Abstract
This chapter focuses on protein kinases that transfer the phosphate group of ATP to the hydroxyl group of a substrate protein. Five hundred eighteen human protein kinases are classified into serine/threonine kinases and tyrosine kinases and individually or synergistically transduce physiologic stimuli into cell to promote cell proliferation or apoptosis, etc. Protein kinases are identified as drug targets because dysfunction of kinases leads to severe diseases such as cancers and autoimmune diseases. A large number of the crystal structures of the protein kinase inhibitor complex are available in Protein Data Bank and facilitated the drug discovery targeting protein kinases. The protein kinase inhibitors are classified into categories, Type-I, Type-II, Type-III, Type-IV, and Type-V, and as a separate class, covalent-type inhibitors. In any type, a protein kinase inhibitor bound to the allosteric region is advantageous in terms of selectivity compared to the traditional ATP-competitive one. In the following sections, the successful and promising examples of the partially or fully allosteric protein kinase inhibitors are illustrated in the following pages.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction
Protein kinases (PKs) catalyze the γ-phosphate group transfer from ATP to the hydroxyl group of substrate proteins requiring magnesium ions. Five hundred eighteen protein kinases had been identified in human genome [23] and divided into tyrosine kinases (TK), serine/threonine kinases (STK), and pseudo-kinases. PK transduces the cellular signals started with the extracellular stimulations to regulate the complicated physiological functions involving cell differentiation, proliferation, and apoptosis. Therefore, collapse of the vital valance sophisticatedly regulated by PKs results in the serious diseases such as cancers. Approximately 30 kinase inhibitors including imatinib and gefitinib have been approved as molecular target drugs by the Food and Drug Administration (FDA) of the USA to date [43]. Modern scientific technologies allow to discover high-potency inhibitors for a target protein kinase associated with the diseases. However, it remains a serious challenge to develop inhibitors to be drugs. Promiscuous kinase inhibitors would suppress a variety of the off-target protein kinases as well as the target kinase and exert the adverse effects on vital. X-ray crystal analyses of the premature inhibitor with the off-target PKs as well as with the target kinase would promote drug discovery process.
PK involves a wide variety of combinations of the functional domains or subunits. About 200 kinds of human protein kinase domain structures are available from the Protein Data Bank (PDB). These crystal structures reveal that the catalytic domain of PK configures a typical kinase fold consisting of an N-lobe involving a α-helix (αC-helix) and five β-strands, a C-lobe with high helicity, and a hinge region connecting these lobes (Fig. 3.1a). The numbering and alphabeting of the β-strands and α-helices have been defined upon the structure of the protein kinase A (Fig. 3.1a) [11]. The hinge region composed of a single strand contributes to the plasticity of the relative positions of the N- and C-lobes and the recognition of the adenine ring of ATP. The flexible regions such as P-loop, A-loop, N- and C-terminals, and insertion loop singly or collaboratively regulate the protein kinase activity, concerted with the regulatory subunit (or protein) binding or detaching and/or chemical modifications such as phosphorylation. The phosphate binding loop (P-loop) with a glycine-rich sequence in the N-lobe binds ATP or ATP-competitive inhibitors. The activation loop (A-loop), which is bracketed by the highly conserved DFG (Asp-Phe-Gly) and APE (Ala-Pro-Glu) motifs, functions as a molecular switch via phosphorylation by the client kinase or itself (autophosphorylation). The A-loop works as a platform for substrate binding in the active state and as a molecular brake in the inactive state. The aspartate residue in the DFG motif binds a magnesium ion that is essential for enzyme activity. Activation by promoting factors such as phosphorylation provokes the configuration changes in the N-lobe position relative to the C-lobe, A-loop, and αC-helix. This structural transition results in the salt-bridge formation between the catalytic lysine residue in the β3-strand and glutamate residue in the αC-helix, which can accommodate the ATP molecule in the transition-state conformation. The active conformation is structurally stabilized by two intramolecular spines, regulatory and catalytic spines (R-spine and C-spine), which consist of the hydrophobic residues, respectively (Fig. 3.1b). The C-spine involves the adenine ring of the ATP molecule, and the R-spine does the benzene ring in the DFG motif (Fig. 3.1b). In case that these structural requirements are prepared, the protein kinase is ready for the catalytic reaction. The catalytic loop (C-loop) contains the catalytic aspartate residue which eliminates the proton from the hydroxyl group in the substrate. Subsequently, the activated hydroxyl group of the substrate makes a nucleophilic attack to the phosphorous atom in the γ-phosphate group of ATP. The N- and C-terminal regions and/or insertion loop represents remarkable diversity in amino acid sequence and size and allosterically regulates the protein kinase activity involving the auto-inhibition, substrate recognition, and subcellular localization. The structural clarification of the allosteric molecular switches eventually provides the valuable clues for producing highly selective protein kinase inhibitors.
