Lengelle J, Defossez E, Vacher C, Robin C, Buée M, Marçais B (2014) Leaf and root-associated fungal assemblages do not follow similar elevational diversity patterns. PLoS One 9:e100668
Coleman JJ, Ghosh S, Okoli I, Mylonakis E (2011) Antifungal activity of microbial secondary metabolites. PLoS One 6:e25321
Compant S, Kaplan H, Sessitsch A, Nowak J, Barka EA, Clement C (2008) Endo-phytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: from the rhizosphere to inflorescence tissues. FEMS Microbiol Ecol 63:84–93
Compant S, Samad A, Faist H, Sessitsch A (2019) A review on the phytobiome: ecology, functions, and emerging trends in microbial application. J Adv Res: 1–9
Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS (2015) Seasonal community succession of the phyllosphere microbiome. Mol Plant-Microbe Interact 28:274–285
Czaran TL, Hoekstra RF, Pagie L (2002) Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci 99:786–790
deBello F, Lavorel S, Diaz S, Harrison PA (2010) Towards an assessment of multiple ecosystem processes and services via functional traits. Biodivers Conserv 19:2873–2893
De Boer W, Leveau JH, Kowalchuk GA, Gunnewiek PJK, Abeln EC, Figge MJ, Sjollema K, Janse JD, van Veen JA (2004) Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int J Syst Evol Microbiol 54:857–864
de Vrieze M, Germanier F, Vuille N, Weisskopf L (2018) Combining different potato associated Pseudomonas strains for improved biocontrol of Phytophthora infestans. Front Microbiol 9:2573
De Weert S, Vermeiren H, Mulders IHM, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, Mot RD, Lugtenberg BJJ (2002) Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact 15:1173–1180
Dekkers LC, Phoelich CC, van der Fits L, Lugtenberg JJ (1998) A site-specific recombinase is required for competitive root colonization by Pseudomonas fluorescens WCS365. Proc Natl Acad Sci 95:7051–7056
Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E (2016) Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229
diCenzo GC, Zamani M, Checcucci A, Fondi M, Griffitts JS, Finan TM, Mengoni A (2019) Multidisciplinary approaches for studying rhizobium–legume symbioses. Can J Microbiol 65:1–33
Dini-Andreote F, van Elsas JD (2013) Back to the basics: the need for ecophysiological insights to enhance our understanding of microbial behaviour in the rhizosphere. Plant Soil 373:1–15
Donn S, Kirkegaard JA, Perera G, Richardson AE, Watt M (2015) Evolution of bacterial communities in the wheat crop rhizosphere. Environ Microbiol 17:610–621
Doornbos R, van Loon L, Bakker P (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron Sustain Dev 32:227–234
Dudenhöffer JH, Scheu S, Jousset A (2016) Systemic enrichment of antifungal traits in the rhizosphere microbiome after pathogen attack. J Ecol 104:1566–1575
Eberl L (1999) N-acyl homoserinelactone-mediated gene regulation in gram-negative bacteria. Syst Appl Microbiol 22:493–506
Edwards J, Johnson C, Santos-medellın C, Lurie E, Kumar N (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci 112:E911–E920
Fitzpatrick CR, Copelandc J, Wang PW, Guttman DS, Peter M, Kotanen PM, Johnson MTJ (2018) Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci 115:E1157–E1165
Flues S, Bass D, Bonkowski M (2017) Grazing of leaf associated Cercomonads (Protists: Rhizaria: Cercozoa) structures bacterial community composition and function. Environ Microbiol 19:3297–3309
Friesen ML, Porter SS, Stark SC, von Wettberg EJ, Sachs JL, Martinez-Romero E (2011) Microbially mediated plant functional traits. Annu Rev Ecol Evol Syst 42:23–46
Garbaye J (1994) Helper bacteria – a new dimension to the mycorrhizal symbiosis. New Phytol 128:197–210
Gimsing AL, Blum J, Dyan FE, Locke MA, Sejer LH, Jacobsen CS (2009) Mineralization of the allelochemical sorgoleone in soil. Chemosphere 76:1041–1047
Glick BR (1999) The enhancement of plant growth by free-living bacteria. Can J Microbiol 41:109–117
Guennoc CM, Rose C, Labbe J, Deveau A (2017) Bacterial biofilm formation on soil fungi: a widespread ability under controls. Biol Res 130740
Gupta VV (2012) Beneficial microorganisms for sustainable agriculture. Microbiol Aust 3:113–115
