Abstract
The diminishing concentration of available fossil fuels and increasing global demand of energy have obligated the need for the production of alternate fuels to current petroleum-based fuels. Microbes have the potential to render renewable and sustainable energy sources that are carbon-neutral to counter the elevated concentration of greenhouse gases in the substantial climate changes. Various advancements in sequencing technologies have enabled the study of the microbial diversity and interpreting the variations within the entire genome of organisms and concluding the most feasible pathway of substrate utilization in a comparatively cheaper and faster way. To completely exploit the biofuel-producing potential of these microbes, various genomes have been sequenced and are now available for study. Computational approaches like functional genomics, genome-scale metabolic engineering, and flux balance analysis can be used to improve the H2-producing efficiencies of microbes. Many microorganisms like Enterobacter sp. IIT-BT 08 are reported to have a high rate of H2 production, and its draft genome was generated at DOE Joint Genome Institute (JGI) using Illumina data. The C. perfringens strain JJC was sequenced using the Illumina MiSeq benchtop sequencer which uses a vast variety of carbohydrates producing acetate, butyrate, lactate, ethanol, H2, and carbon dioxide and has various industrial applications. Access to multiple microalgal genome sequences now provides opportunities for application of “omic” approaches to decipher algal lipid metabolism and identify gene targets for the development of potentially engineered strains with optimized lipid content from which biofuel can be produced.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
14.1 Introduction
Microorganisms are the most abundant and diverse forms of life and found in most habitats on the Earth including those conducive to extreme environments like hot springs, glaciers, miles beneath the soils, etc. The genetic, metabolic, and physiological diversity of microbial species is far greater than that found in plants and animals. The enormity of microbial species was estimated to be approximately 1011 to 1012 out of which most of the microbial species are still unknown. Of those species that have been described, their biological diversity is extraordinary, having adapted to grow under extreme temperature, pH, salt concentration, and oxygen levels. Currently, various advancements in sequencing technologies have enabled the study of microbial diversity and interpreting the variations within the entire genome of organisms and concluding the most feasible pathway of substrate utilization in a comparatively cheaper and faster way (Shendure and Ji 2008). The first bacterial genome of Haemophilus influenzae was sequenced in 1995 and took more than 13 months of effort to complete. Today, the entire genome of a microbial species can be sequenced in a very short span of time and take less than 30 h to sequence the entire genome. For example, presently sequencer like MiSeq produced by Illumina was delineated as a fast, personal benchtop sequencer, with very less run time as short as 10 h and outputs planned for targeted sequencing and small genome sequencing (Reuter et al. 2015). The whole genome sequencing of DNA extracted from culturable microorganisms or metagenome (genetic material isolated directly from environmental samples) unveils preliminary idea of the gene associated in numerous pathways related to energy production, metabolism, carbon sequestration, etc. (Fig. 14.1) (Yadav and Dubey 2018).
The key aim of carbon management is to develop options and mechanisms to reduce the causes and effects of climate change, including minimizing emissions and removal of various GHGs through natural and anthropogenic methods. Microbes have played a vital role in regulating the concentration of atmospheric GHGs (e.g., methane, carbon dioxide, nitrous oxide) that impact climate change. Study of microbial diversity involved in cycling of GHGs and integration of microbial genomics to global carbon management could enhance our capability to develop and evaluate microbial strategies for capturing and sequestering atmospheric CO2. Autotrophic microorganisms and macroalgae are known to contribute significantly to CO2 assimilation in aquatic systems such as the oceans and wetlands but have not generally been thought to have a key role in CO2 fixation and sequestration in soils. This is despite the fact that microbial autotrophs have been reported in a number of soil studies.