A typical protein kinase fold. (a) The numbering and alphabeting of the secondary structure and the nomenclatures of the sub-domains are shown on the protein kinase A. (b) Hydrophobic intramolecular spines. The regulatory spine (R-spine, brown) and catalytic spine (C-spine, green) consisting of the hydrophobic amino acid residues stabilize the protein kinase fold. The phenylalanine residue in the DFG motif is jointed to the R-spine
3.2 Classification of Protein Kinase Inhibitors
Approximately 3000 crystal structures of the protein kinase-inhibitor complex have been registered in the Protein Data Bank (PDB) and facilitated the anti-protein kinase drug discovery. Kinase inhibitors are classified into categories Type-I, Type-II, Type-III, Type-IV, and Type-V and, as a separate class, covalent-type inhibitors as shown in Table. 3.1 [24]. Type-I inhibitor binds mainly to the ATP binding region, which is located at the hydrophobic slot between the N- and C-lobes. The structural requirements regarding ATP binding in this region are well-conserved among protein kinases. The main chain atoms in the hinge region make the hydrogen bonds with the adenine moiety of ATP. The hydrophobic residues participated in the R-spine bracket the adenine ring of ATP (Fig. 3.1b). Thus, a number of Type-I inhibitors involve the hetero-aromatic ring as a substitute for the adenine moiety. The lysine and magnesium ion ligated at the aspartate residue in the DFG motif, which are conserved in the ATP binding site, largely contribute to recognize the phosphate groups. The hydrogen bonding with these lysine and aspartate residues is essential for the potential inhibitors. Type-II inhibitor binds to the so-called DFG-out conformation, which is caused by the flipping motion of the DFG motif in the N-terminal end of the A-loop. This motion results in the emergence of the hydrophobic pocket, referred to as DFG-out region, a poorly conserved region among protein kinases. Type-II inhibitors bind to the DFG-out region as well as the ATP binding site and thus present the rather high selectivity against the off-target kinases. Type-III inhibitor binds to the near-allosteric region such as the DFG-out region but not to the ATP binding region. Type-IV inhibitor binds to the far-allosteric region, distant to the ATP site, involving the substrate binding or autoregulatory regions. Type-V inhibitor is composed of the ATP analogue and peptide derived from substrate protein and covers almost all of the enzyme reaction sites [31]. However, Type-V inhibitor is apt to display the low inhibitory activity because the peptide moiety causes the entropy loss due to its conformational flexibility. Further, the peptide part contains not less than ten residues by which the molecular weight of the Type-V inhibitors greatly exceeds to 1000 daltons. The peptide sequence around the phosphorylation site configures the extended formation and forms the β-sheet structure with the A-loop. Namely, protein kinases request the rather low sequence specificity for the substrate recognition. These obstacles likely put a damper on the development of the Type-V inhibitors. Recently, it had reported that the covalent-type inhibitor was effective for a kind of protein kinases that possessed the cysteine residues around the ATP binding site [22]. The cysteine residues are observed in the hinge region, gatekeeper, P-loop, A-loop, and the other allosteric sites in protein kinases, but not conserved among all of them. In practice, these cysteine residues were significant for developing the highly selective drugs such as afatinib and ibrutinib (Table. 3.1). Crystal structure showed that the α,β-unsaturated ketone moiety of afatinib covalently bound to Cys797 in the hinge region end of epidermal growth factor receptor (EGFR) [37]. In addition of the covalent bond, the 2-chloro-3-fluorobenzene moiety bound into the deep ATP site which was occupied by the methylbenzene moiety of erlotinib (described in detail in the next section), and the quinazoline ring forms the hydrogen bonds with the main chain atoms in the hinge region. Afatinib is classified into the Type-I inhibitor with the covalent bonding. Finally, the covalent bonding character could be introduced to all types of protein kinase inhibitors.