Guyomar C, Legeai F, Jousselin E, Mougel C, Lemaitre C, Simon J-C (2018) Multi-scale characterization of symbiont diversity in the pea aphid complex through metagenomic approaches. Microbiome 6(1):181
Hacquard S (2016) Disentangling the factors shaping microbiota composition across the plant holobiont. New Phytol 209:454–457
Hacquard S, Garrido-Oter R, González A, Spaepen S, Ackermann G, Lebeis S, McHardy AC, Dangl JL, Knight R, Ley R, Schulze-Lefert P (2015) Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17:603–616
Haney CH, Samuel BS, Bush J, Ausubel FM (2015) Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plant 1:15051
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471
Hardoim PR, van Overbeek LS, Berg G, Pirttila AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320
Haron MH, Tyler HL, Chandra S, Moraes RM, Jackson CR, Pugh ND, Pasco DS (2019) Plant microbiome-dependent immune enhancing action of echinacea purpurea is enhanced by soil organic matter content. Sci Rep 9(1):136
Hartman K, Van der Heijden MGA, Wittwer RA, Banerjee S, Walser JC, Schlaeppi K (2018) Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6:14
Hassani MA, Durán P, Hacquard S (2018) Microbial interactions within the plant holobiont. Microbiome 6:58
Hendgen M, Hoppe B, Döring J, Friedel M, Kauer R, Frisch M, Dahl A, Kellner H (2018) Effects of different management regimes on microbial biodiversity in vineyard soils. Science Report 8:9393
Hiltner L (1904) Uber neuere erfahrungen und probleme auf dem gebiet der bodenbakteriologie und unter besonderer berucksichtigung der grundungung und brache. Arbeiten der Deutschen Landwirtschaftlichen Gesellschaft 98:59–78
Hirsch PR, Mauchline TH (2012) Who’s who in the plant root microbiome? Nat Biotechnol 30:961–962
Honma M, Shimomura T (1978) Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric Biol Chem 42:1825–1831
Horton MW, Bodenhausen N, Beilsmith K, Meng DZ, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, Bergelson J (2014) Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun 5:5320
Howard JB, Rees DC (1996) Structural basis of biological nitrogen fixation. Chem Rev 96:2965–2982
Hu J, Wei Z, Friman VP, Gu SH, Wang XF, Eisenhauer N, Yang TJ, Ma J, Shen QR, Xu YC, Jousset A (2016) Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 7(6):1790–1816
Huang F, Chaparro JM, Reardon KF et al (2014) Rhizosphere interactions: root exudates, microbes, and microbial communities 1 Xing. Botany 92:267–275. https://doi.org/10.1139/cjb-2013-0225
Jaeger CHIII, Lindow SE, Miller W, Clark E, Firestone MK (1999) Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol 65:2685–2690
Kagan IA, Rimando AM, Dyan FE (2003) Elucidation of the biosynthetic pathway of the allelochemical sorgoleone using retro biosynthetic NMR analysis. J Biol Chem 278:28607–28611
Kaplan D, Maymon M, Agapakis CM, Lee A, Wang A, Prigge BA, Volkogon M, Hirsch AM (2013) A survey of the microbial community in the rhizosphere of two dominant shrubs of the Negev Desert highlands, Zygophyllum dumosum Boiss. and Atriplex halimus, using cultivation-dependent and -independent methods. Am J Bot 100:1713–1725
Kavamura VN, Santos SN, Silva JL, Parma MM, Avila LA, Visconti A, Zucchi TD, Taketani RG, Andreote FD, Melo IS (2013) Screening of Brazilian cacti rhizobacteria for plant growth promotion under drought. Microbiol Res 168:183–191
Keiblinger KM, Wilhartitz IC, Schneider T, Roschitzki B, Schmid E, Eberl L, Riedel K, Zechmeister-Boltenstern S (2012) Soil metaproteomics: comparative evaluation of protein extraction protocols. Soil Biol Biochem 54:14–24
Kishore GK, Pande S, Podile AR (2005) Biological control of late leaf spot of peanut (Arachis hypogaea) with chitinolytic bacteria. Phytopathology 95:1157–1165
Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417:67–70
Kolmeder CA, de Vos WM (2014) Metaproteomics of our microbiome: developing insight in function and activity in man and model systems. J Proteome 97:3–16
Krober M, Wibberg D, Grosch R, Eikmeyer F, Verwaaijen B, Chowdhury SP, Hartmann A, Pühler A, Schlüter A (2014) Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front Microbiol 5:252