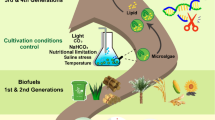
The world is presently focusing on the development of sustainable and nonpolluting energy sources, which will restore fossil fuels in the post-fossil fuel era (Rittmann et al. 2008). There are many alternative future fuels (e.g., hydrogen, methane, ethanol, methanol, gasoline, etc.) among which bio-hydrogen seems to be the most promising because it burns to water, which can be re-used in an environment-friendly manner (Nielsen et al. 2001). The preliminary idea for the production of renewable energy with microorganisms involves using communities of anaerobic microorganisms to transform the energy value in biomass to useful forms of energy. Waste and residues of agriculture, food processing, and other industries contain a large amount of biomass (Pfaltzgraff et al. 2013). Converting the biomass in these wastes in the form of energy provides two advantages at a time: first the generation of renewable energy and second the minimization of environmental pollution. The conversion of biomass to three valuable energy outputs (e.g., methane gas (CH4), hydrogen gas (H2), and electrons from bioelectricity) that are produced by a microbial fuel cell (MFC) can be achieved by various communities of anaerobic microorganisms (Logan 2004). Methanogenesis is already in widespread use today, and microbial sources of H2 and electricity are being intensively investigated. The second approach exploits photoautotrophic microorganisms (e.g., cyanobacteria and eukaryotic algae) that capture the energy of sunlight to grow and thereby produce biomass that can be harvested to augment the biomass produced from nature and agriculture (Fig. 14.2) (Liao et al. 2016). Owing to their high specific growth rates, year-round harvesting, and homogeneity, photosynthetic microorganisms can produce larger (by 100-fold or more) biomass-based energy stocks than plants (Misra et al. 2013). As a result, it might be feasible in production of sufficient microbial biomass to replace fossil fuels. Genomic-based study of microorganisms associated in energy production validated the base sequence of the whole DNA, and all the vital biological reactions of microorganisms can be decoded by the complete genome. On the basis of genomic data, the various metabolic pathways of microbes have been finished with speedy evolution in developing genome projects, a number of microorganisms have been sequenced completely and some are partially sequenced, and annotation of genes from the sequence information is done using bioinformatics (see http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genome_table_cgi). The enzymes that can be coded by their genomes are discovered from the annotated sequence data, for example, the relation of hydrogenases to the redox proteins and enzymes has been demonstrated by biochemical work, and perhaps in the future through the compilation of sequence data. Currently, more than 100 sequences of hydrogenases are accessible, and genomes of 80 microbes have been sequenced (Vignais et al. 2001). The imminent requirement to face the challenges associated in enabling these microorganisms to more realistic alternative for replacing the conventional fossil fuels using different omics-based techniques has been conferred in this chapter with possible future directions.
14.2 Carbon Management
To mitigate the effects of global environmental changes due to increasing emissions of greenhouse gases (GHGs), mainly CO2 is one of the major problems of the twenty-first century (Schmidt et al. 2011). The carbon (C) emitted into the atmosphere was estimated to be 405 ± 30 Gigatons and primarily due to anthropogenic activities over the past 200 years, and as a result, the global atmospheric concentration of CO2 has risen from 280 to 382 ppm in 2007, with a current annual increase of 0.88 ppm (Canadell et al. 2007). Thus to ensure global environmental security and to mitigate the effect of climate change, there is an immediate need to find cost-effective strategies for minimizing anthropogenic CO2 emissions. There are three main methods that can be used to manage carbon in various stages of discovery and development: (i) near-term storage in the terrestrial biosphere where vegetation would fix the CO2 and store it in biomass and soil; (ii) long-term storage in the earth’s soil by pumping CO2 into existing or drilled/excavated sub-surface reservoirs; and (iii) long-term storage in the earth’s oceans where CO2 would be injected thousands of feet deep and trapped by the water. Terrestrial carbon sequestration is an important step towards mitigating anthropogenic CO2 emissions. Increased CO2 concentrations in the atmosphere are thought to be partly contributed by the soil under agriculture. According to an approximation, soils have contributed 55–878 billion tons (GT) of carbon to the total atmospheric CO2 (Kimble et al. 2002). Soil microorganisms are of great importance in soil carbon cycling. Carbon sources are added externally to soil as crop residues undergo microbial decomposition which results in loss of 2/3 of the carbon. A small portion of it is absorbed as the microbial biomass, and a major part of it is released into the environment as CO2. As a result of this process accumulation of comparatively resistant SOC fraction, this is slightly transformed and may be attacked in the future by the microbial population. Carbon management requires the channelizing of CO2 in the atmosphere into long-lived pools to mitigate or reduce their immediate remittance. Soil is a reservoir of huge stock of potentially volatile C which act both as a buffer against atmospheric CO2 increase and as a possible sink for additional C depending on the balance between photosynthesis, the respiration of decomposer organisms, and stabilization of C in soils (Lal 2004; Woodward et al. 2009). Soil C sequestration can significantly contribute to the idea of mitigation and potentially offset a remarkable amount of diffuse CO2 sources for which direct capture is not yet reasonable (King 2011). In terrestrial habitats, plants dominate the uptake of CO2 from the atmosphere by net primary production (NPP), but microorganisms contribute largely to ecosystem C budgets with their roles as decomposers, plant symbionts, or pathogens, hence modifying nutrient availability and affecting C turnover and absorption in soil (King 2011; Lal 2004). Methane (CH4), the second most important greenhouse gas after CO2, contributes to about 20% of the warming effects. Methane oxidation in aerobic soils is mediated by methane-oxidizing bacteria (MOB; methanotrophs), a subset of a physiological group of bacteria (methylotrophs). These bacteria utilize CH4 as the sole carbon and energy source (Dubey 2005; Hanson and Hanson 1996). The role of methanotrophs can never be underestimated as these are the important contributors to attenuate CH4 flux at the oxic-anoxic interfaces (Dutaur and Verchot 2007). The whole genome sequences of methanotrophic bacteria such as Methylocystis sp. Strain Rockwell (ATCC 49242) and Methylomonas methanica MC09 provide insights into the genomic and physiological information that can utilize further to optimize the use of these bacteria in industry or biotechnological purposes (Boden et al. 2011; Stein et al. 2011).