The comparison in the amino acid sequence and crystal structure suggests that the protein kinase inhibitor bound to the allosteric region is advantageous in terms of selectivity compared to the traditional ATP-competitive one such as staurosporine. The successful and promising examples of the partially or fully allosteric kinase inhibitor are illustrated in the following pages.
3.3 Utilization of the ATP Back Pocket, a Near-Allosteric Site/Type-I
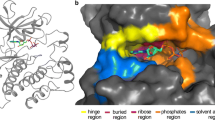
Due to binding to the highly conserved ATP binding region, the Type-I inhibitors display a low selectivity against the off-target kinases. However, the extensive structural dissections in the deep-inside and/or peripheral region of the ATP binding site clarified the subtle but unique structural differences among protein kinases and promoted to produce the highly selective Type-I inhibitors [21]. The gatekeeper residue and N-terminal neighboring one of the DFG motif (DFG-1) in front of the hydrophobic pocket in the deep ATP site, referred to as ATP back pocket, are the key residues for the definition of pharmacophore for protein kinase inhibitors (Fig. 3.2) [46]. This ATP back pocket is divergent among protein kinases and has been utilized for drug discovery of selective protein kinase inhibitors such as gefitinib and erlotinib (Table. 3.1). Erlotinib, an anticancer drug, targets epidermal growth factor receptor (EGFR). The threonine residues as the gatekeeper and DFG-1 cause the large hydrophobic back pocket deep in the ATP binding site, thus defined as a near-allosteric site (Fig. 3.2). The acetylenyl benzene moiety of erlotinib binds to the ATP back pocket (Fig. 3.2) [32]. This characteristic interaction allows erlotinib to be highly selective for EGFR. However, the clinical efficacy of gefitinib and erlotinib is ultimately reduced by the emergence of the acquired drug resistance such as by mutation of the gatekeeper Thr790 residue of EGFR (T790M), which is detected in half of the drug-administrated patients. The resulting Met790 buried the back pocket in the deep ATP site essential for the recognition of the acetylenyl benzene moiety of erlotinib and significantly decreased the binding affinity of erlotinib with the T790M mutant. Zhou et al. discovered a novel compound effective to the T790M mutant and showed that this inhibitor fitted into the modified ATP binding site [45]. On the other hand, the clinical activity of the Type-I inhibitor entrectinib was greatly limited by the acquired resistance by the mutation at the DFG-1 position of tropomyosin receptor kinase A (TrkA) (G667C). The resulting Cys667 works as a steric hindrance for entrectinib binding to the ATP back pocket of the G667C mutant, while foretinib suppressed both of the wild-type and G667C mutant [28]. The molecular modification matched for the drug-resistant mutant could be implemented, but cancer cells would acquire the resistance against the new drug. The cat-and-mouse game between the cancer cells and human wisdom will be permanently continued. Nevertheless, the ATP back pocket plays a key role in producing highly selective Type-I protein kinase inhibitors.
The ATP back pocket of the epidermal growth factor receptor (EGFR) kinase domain. The acetylenyl benzene group of erlotinib (orange) intrudes into the ATP back pocket (enclosed black dot line). The gatekeeper (Thr766, shown in red) and DFG-1 (Thr830, shown in green) residues are located in front of the ATP back pocket
3.4 Utilization of the DFG-out Region, a Near-Allosteric Site/Type-II
The DFG-out region of protein kinase has appeared adjacent to the ATP binding site along with the flip-flop motion in the DFG motif. In the innate DFG-out conformation (DFG-in), the side chain of the aspartate residue is faced into the ATP site and that of the phenylalanine residue is involved in the R-spine outside of the ATP site. In the DFG-out conformation, the side chains of aspartate and phenylalanine residues moved outside and inside of the ATP binding site, respectively. Consequently, the DFG-out region retains the hydrophobic slot occupied by phenylalanine in the DFG-in conformation. The DFG-out region represents the structural divergence and connects to the ATP back pocket. Type-II inhibitor binds to the DFG-out region as well as the ATP binding site via the ATP back pocket and thus tends to be highly selective compared with Type-I inhibitors. Imatinib, an approved drug for chronic myeloid leukemia, binds