Kumari B, Mallick MA, Solanki MK et al (2019) Plant growth-promoting rhizobacteria (PGPR): modern prospects for sustainable agriculture. In: Ansari RA, Mahmood I (eds) Plant health under biotic stress, vol II. Springer Netherlands, Dordrecht
Lakshmanan V, Selvaraj G, Bais HP (2014) Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol 166:689–700
Lambais MR, Crowley DE, Cury JC, Bull RC, Rodrigues RR (2006) Bacterial diversity in tree canopies of the Atlantic forest. Science 312:1917
Lambais MR, Barrera SE, Santos EC, Crowley DE, Jumpponen A (2017) Phyllosphere metaproteomes of trees from the Brazilian Atlantic forest show high levels of functional redundancy. Microb Ecol 73:123–134
Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, del Rio TG, Jones CD, Tringe SG, Dangl JL (2015) PHYTOBIOME: salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349(6250):860–864
Lederberg J, McCray AT (2001) ‘Ome sweet omics’ - a genealogical treasury of words. Scientist 15:8
Leff JW, Del Tredici P, Friedman WE, Fierer N (2015) Spatial structuring of bacterial communities within individual Ginkgo biloba trees. Environ Microbiol 17:2352–2361
Lemanceau P, Barret M, Mazurier S, Mondy S, Pivato B, Fort T, Vacher C (2017) Plant communication with associated microbiota in the spermosphere, rhizosphere and phyllosphere. In: Beards G (ed) How plants communicate with their biotic environment. Academic, London, pp 101–133
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883
Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J (2008) Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol 62:375–401
Longa CMO, Nicola L, Antonielli L, Mescalchin E, Zanzotti R, Turco E, Pertot I (2017) Soil microbiota respond to green manure in organic vineyards. J Appl Microbiol 123:1547–1560
Loper JE, Schroth MN (1986) Influence of bacterial source of indole-3-acetic acid on root elongation of sugar beet. Phytopathology 76:386–389
Lücking R, Huhndorf S, Pfister DH, Plata ER, Lumbsch HT (2009) Fungi evolved right on track. Mycologia 101:810–822
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Mader U, Antelmann H, Buder T, Dahl MK, Hecker M, Homuth G (2002) Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol Genet Genomics 268:455–467
Maida I, Chiellini C, Mengoni A, Bosi E, Firenzuoli F, Fondi M, Fani R (2016) Antagonistic interactions between endophytic cultivable bacterial communities isolated from the medicinal plant Echinacea purpurea. Environ Microbiol 18:2357–2365
Malviya MK, Solanki MK, Li C-N et al (2019) Beneficial linkages of endophytic Burkholderia anthina MYSP113 towards sugarcane growth promotion. Sugar Tech:1–12. https://doi.org/10.1007/s12355-019-00703-2
Martinez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CM, Pozo MJ, Ton J, van Dam NM, Conrath U (2016) Recognizing plant defense priming. Trends Plant Sci 21:818–822
Martinez-Romero E (2006) Dinitrogen-fixing prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 793–817
Mavrodi DV, Mavrodi OV, Parejko JA, Weller DM, Thomashow LS (2011) The role of 2,4-diacetylphloroglucinol- and phenazine-1-carboxylic acid-producing Pseudomonas spp. in natural protection of wheat from soilborne pathogens. In: Maheshwari DK (ed) Bacteria in agrobiology: plant nutrient management. Springer, Berlin, pp 60–63
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37(5):634–663
Meng A, Marchet C, Corre E, Peterlongo P, Alberti A, Da Silva C, Wincker P, Pelletier E, Probert I, Decelle J, Le Crom S, Not F, Bittner L (2018) A de novo approach to disentangle partner identity and function in holobiont systems. Microbiome 6(1):105
Mercado-Blanco J, Bakker PAHM (2007) Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 92:367–389
Molina-Romero D, Baez A, Quintero-Hernández V, Castañeda-Lucio M, Fuentes-Ramírez LE, Bustillos-Cristales MDR, Rodríguez-Andrade O, Morales-García YE, Munive A, Muñoz-Rojas J (2017) Compatible bacterial mixture, tolerant to desiccation, improves maize plant growth. PLoS One 12(11):e0187913
Moreno R, Martines-Gomariz M, Yuste L, Gil C, Rojo F (2009) The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9:2910–2928
Mousa WK, Shearer C, Limay-Rios V, Ettinger CL, Eisen JA, Raizada MN (2016) Root-hair endophyte stacking in finger millet creates a physicochemical barrier to trap the fungal pathogen Fusarium graminearum. Nat Microbiol 1:16167