Land management and land use practices can manipulate the terrestrial ecosystem development of distinct microbial communities that support C sequestration (Bardgett et al. 2008; Singh et al. 2010). The fungal to bacterial ratio in soils has been related with C sequestration capability with greater fungal population being related to greater C storage. Recently, a cross-biome metagenomic research has revealed that the ratio of fungal/bacterial rRNA reads showed variation across different soils, among temperate and boreal forests having the highest fungal/bacterial ratios (Fierer et al. 2007). Higher C storage in fungal-dominated soils can be attributed to higher C use efficiency; longer retention of C in living biomass; and recalcitrant necromass resulting in longer resident time of C (Strickland and Rousk 2010). Challenges in manipulating microbial community for enhanced C sequestration arise from the enormous diversity and unculturability of soil microbial communities, which have precluded their comprehensive characterization and limited our understanding on their ecological functions. The new generation of omics methods (e.g., genomics, transcriptomics, proteomics, metabolomics, metagenomics) is proving instrumental in providing valuable information about the taxonomic, genetic, and functional properties of soil microbial communities. These techniques have begun to allow investigation of functional processes of terrestrial microbial communities involved in C cycling that can be incorporated into mechanistic and predictive ecological models (Larsen et al. 2012).
Based on the information gathered from full genome sequences, we infer that bacteria belonging to Acidobacteria and Actinobacteria possess an impressive array of genes allowing breakdown, utilization, and biosynthesis of diverse structural and storage polysaccharides and resilience to stressful soil conditions making them truly ubiquitous in terrestrial ecosystems. This finding supports the metagenomic evidence of higher SOC in Acidobacteria- and Actinobacteria-dominated communities and suggest that these groups promote soil C storage not only due to lifestyle (slow growth and lower metabolic activities) but also by producing polysaccharides for soil structural stability (Singh et al. 2010).
Metagenomic analysis has revealed that structure of microbial communities was markedly different between ambient CO2 (aCO2) and elevated CO2 (eCO2) as indicated by detrended correspondence analysis (DCA) of gene-based pyrosequencing data and functional gene array data. While the abundance of genes involved in decomposing recalcitrant C remained unchanged, those involved in labile C degradation and C and N fixation were significantly increased under eCO2. Here, using metagenomic technologies, we showed that 10 years of field exposure of a grassland ecosystem to eCO2 dramatically altered the structural and functional potential of soil microbial communities (He et al. 2010).
Microbial communities living near the surface layers of oceans are the primary photosynthetic organisms driving the biological pump. Absorbing CO2 and sunlight to produce most oceanic organic materials, the organisms make up the foundation of the marine food chain. Photosynthesis of phytoplankton such as diatoms, dinoflagellates, and cyanobacteria converts about as much atmospheric carbon to organic carbon in the ocean as plant photosynthesis does on land. Large variations in phytoplankton abundance, therefore, can greatly impact the oceans’ ability to take up atmospheric carbon. Phytoplankton photosynthesis (Rivkin and Legendre 2001) fixes approximately 45 Pg C year−1 (Falkowski et al. 2000). Dominant organisms in surface waters include such as cyanobacteria as Synechococcus sp. and Prochlorococcus marinus, which capture CO2 and light to carry out photosynthesis. Prochlorococci now are thought to be the most abundant photosynthetic organisms on earth. Eukaryotic diatoms such as the recently sequenced Thalassiosira pseudonana also live in surface waters and convert CO2 and other nutrients into hard silicates. This process carries organically complexed carbon to ocean depths, thus converting its relatively rapid cycling in surface waters (where it is returned to the atmosphere) to a considerably slower one in ocean sediments. The main goal of ocean carbon sequestration is to increase the export of deep ocean inventory of CO2. Two approaches are taken into account: direct injection of a CO2 stream into the ocean depths and iron fertilization to increase photosynthesis by phytoplankton in the biological pump and thus enhance the uptake of carbon.