to the ATP binding site and DFG-out region of Bcr-Abl, a chimera protein kinase resulting from the gene translocation [35]. Therefore, imatinib is classified as a Type-II kinase inhibitor. The pyridine, pyrimidine, and methyl benzene moieties of imatinib bound to the ATP binding site and the other portion of the inhibitor occupied the DFG-out region. The nitrogen atom in the pyridine ring forms a hydrogen bond with the main chain NH of the hinge region in the ATP binding site. The three rings are bracketed by the hydrophobic residues in the N- and C-lobes. These hydrophobic interactions complete the C-spine. The interactions like these are observed in binding of the typical Type-I inhibitor to the ATP site. The pyridine moiety of imatinib fitted into the hydrophobic space surrounded in the DFG-out region of Bcr-Abl. The pyridine ring inserts and completes the R-spine of Bcr-Abl. However, the DFG-out region was not the ultimate target for gaining the complete selectivity. The post-marketing surveillance studies revealed that imatinib also inhibited c-Kit, a receptor-type tyrosine kinase, although it was at first identified as a selective Bcr-Abl inhibitor. To date, the DFG-out configuration has been observed in a large number of protein kinases. The crystal structures of protein kinases indicated that the size of the gatekeeper residue correlates with the propensity to be DFG-out. The small gatekeeper residue probably tolerates DFG-out motion but the large one does not. Imatinib also binds to the DFG-in conformation of spleen tyrosine kinase (Syk) that has the methionine residue at the gatekeeper position although it binds to the DFG-out conformation of the target kinases with the threonine gatekeeper [3]. This gatekeeper rule is applied to the other protein kinases such as mitogen-activated protein kinases (MAPKs). The DFG-out configuration was often observed in p38α MAPK but little or no in extracellular-regulated kinase 2 (ERK2) nor c-Jun N-terminal kinase 1 (JNK1) to date. The gatekeeper residues are threonine, glutamine, and methionine in p38α MAPK, ERK2, and JNK1, respectively.
Finally, Type-II inhibitor utilizing the ATP site and DFG-out region as a near-allosteric region could not achieve the complete selectivity. The following illustrates about the unique allosteric binding sites distinct from the regions utilized by the Types-I and Type-II inhibitors.
3.5 Characteristic p38α MAPK Inhibitors/Type-I½
p38α mitogen-activated protein kinase (MAPK) plays a crucial role in the regulation of pro-inflammatory cytokine production such as IL-6 and is an attractive drug target for inflammatory diseases including rheumatoid arthritis and psoriasis. The Type-I½ inhibitors for p38α MAPK displayed high potency and selectivity [41]. The classification of Type-I½ was descended from the combination of the Type-I and Type-II features. The most deterministic factor in the discrimination between Type-I and Type-II is the inhibitor-binding conformation in the DFG motif. The Type-I inhibitor binds to the DFG-in conformation and has no involvement in the R-spine, while the Type-II inhibitor binds to the DFG-out conformation and the hydrophobic moiety of the inhibitor takes part in forming the R-spine. The Type-I½ inhibitor binds to the DFG-in conformation, but the hydrophobic moiety of the inhibitor interacts with and stabilizes the R-spine, moving the phenylalanine residue of the DFG motif a little. Consequently, the near-allosteric pharmacophore around the ATP back pocket of p38α MAPK represents a quite unique shape among protein kinases and is pivotal for producing highly selective inhibitors. Furthermore, the Type-I½ configuration of p38α MAPK is often accompanied with the flip motion at Gly110 in the hinge region and resultantly possesses the unique shape in the ATP binding site, which accommodates atypical moieties dissimilar to the adenine moiety. The flipping point observed in p38α MAPK is not conserved among the MAPK family: ERK2 and JNK1 possess the glutamate and aspartate residues at the Gly110 position, respectively. Our structural inspections for MAPKs revealed that the Type-I½ configuration of p38α MAPK was quite unique and thus a beneficial target for elaborating highly selective inhibitors. Moreover, the structural flexibility of p38α MAPK offers the great potential to configure an unidentified conformation involving a novel allosteric pocket useful for producing potent and selective inhibitors.