Müller H, Berg C, Landa BB, Auerbach A, Moissl-Eichinger C, Berg G (2015) Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front Microbiol 6:138
Mullis KB, Erlich HA, Arnheim N, Horn GT, Saiki RK, Scharf SJ (1987) Process for amplifying, detecting, and/or-cloning nucleic acid sequences. US Patent Application No. US4683195A
Nadell CD, Xavier JB, Foster KR (2009) The sociobiology of biofilms. FEMS Microbiol Rev 33:206–224
Neal AL, Ahmad S, Gordon-Weeks R, Ton J (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7:e35498
Netzker T, Fischer J, Weber J, Mattern DJ, Konig CC, Valiante V, Schroeckh V, Brakhage AA (2015) Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol 6:299
Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D (2014) Niche and host-associated functional signatures of the root surface microbiome. Nat Commun 5:4950
Oh KB, Miyazawa H, Naito T, Matsuoka H (2001) Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans. Proc Natl Acad Sci 98:4664–4668
Palumbo JD, Kado CI, Phillips DA (1998) An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J Bacteriol 180:3107–3113
Paungfoo-Lonhienne C, Lonhienne TGA, Yeoh YK, Donose BC, Webb RI, Parsons J, Liao W, Sagulenko E, Lakshmanan P, Hugenholtz P, Schmidt S, Ragan MA (2016) Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci Rep 6:37389
Peay KG, Kennedy PG, Talbot JM (2016) Dimensions of biodiversity in the earth mycobiome. Nat Rev Microbiol 14:434
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci 110(16):6548–6653
Phillips DA, Fow TC, Six J (2006) Root exudation (net efflux of amino acids) may increase rhizodeposition under elevated CO2. Glob Chang Biol 12:561–567
Piel J (2011) Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Annu Rev Microbiol 65:431–453
Pineda A, Zheng S, van Loon JJA, Pieterse CMJ, Dicke M (2010) Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci 15:507–514
Ploch S, Rose LE, Bass D, Bonkowski M (2016) High diversity revealed in leaf-associated Protists (Rhizaria: Cercozoa) of Brassicaceae. J Eukaryot Microbiol 63:635–641
Raaijmakers JM, Mazzola M (2012) Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol 50:403–424
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moenne-Loccoz Y (2009) The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Radzki W, Gutierrez Mañero FJ, Algar E, Lucas García JA, García-Villaraco A, Ramos Solano B (2013) Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek 104:321–330
Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JHJ (2012) Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J 6:1812–1822
Raymond J, Siefert JL, Staples CR, Blankenship RE (2004) The natural history of nitrogen fixation. Mol Biol Evol 21:541–554
Reading NC, Sperandio V (2006) Quorum sensing: the many languages of bacteria. FEMS Microbial Lett 254:1–11
Records AR (2011) The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant-Microbe Interact 24:751–757
Riehl C, Frederickson ME (2016) Cheating and punishment in cooperative animal societies. Philos Trans R Soc Lond Ser B Biol Sci 371:20150090
Riera N, Handique U, Zhang Y, Dewdney MM, Wang N (2017) Characterization of antimicrobial-producing beneficial bacteria isolated from Huanglongbing escape citrus trees. Front Microbiol 8:2415
Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Metraux JP, L’Haridon F (2016) The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol 210:1033–1043
Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R, Pierotti CF, Borin S, Sorlini C, Zocchi G, Daffonchio D (2015) Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol 17:316–331
Rosenblueth M, Martínez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 19:827–837
Rout ME, Chrzanowski TH (2009) The invasive Sorghum halepense harbors endophytic N2-fixing bacteria and alters soil biogeochemistry. Plant Soil 315:163–172
Rout ME, Chrzanowski TH, DeLuca TH, Westlie TK, Callaway RM, Holben WE (2013) Bacterial endophytes enhance invasive plant competition. Am J Bot 100:1726–1737
Rudgers JA, Afkhami MA, Rua MA, Davitt SH, Hammer S, Huguet VM (2009) A fungus among us: broad pattern of endophyte distribution in the grasses. Ecology 90:1531–1539