14.3 Energy Production
To mitigate the increased discharge of greenhouse gases in the atmosphere and to fulfill the mounting global demand for energy to counter the decreasing concentration of fossil fuels have necessitated the production of alternate environment-friendly fuels. Different biological processes for the production of fuels such as ethanol, diesel, hydrogen (H2), methane, etc. have capability to furnish sustainable energy system for the betterment of society (Angenent et al. 2004). In recent years, the interest in the production of different kinds of biofuels by exploiting microorganisms has been increasing steadily (Liao et al. 2016) especially because of the metabolic variety of different microorganisms that makes possible the production of biofuels from different substrates. For example, most of the bacteria can effortlessly transform sugars into ethanol, and cellulolytic microbes can use plant-driven substrates in the production of biofuels. Cyanobacteria and microalgae possess the capability to reduce the atmospheric CO2 into biofuels photosynthetically, and methanotrophs can utilize methane to produce methanol (Liao et al. 2016). The genomic data of sequenced microbes can be connected to biofuel production yields. Genome sequence and metabolic pathway databases can be utilized for screening microbes. The National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp) databases greatly facilitate such analyses. Sequence analysis and pathway alignment of hydrogen metabolism in complete and incomplete genomes have led to the identification of potential hydrogen producers (Kalia et al. 2003) (Table 14.1). The work of Carere et al. (2012) demonstrated that the presence or absence of specific genes dictating carbon and electron flow towards end products may be used to infer end-product synthesis patterns and help to develop informed metabolic engineering strategies for optimization of H2 and ethanol yields. Furthermore, certain genes may be used as suitable biomarkers for screening novel microorganisms’ capability of producing optimal H2 or ethanol and may be suitable targets for metabolic engineering strategies for optimization of either ethanol or H2 yields.
14.3.1 Biological Hydrogen Production
The biological hydrogen production procedure makes utilization of microorganisms that tend to produce H2 from lignocellulosic biomass and waste material (Bakonyi et al. 2014; Kumar et al. 2015). These materials are excellent source of fermentable sugar and are present in complex form and hardly digestible (Kumar et al. 2008). Direct or indirect biophotolysis, photo-fermentation, and dark fermentation methods are exploited for biological hydrogen production. The lower level yield of H2 by biological hydrogen production methods is one of the major challenges that need to be addressed before it can be used for industrial purpose. Apart from wet lab experiments, in silico approaches which include functional genomics, genome-scale metabolic engineering, and flux balance analysis can be used to improve the H2-producing capabilities. Many microorganisms are being explored for future biohydrogen generation at industrial scales. Among them, Enterobacter sp. IIT-BT 08 is reported to have a high rate of H2 production, and its draft genome was generated at DOE Joint Genome Institute (JGI) using Illumina data. A complete genome sequence analysis was carried out for further enhancement of H2 production by strain development (Khanna et al. 2013). Halanaerobium hydrogeniformans isolated from the haloalkaline environment is an obligately anaerobic, Gram-negative, nonmotile, nonsporulating, elongated rod. It can ferment a vast range of carbohydrates with optimal growth at pH 11, and 33°C, and produce acetate, formate, and H2 as major metabolic end products. The H. hydrogeniformans genome was sequenced using a combination of Illumina and 454 technologies to improve assessment of its metabolic and bioenergy potential for robust H2 production (Brown et al. 2011). Similarly, Clostridium perfringens, a Gram-positive and spore-forming strict anaerobe, can successfully utilize a vast variety of carbohydrates producing acetate, butyrate, lactate, ethanol, H2, and carbon dioxide, which have industrial applications. The genome sequencing of C. perfringens strain JJC was performed using the Illumina MiSeq benchtop sequencer (2150-bp paired-end sequencing). The whole-genome shotgun project of C. perfringens strain JJC containing its assembly and annotation has been deposited at DDBJ/EMBL/Gene Bank under the accession no.AWRZ00000000 (Wong et al. 2014) for further applications.
14.3.2 Liquid Biofuels
Liquid biofuels from plants and microalgae feedstock represent a renewable sustainable alternative to petroleum energy. The greatly minimized acreage estimates, high lipid or starch content, and biomass production rates that surpass those of terrestrial plants suggest that biodiesel or ethanol derived from lipids or starch produced by microalgae may circumvent many of the limitations ascribed to petroleum fuel and first-generation plant-based biofuels. An in-depth knowledge of microalgae genomics precludes these necessary increases in biological efficiency. Access to multiple microalgal genome sequences now provides a wealth of opportunities for application of “omic” approaches to unravel algal lipid metabolism and identify gene targets for the development of potentially engineered strains with optimized lipid content (Beer et al. 2009; Georgianna and Mayfield 2012; Mukhopadhyay et al. 2008; Rodríguez-Moyá and Gonzalez 2010; Yu et al. 2011). Bio-oil from microalgae can be used directly as fuel or chemically trans-esterified into biodiesel. Microalgae seem to be an attractive way to produce biofuel due to their ability to accumulate lipids and their very high actual photosynthetic yields; about 3–8% of solar energy can be converted to biomass, whereas observed yields for terrestrial plants are about 0.5%. The genetic information of the sequenced organisms has enabled the metabolic pathways for the lipid synthesis and which can be used in genetic engineering process efforts directed towards augmenting lipid accumulation in microalgae (Georgianna and Mayfield 2012). KEGG (Kyoto Encyclopedia of Genes and Genomes) (http://www.genome.jp/kegg/) is one of the most widely used comprehensive resources of metabolic pathways including for several organisms (Kanehisa et al. 2010). Currently, genome-wide studies have employed KEGG pathway database to identify genes and reconstruct major lipid biosynthetic pathways in various oleaginous microalgal species (Hashimoto et al. 2008; Misra et al. 2013; Rismani-Yazdi et al. 2011; Smith et al. 2012). Most of the omic-based studies undertaken so far have primarily addressed identification of gene targets for improving lipid production in microalgae. Now it is apparent that modification of the fatty acid profile to include more stearic acid (C18:0) and oleic acid (C18:1) is also indispensable for improving the algal-derived biofuel properties (Knothe 2009).