3.6 Potential of the Allosteric Regions for the CK2α Inhibition/Type-I and Type-IV
Casein kinase 2 (CK2) is a serine/threonine kinase that promotes cellular growth, proliferation, and survival. CK2 is an important target protein for cancer and glomerulonephritis therapies. The CK2 holoenzyme is constitutively active without phosphorylation and consists of two catalytic subunits (CK2α) and two regulatory subunits (CK2β). CK2 uses either ATP or GTP as a phosphate group donor in the protein kinase reaction. This unusual dual specificity for nucleosides was underpinned by the particularly large space near the ribose binding site, as well as the flexibility in the αD-helix connected to the hinge region (Fig. 3.3a) [27]. CK2α displays a constitutive enzyme activity regardless of whether or not it is phosphorylated. Instead, the N-terminal segment stabilizes the active conformation of CK2α via the hydrogen bonds and hydrophobic interaction. The salt bridge between Lys68 in the β3-strand and Glu81 in the αC-helix is complete in the constitutive active conformation. The N-terminal segment, the phenylalanine residue (Phe113) at the gatekeeper position, and the tryptophan residue (Trp176) replaced by phenylalanine in the DFG motif of CK2α structurally stabilize the DFG-in configuration, i.e., likely impede transition to the DFG-out configuration. The phenylalanine residue (Phe113) at the gatekeeper position limits the accessibility to the ATP back pocket. The crystal structures of the inhibitor-bound CK2α conferred a highly conserved water molecule hydrogen bonding with the side chains of Glu81 in the ATP back pocket and main chain NH of Trp176 in the DWG motif. This structural insight is helpful for developing selective Type-I CK2α inhibitors. The carboxylic group of the several CK2α inhibitors forms a hydrogen bond with the conserved water molecule [18]. Further, the ATP binding site is narrow compared with the other kinases owing to the bulky residues involving Phe113, Leu45, Val53, Met163, and Ile174. Several planar compounds have been identified as the Type-I inhibitor for CK2 by the structural analyses. Among them, silmitasertib (CX-4945) is in preclinical development for cancer therapy. The low flexibility except for the αD-helix constrains the pharmacophore for CK2α and thus enabled to easily develop potent inhibitors based upon the crystal structures.
Near- and far-allosteric sites in the protein kinase domains of CK2α and ERK2. The ATP binding and phosphorylation sites are shown in yellow and pink, respectively. (a) CK2α equips the αD-helix pocket and CK2β binding interface for the allosteric binding (orange circles). The αD-helix pocket is located adjacent to the ATP binding site, and the CK2β binding interface is far from the ATP site. (b) ERK2 involves the KIM and FXFP sites essential for the substrate recognition and dimer interface essential for activity in cytosol (green circles)
The high-resolution crystal structure manifested two novel allosteric regions, the hydrophobic pocket near the αD-helix and CK2β binding interface (Fig. 3.3a) [17]. An ethylene glycol molecule, a precipitant reagent for the crystallization, bound in the αD-helix pocket, and the other ethylene glycol molecules occupied the CK2β interface. Recently, several compounds bound to the αD-helix pocket were discovered and merged with the low-selective Type-I inhibitors on the basis of several crystal structures [14]. The αD-helix pocket located in the C-lobe is a unique character of CK2 and large enough to accommodate fused-ring heterocyclic compounds such as the indole moiety. Therefore, this fragment-merge strategy is useful for producing the highly selective CK2α inhibitors. On the other hand, Raaf et al. indicated that 5,6-dichloro-1-b-d-ribofuranosyl benzimidazol (DRB), an ATP-competitive CK2 inhibitor, binds to the CK2β interface as well as the ATP binding site, and this conferred the potential to interfere with the CK2α/CK2β interaction [33]. Several compounds involving DRB bind to the CK2α/CK2β interface located at the top of the N-lobe (Fig. 3.3a) and inhibit the enzyme activity with a non-ATP competitive manner [6]. These structurally unique regions, the αD-helix pocket and CK2α/CK2β interface, are beneficial to develop CK2α-specific drugs as Type-I or Type-IV.