Rudnick MB, van Veen JA, de Boer W (2015) Baiting of rhizosphere bacteria with hyphae of common soil fungi reveals a diverse group of potentially mycophagous secondary consumers. Soil Biol Biochem 88:73–82
Rudrappa T, Czymmek KJ, Pare PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148:1547–1556
Ruggiero MA, Gordon DP, Orrell TM, Bailly N, Bourgoin T, Brusca RC, Cavalier-Smith T, Guiry MD, Kirk PM (2015) A higher-level classification of all living organisms. PLoS One 10:e0119248
Sachs JL, Mueller UG, Wilcox TP, Bull JJ (2004) The evolution of cooperation. Q Rev Biol 79:135–160
Santhanam R, Van LT, Weinhold A, Goldberg J, Oh Y, Baldwin IT (2015) Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci 112:5013–5020
Sapp M, Ploch S, Fiore-Donno AM, Bonkowski M, Rose LE (2018) Protists are an integral part of the Arabidopsis thaliana microbiome. Environ Microbiol 20:30–43
Sarwar M, Kremer RJ (1995) Enhanced suppression of plant growth through production of L-tryptophan-derived compounds by deleterious rhizobacteria. Plant Soil 172:261–269
Schade J, Hobbie SE (2005) Spatial and temporal variation in the islands of fertility in the Sonoran Desert. Biogeochemistry 73:541–553
Schmidt R, Etalo DW, de Jager V, Gerards S, Zweers H, de Boer W, Garbeva P (2016) Microbial small talk: volatiles in fungal-bacterial interactions. Front Microbiol 6:1495
Schreiter S, Sandmann M, Smalla K, Grosch R (2014) Soil type dependent rhizosphere competence and biocontrol of two bacterial inoculant strains and their effects on the rhizosphere microbial community of field-grown lettuce. PLoS One 9:e103726
Shaharoona B, Naveed M, Arshad M, Zahir ZA (2008) Fertilizer-dependent efficiency of pseudomonads for improving growth, yield and nutrient use efficiency of wheat (Triticum aestivum L.). Appl Microbiol Biotechnol 79:147–155
Sharma M, Prasad R (2011) The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in Candida albicans. Antimicrob Agents Chemother 55:4834–4843
Shi SJ, Nuccio E, Herman DJ, Rijkers R, Estera K, Li JB, da Rocha UN, He Z, Pett-Ridge J, Brodie EL, Zhou J, Firestone M (2015) Successional trajectories of rhizosphere bacterial communities over consecutive seasons. mBio 6:e00746-15
Solanki MK, Kumar S, Pandey AK et al (2012) Diversity and antagonistic potential of Bacillus spp. associated to the rhizosphere of tomato for the management of Rhizoctonia solani. Biocontrol Sci Tech 22:203–217. https://doi.org/10.1080/09583157.2011.649713
Solanki MK, Wang F-Y, Wang Z et al (2019) Rhizospheric and endospheric diazotrophs mediated soil fertility intensification in sugarcane-legume intercropping systems. J Soils Sediments 19:1911–1927. https://doi.org/10.1007/s11368-018-2156-3
Stevenson FJ, Cole MA (1999) Cycles of soil: carbon, nitrogen phosphorus, sulphur and micronutrients, 2nd edn. Wiley, New York
Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209
Syed Ab Rahman SF, Singh E, Pieterse CMJ, Schenk PM (2018) Emerging microbial biocontrol strategies for plant pathogens. Plant Sci 267:102–111
Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI, Erlandson S, Vilgalys R, Liao HL, Smith ME, Peay KG (2014) Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci 111:6341–6346
Taulé C, Mareque C, Barlocco C, Hackembruch F, Reis VM, Sicardi M, Battistoni F (2012) The contribution of nitrogen fixation to sugarcane (Saccharum officinarum L.), and the identification and characterization of part of the associated diazotrophic bacterial community. Plant Soil 356:35–49
Teplitski M, Robinson JB, Bauer WD (2000) Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant-Microbe Interact 13:637–648
Thomson BC, Tisserant E, Plassart P, Uroz S, Griffiths RI, Hannula SE, Buee M, Mougel C, Ranjard L, Van Veen JA, Martin F, Bailey MJ, Lemanceau P (2015) Soil conditions and land use intensification effects on soil microbial communities across a range of European field sites. Soil Biol Biochem 88:403–413
Thrall PH, Hochberg ME, Burdon JJ, Bever JD (2007) Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol Evol 22:120–126
Tkacz A, Cheema J, Chandra G, Grant A, Poole PS (2015) Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J 9:2349–2359