To date, ethanol accounts for up to 75% of the total biofuel use. Bioethanol dominates the market with a sale of 58 billion dollars per year. Nearly 50% of global sugar is utilized for ethanol production, and approximately 86,000 k ton/year ethanol so produced is majorly used for biofuel application (Burk 2010). The United States and Brazil are the leading producers dominantly using simple substrates such as corn and sugarcane, respectively (Aro 2016). Even though India is the second largest producer of sugarcane in the world, it contributes to only 2% of the global bioethanol production. Despite the abundant supply of lignocelluloses, their commercial conversion to ethanol is limited to their recalcitrance (due to lignin sheath) to degradation and unique chemical composition (Zhao et al. 2012). In order to produce bioethanol from lignocellulosic biomass that is economically feasible, sustainable, and competitive with petroleum-based fuels, conventional process steps need to be integrated into the consolidated process to avoid maximum production of inhibitory sugar derivatives and achieve high ethanol titers. Environmental stresses and inhibitors encountered by Saccharomyces cerevisiae strains are the main limiting factors in bioethanol fermentation. Strains with different genetic backgrounds usually show diverse stress tolerance responses. An understanding of the mechanisms underlying these phenotypic diversities within S. cerevisiae populations could guide the construction of strains with desired traits. This kind of study, provided novel transcriptomic information on microbes and their RNA-seq data were useful in targeting genes involved in ethanol production for future genetic engineering (Wang et al. 2016).
14.3.3 Microbial Fuel Cell (MFC)
MFC is a biochemical-catalyzed system, which generates electricity by oxidizing soluble or dissolved organic wastes in the presence of either fermentative bacteria or enzyme. MFC technology relies on the electrogenic nature of certain bacteria while treating different wastewater and producing electrical energy. The microorganism generally presents in the anode chamber of fuel cell act as biocatalyst and generates electrons (e−) and protons (H+) by way of anaerobic respiration of organic substrate. The electron transfer through the anode is integrated with an external circuit to cathode and protons through the proton exchange membrane (which separates cathode and anode chamber) into the cathode chamber where they combine with the help of a mediator. The potential between the respiratory system and electron acceptor generates the current and voltage needed to make electricity (Logan 2004). Recently, a number of bacteria such as Shewanella putrefaciens, family of Geobacteraceae, Rhodoferax ferrireducens, Bacillus subtilis, Geobacter sulfurreducens, and Escherichia coli were reported in the literature and have the ability to transfer produced electrons from oxidized fuel (substrate) to the electrode without using artificial mediator, making it possible to establish mediator-less MFCs (Kaufmann and Lovley 2001; Kim et al. 1999). The 16S rDNA analysis of anode biofilm and suspended cells reveals predominance bacterial community involved in electron transfer. Patil et al. (2009) studied the activated sludge-based microbial fuel cell and analyzed the developed microbial community in the anode chamber and reported the predominance of β-Proteobacteria clones with 50.6% followed by unclassified bacteria (9.9%), α-Proteobacteria (9.1%), other Proteobacteria (9%), Planctomycetes (5.8%), Firmicutes (4.9%), Nitrospora (3.3%), Spirochaetes (3.3%), Bacteroides (2.4%), and γ-Proteobacteria (0.8%). Diverse bacterial groups represented as members of the anode chamber community. They suggest that chocolate wastewater has a potential for future MFC practical applications as it can provide a readily biodegradable waste source for electricity generation.
14.4 Conclusion and Future Prospective
Microbes have the potential to transform biomass into eco-friendly biofuels like ethanol, H2, etc. through bioprocessing. To address the global energy crisis, genomics shall play an important role. Through knowledge of different omics technology, it will be easy to develop a better understanding to harness different renewable and carbon-neutral energy sources like lignocellulosic biomass, microalgae, and cyanobacteria. Moreover, the genetic engineering of different enzymes will be a vital factor in optimizing the development of sustainable energy in the form of biofuel. In the coming years, researchers will continue to look to nature for solutions to the global energy crisis. By applying genomic research and engineering to renewable fuel stocks and the bacteria and enzymes that convert those sources to energy, scientists can optimize billions of years of evolution to meet our growing energy needs in an environmentally friendly way.
References
Angenent LT, Karim K, Al-Dahhan MH, Wrenn BA, Domíguez-Espinosa R (2004) Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol 22(9):477–485
Aro EM (2016) From first generation biofuels to advanced solar biofuels. Ambio 45:24–31
Bakonyi P, Nemestóthy N, Simon V, Bélafi-Bakó K (2014) Review on the start-up experiences of continuous fermentative hydrogen producing bioreactors. Renew Sust Energ Rev 40:806–813
Bao G, Wang R, Zhu Y, Dong H, Mao S, Zhang Y, Chen Z, Li Y, Ma Y (2011) Complete genome sequence of Clostridium acetobutylicum DSM 1731, a solvent-producing strain with multireplicon genome architecture. J Bacteriol 193(18):5007–5008
Bardgett RD, Freeman C, Ostle NJ (2008) Microbial contributions to climate change through carbon cycle feedbacks. ISME J 2:805
Beer LL, Boyd ES, Peters JW, Posewitz MC (2009) Engineering algae for biohydrogen and biofuel production. Curr Opin Biotechnol 20(3):264–271
Boden R, Cunliffe M, Scanlan J, Moussard H, Kits KD, Klotz MG, Jetten MSM, Vuilleumier S, Han J, Peters L, Mikhailova N, Teshima H, Tapia R, Kyrpides N, Ivanova N, Pagani I, Cheng J-F, Goodwin L, Han C, Hauser L, Land ML, Lapidus A, Lucas S, Pitluck S, Woyke T, Stein L, Murrell JC (2011) Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J Bacteriol 193:7001
Brown SD, Begemann MB, Mormile MR, Wall JD, Han CS, Goodwin LA, Pitluck S, Land ML, Hauser LJ, Elias DA (2011) Complete genome sequence of the haloalkaliphilic, hydrogen-producing bacterium Halanaerobium hydrogeniformans. J Bacteriol 193:3682–3683
Burk MJ (2010) Sustainable production of industrial chemicals from sugars. Int Sugar J 112:30–35
Canadell JG, Le Quéré C, Raupach MR, Field CB, Buitenhuis ET, Ciais P, Conway TJ, Gillett NP, Houghton RA, Marland G (2007) Contributions to accelerating atmospheric CO2 growth from economic activity, carbon intensity, and efficiency of natural sinks. Proc Natl Acad Sci 104:18866–18870
Carere CR, Rydzak T, Verbeke TJ, Cicek N, Levin DB, Sparling R (2012) Linking genome content to biofuel production yields: a meta-analysis of major catabolic pathways among select H2 and ethanol-producing bacteria. BMC Microbiol 12:295
de Vrije T, Mars AE, Budde MA, Lai MH, Dijkema C, de Waard P, Claassen PA (2007) Glycolytic pathway and hydrogen yield studies of the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Microbiol Biotechnol 74:1358–1367
Desiniotis A, Kouvelis VN, Davenport K, Bruce D, Detter C, Tapia R, Han C, Goodwin LA, Woyke T, Kyrpides NC, Typas MA, Pappas KM (2012) Complete genome sequence of the ethanol-producing Zymomonas mobilis subsp. mobilis centrotype ATCC 29191. J Bacteriol 194(21):5966–5967
Dubey S (2005) Microbial ecology of methane emission in rice Agroecosystem: a review. Appl Ecol Environ Res 3:1–27
Dutaur L, Verchot LV (2007) A global inventory of the soil CH4 sink. Glob Biogeochem Cy 21
Falkowski P, Scholes RJ, Boyle E, Canadell J, Canfield D, Elser J, Gruber N, Hibbard K, Högberg P, Linder S, Mackenzie FT, Moore B III, Pedersen T, Rosenthal Y, Seitzinger S, Smetacek V, Steffen W (2000) The Global Carbon Cycle: A Test of Our Knowledge of Earth as a System. Science 290:291–296
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Georgianna DR, Mayfield SP (2012) Exploiting diversity and synthetic biology for the production of algal biofuels. Nature
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 60:439–471
Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M (2008) The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res 49(1):183–191
He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, Van Nostrand JD, Hobbie SE, Reich PB, Zhou J (2010) Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett 13(5):564–575
Kalia VC, Lal S, Ghai R, Mandal M, Chauhan A (2003) Mining genomic databases to identify novel hydrogen producers. Trends Biotechnol 21:152–156
Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38(Database issue):D355–D360