Toju H, Yamamoto S, Sato H, Tanabe AS (2013) Sharing of diverse mycorrhizal and root-endophytic fungi among plant species in an oak-dominated cool-temperate forest. PLoS One 8:e78248
Toussaint JP, Pham TTM, Barriault D, Sylvestre M (2012) Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl Microbiol Biotechnol 95:1589–1603
Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, Osbourn A, Grant A, Poole PS (2013) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7:2248–2258
Uhlik O, Leewis MC, Strejcek M, Musilova L, Mackova M, Leigh MB, Macek T (2013) Stable isotope probing in the metagenomics era: a bridge towards improved bioremediation. Biotechnol Adv 31:154–165
Van Acker H, Van Dijck P, Coenye T (2014) Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333
Van Buyten E, Hofte M (2013) Pythium species from rice roots differ in virulence, host colonization and nutritional profile. BMC Plant Biol 13:203
Van der Ent S, Van Hulten M, Pozo MJ, Czechowski T, Udvardi MK, Pieterse CM, Ton J (2009) Priming of plant innate immunity by rhizobacteria and beta-aminobutyric acid: differences and similarities in regulation. New Phytol 183:419–431
Van der Heijden MG, de Bruin S, Luckerhoff L, van Logtestijn RS, Schlaeppi K (2016) A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J 10:389–399
Van Overbeek L, van Elsas JD (2008) Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol Ecol 64:283–296
Vorholt JA (2012) Microbial life in the phyllosphere. Nat Rev 10:828–840
Wagner MR, Lundberg DS, del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T (2016) Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun 7:12151
Wang JJ, Shen J, Wu YC, Tu C, Soininen J, Stegen JC, He J, Liu X, Zhang L, Zhang E (2013) Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J 7:1310–1321
Wang Z, Solanki MK, Pang F et al (2017) Identification and efficiency of a nitrogen-fixing endophytic actinobacterial strain from sugarcane. Sugar Tech 19:492–500. https://doi.org/10.1007/s12355-016-0498-y
Wang Z, Solanki MK, Yu Z-X et al (2018) Draft genome analysis offers insights into the mechanism by which Streptomyces chartreusis WZS021 increases drought tolerance in sugarcane. Front Microbiol 9:3262. https://doi.org/10.3389/fmicb.2018.03262
Wei Z, Yang T, Friman VP, Xu Y, Shen Q, Jousset A (2015) Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun 6:8413
Yergeau E, Sanschagrin S, Maynard C, St-Arnaud M, Green CW (2014) Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J 8(2):344–358
Yunshi L, Xiukun W, Tuo C, Wanfu W, Guangxiu L, Wei Z, Shiweng L, Minghao W, Changming Z, Huaizhe Z, Gaosen Z (2018) Plant phenotypic traits eventually shape its microbiota: a common garden test. Front Microbiol 9:2479
Zaidi A, Khan MS (2005) Interactive effect of rhizospheric microorganisms on growth, yield and nutrient uptake of wheat. J Plant Nutr 28:2079–2092
Zamioudis C, Korteland J, Van Pelt JA, van Hamersveld M, Dombrowski N, Bai Y, Hanson J, Van Verk MC, Ling HQ, Schulze-Lefert P, Pieterse CM (2015) Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses. Plant J 84:309–322
Zehr JP, Jenkins BD, Short SM, Steward GF (2003) Nitrogenase gene diversity and microbial community structure: A cross-system comparison. Environ Microbiol 5:539–554
Zhang M, Silva MCP, De Mares MC, van Elsas J (2014) The mycosphere constitutes an arena for horizontal gene transfer with strong evolutionary implications for bacterial-fungal interactions. FEMS Microbiol Ecol 89:516–526
Zhang Y, Xu J, Riera N, Jin T, Li J, Wang N (2017) Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 5:97
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Singh, M.P., Singh, P., Singh, R.K., Solanki, M.K., Bazzer, S.K. (2020). Plant Microbiomes: Understanding the Aboveground Benefits. In: Solanki, M., Kashyap, P., Kumari, B. (eds) Phytobiomes: Current Insights and Future Vistas. Springer, Singapore. https://doi.org/10.1007/978-981-15-3151-4_3
Download citation
DOI: https://doi.org/10.1007/978-981-15-3151-4_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-3150-7
Online ISBN: 978-981-15-3151-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)