Kataeva IA, Yang SJ, Dam P, Poole FL 2nd, Yin Y, Zhou F, Chou W-C, Xu Y, Goodwin L, Sims DR, Detter JC, Hauser LJ, Westpheling J, Adams MW (2009) Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium “Anaerocellum thermophilum” DSM 6725. J Bacteriol 191(11):3760–3761
Kaufmann F, Lovley DR (2001) Isolation and characterization of a soluble NADPH-dependent Fe(III) reductase from Geobacter sulfurreducens. J Bacteriol 183:4468
Khanna N, Ghosh AK, Huntemann M, Deshpande S, Han J, Chen A, Kyrpides N, Mavrommatis K, Szeto E, Markowitz V, Ivanova N, Pagani I, Pati A, Pitluck S, Nolan M, Woyke T, Teshima H, Chertkov O, Daligault H, Davenport K, Gu W, Munk C, Zhang X, Bruce D, Detter C, Xu Y, Quintana B, Reitenga K, Kunde Y, Green L, Erkkila T, Han C, Brambilla E-M, Lang E, Klenk H-P, Goodwin L, Chain P, Das D (2013) Complete genome sequence of Enterobacter sp. IIT-BT 08: A potential microbial strain for high rate hydrogen production. Stand Genomic Sci 9:359–369
Kim BH, Kim HJ, Hyun MS, Park DH (1999) Direct electrode reaction of Fe(III)-reducing bacterium, Shewanella putrefaciens. J Microbiol Biotechnol 9:127–131
Kimble JM, Lal R, Follett RF (2002) Agricultural practices and policies for carbon sequestration in soil. CRC Press, Boca Raton
King GM (2011) Enhancing soil carbon storage for carbon remediation: potential contributions and constraints by microbes. Trends Microbiol 19:75–84
Knothe G (2009) Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ Sci 2:759–766
Kumar R, Singh S, Singh OV (2008) Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol 35(5):377–391
Kumar G, Bakonyi P, Periyasamy S, Kim SH, Nemestóthy N, Bélafi-Bakó K (2015) Lignocellulose biohydrogen: Practical challenges and recent progress. Renew Sust Energ Rev 44:728–737
Lal R (2004) Soil carbon sequestration impacts on global climate change and food security. Science 304:1623–1627
Larsen PE, Field D, Gilbert JA (2012) Predicting bacterial community assemblages using an artificial neural network approach. Nat Methods 9:621
Li X, Huang S, Yu J, Wang Q, Wu S (2013) Improvement of hydrogen production of Chlamydomonas reinhardtii by co-cultivation with isolated bacteria. Int J Hydrog Energy 38:10779–10787
Liao JC, Mi L, Pontrelli S, Luo S (2016) Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat Rev Microbiol 14(5):288–304
Logan BE (2004) Peer reviewed: extracting hydrogen and electricity from renewable resources. Environ Sci Technol 38:160A–167A
Misra N, Panda PK, Parida BK (2013) Agrigenomics for microalgal biofuel production: an overview of various bioinformatics resources and recent studies to link OMICS to bioenergy and bioeconomy. OMICS 17(11):537–549
Mukhopadhyay A, Redding AM, Rutherford BJ, Keasling JD (2008) Importance of systems biology in engineering microbes for biofuel production. Curr Opin Biotechnol 19(3):228–234
Nielsen AT, Amandusson H, Bjorklund R, Dannetun H, Ejlertsson J, Ekedahl L-G, Lundström I, Svensson BH (2001) Hydrogen production from organic waste. Int J Hydrog Energy 26:547–550
O-Thong S, Khongkliang P, Mamimin C, Singkhala A, Prasertsan P, Birkeland NK (2017) Draft genome sequence of Thermoanaerobacterium sp. strain PSU-2 isolated from thermophilic hydrogen producing reactor. Genom Data 12:49–51
Ouhib-Jacobs O, Lindley ND, Schmitt P, Clavel T (2009) Fructose and glucose mediates enterotoxin production and anaerobic metabolism of Bacillus cereus ATCC14579(T). J Appl Microbiol 107(3):821–829
Patil SA, Surakasi VP, Koul S, Ijmulwar S, Vivek A, Shouche YS, Kapadnis BP (2009) Electricity generation using chocolate industry wastewater and its treatment in activated sludge based microbial fuel cell and analysis of developed microbial community in the anode chamber. Bioresour Technol 100:5132–5139
Pfaltzgraff LA, De bruyn M, Cooper EC, Budarin V, Clark JH (2013) Food waste biomass: a resource for high-value chemicals. Green Chem 15:307–314
Reuter JA, Spacek DV, Snyder MP (2015) High-throughput sequencing technologies. Mol Cell 58(4):586–597
Rismani-Yazdi H, Haznedaroglu BZ, Bibby K, Peccia J (2011) Transcriptome sequencing and annotation of the microalgae Dunaliella tertiolecta: Pathway description and gene discovery for production of next-generation biofuels. BMC Genomics 12:148
Rittmann BE, Krajmalnik-Brown R, Halden RU (2008) Pre-genomic, genomic and post-genomic study of microbial communities involved in bioenergy. Nat Rev Microbiol 6:604
Rivkin RB, Legendre L (2001) Biogenic carbon cycling in the upper ocean: effects of microbial respiration. Science 291(5512):2398–2400
Rodríguez-Moyá M, Gonzalez R (2010) Systems biology approaches for the microbial production of biofuels. Biofuels 1:291–310
Roh H, Ko H-J, Kim D, Choi DG, Park S, Kim S, Chang IS, Choi I-G (2011) Complete Genome Sequence of a Carbon Monoxide-Utilizing Acetogen, Eubacterium limosum KIST612. J Bacteriol 193:307–308
Rydzak T, Levin DB, Cicek N, Sparling R (2009) Growth phase-dependant enzyme profile of pyruvate catabolism and end-product formation in Clostridium thermocellum ATCC 27405. J Biotechnol 140(3–4):169–175
Schmidt MWI, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, Kleber M, Kögel-Knabner I, Lehmann J, Manning DAC, Nannipieri P, Rasse DP, Weiner S, Trumbore SE (2011) Persistence of soil organic matter as an ecosystem property. Nature 478:49
Shendure J, Ji HL (2008) Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145
Singh BK, Bardgett RD, Smith P, Reay DS (2010) Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8:779–790
Smith SR, Abbriano RM, Hildebrand M (2012) Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon partitioning pathways. Algal Res 1:2–16
Stein LY, Bringel F, DiSpirito AA, Han S, Jetten MSM, Kalyuzhnaya MG, Kits KD, Klotz MG, Op den Camp HJM, Semrau JD, Vuilleumier S, Bruce DC, Cheng J-F, Davenport KW, Goodwin L, Han S, Hauser L, Lajus A, Land ML, Lapidus A, Lucas S, Médigue C, Pitluck S, Woyke T (2011) Genome sequence of the methanotrophic Alphaproteobacterium Methylocystis sp. Strain Rockwell (ATCC 49242). J Bacteriol 193:2668
Strickland MS, Rousk J (2010) Considering fungal:bacterial dominance in soils – Methods, controls, and ecosystem implications. Soil Biol Biochem 42:1385–1395
Su H, Zhang T, Bao M, Jiang Y, Wang Y, Tan T (2014) Genome Sequence of a Promising Hydrogen-Producing Facultative Anaerobic Bacterium, Brevundimonas naejangsanensis Strain B1. LID - 10.1128/genomeA.00542-14 [doi] LID - e00542-14 [pii]. Genome, Announc
Vignais PM, Billoud B, Meyer J (2001) Classification and phylogeny of hydrogenases. FEMS Microbiol Rev 25(4):455–501
Wang J, Suzuki T, Dohra H, Takigami S, Kako H, Soga A, Kamei I, Mori T, Kawagishi H, Hirai H (2016) Analysis of ethanol fermentation mechanism of ethanol-producing white-rot fungus Phlebia sp. MG-60 by RNA-seq. BMC Genomics 17(1):616
Wong YM, Juan JC, Gan HM, Austin CM (2014) Draft Genome Sequence of Clostridium perfringens Strain JJC, a Highly Efficient Hydrogen Producer Isolated from Landfill Leachate Sludge. Genome Announc 2:e00064–e00014
Woodward FI, Bardgett RD, Raven JA, Hetherington AM (2009) Biological approaches to global environment change mitigation and remediation. Curr Biol 19:R615–R623
Yadav S, Dubey SK (2018) Cellulose degradation potential of Paenibacillus lautus strain BHU3 and its whole genome sequence. Bioresour Technol 262:124–131
Yu W-L, Ansari W, Schoepp NG, Hannon MJ, Mayfield SP, Burkart MD (2011) Modifications of the metabolic pathways of lipid and triacylglycerol production in microalgae. Microb Cell Factories 10:91
Zhao XQ, Zi LH, Bai FW, Lin HL, Hao XM, Yue GJ, Ho NWY (2012) Bioethanol from lignocellulosic biomass. Adv Biochem Eng Biotechnol 128:25–51
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Neha, Singh, A., Yadav, S., Bhardwaj, Y. (2019). Linking Microbial Genomics to Renewable Energy Production and Global Carbon Management. In: Tripathi, V., Kumar, P., Tripathi, P., Kishore, A. (eds) Microbial Genomics in Sustainable Agroecosystems. Springer, Singapore. https://doi.org/10.1007/978-981-13-8739-5_14
Download citation
DOI: https://doi.org/10.1007/978-981-13-8739-5_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8738-8
Online ISBN: 978-981-13-8739-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)