3.7 Unique Allosteric Binding Sites of MAP 2K/Type-III and Type-IV
Mitogen-activated protein kinase kinase 1 (MAP 2K1) functions as cell proliferation and survival and is defined as an attractive drug target of cancers. The crystal structure of the MAP 2K1 with U0126, a non-ATP-competitive inhibitor, unveiled a highly unique region adjacent to the ATP binding site [30]. In this structure, the inhibitor is bound to the unique site without interfering the ATP binding. Further investigation indicated that the unique region appeared in an auto-inhibition state adopted by the N-terminal regulatory domain (NRD) consisting of a single helix at the N-terminal of the kinase domain (Fig. 3.4) ([8]). The NRD region bound to the back side of the hinge region and pulled the N-lobe toward the hinge region. The gain-of-function mutations in the NRD region found in the cancer cells likely cancel the hydrogen bond network between the NRD region and N- and C-lobes in the kinase domain. Subsequently, the αC-helix moved out from the ATP site in the auto-inhibition state, and this αC-out motion resulted in the emergence of the unique region, which accommodates a variety of non-ATP-competitive inhibitors involving U0126. Consequently, these structural insights indicated that this unique region was available for producing the Type-III MAP 2K1 inhibitor, defined as a near-allosteric site binder. Trametinib had been approved as an anticancer drug and displayed a high selectivity for MAP 2K1 and MAP 2K2 against the other MAP 2K [40]. MAP 2K2 presents a similar physiological role with MAP 2K1 and conserves the NRD and trametinib binding region [30]. The trametinib binding region of auto-inhibited MAP 2K1 is structurally unique and likely has potential to produce highly selective Type-III inhibitors. The strong point of the Type-III inhibitor is that this binds to the target kinase collaboratively with the ATP molecule, while the Type-I or Type-II inhibitor must eliminate the ATP molecule from the ATP binding site, resulting in a large enthalpy loss. Several Type-III inhibitors for MAP 2K1 bound to the unique region, making the hydrogen bonds with the phosphate group of ATP. The allosteric pockets similar to MAP 2K1 have been observed in with-no-lysine kinase 1 (WNK1) and cyclin-dependent kinase 2 (CDK2) [44] [5], although these are distinguishable based upon their detailed structures. In the near future, the structural mechanism for forming the MAP 2K1-type allosteric pocket along with the αC-out motion would be clarified based upon the crystal structure analyses, and this likely facilitates the development of selective inhibitors for these protein kinases.
Auto-inhibition mechanisms of the MAP 2K family kinases. MAP 2K1 is autoregulated by the N-terminal helix, which fixes the hinge motion between the N- and C-lobes. MAP 2K4 is autoregulated by binding the substrate in the N-lobe groove and configuring a long helix in the A-loop region. MAP 2K6 is autoregulated by configuring the three helices in the A-loop region. Commonly, the A-loop configurations of MAP 2K1, MAP 2K4, and MAP 2K6 in auto-inhibition state are not ready for substrate binding. The auto-inhibition state of MAP 2K7 is formed by an interaction of P-loop with the C-lobe, interfering the ATP binding
On the other hand, crystal structures of the MAP 2K1 homologues, MAP 2K4, MAP 2K6, and MAP 2K7, in the auto-inhibition state indicated that these three MAP 2Ks owned no NRD, but each has a characteristic auto-inhibition mechanism (Fig. 3.4) [25, 26, 36]. MAP 2K4 is a client kinase of p38 mitogen-activated protein kinases (p38 MAPKs) and c-Jun N-terminal kinases (JNKs), and its dysregulation occurs in the diseases such as cancers. The substrate binding to a deep cleft in the top of the N-lobe induces an auto-inhibition conformation of MAP 2K4, in which the A-loop region configures a long helix, interfering substrate binding (Fig. 3.4) [25]. Therefore, the N-lobe cleft and vacant space caused by the structural rearrangement in A-loop are likely available for producing allosteric inhibitors of MAP 2K4. MAP 2K6 is a client kinase of p38 MAPKs and an attractive drug target for inflammation and autoimmune diseases. The ATP binding triggers an auto-inhibition conformation of MAP 2K6, in which the A-loop segment was converted into three short helices, interfering substrate binding (Fig. 3.4) [26]. The hydrophobic space located at the back side of these helices is perhaps available as an allosteric site for discovering novel MAP 2K6 inhibitors. MAP 2K7 is involved in the JNK signal cascade and a significant drug discovery target against arthritis, cardiac hypertrophy, and so on. The n-σ∗ interaction between Cys218 at the N-terminal of the αD-helix and Gly145 in the P-loop of MAP 2K7 shaped a closed structure, interfering with the ATP molecule binding (Fig. 3.4) [36]. The resulting structure presents the large and unique hydrophobic space in the DFG-out region sequestered by the ATP binding site. These allosteric spaces discovered in the auto-inhibition state of MAP 2K4, MAP 2K6, and MAP 2K7 are not conserved among the MAP 2K family kinases and thus noteworthy for the development of highly selective Type-III inhibitors.
High-resolution X-ray analysis, which was achieved through the experiments under the microgravity environment, conferred the alternative auto-inhibition state of MAP 2K7, regulating with an intermolecular manner [16]. The C-terminal fragment of Gly424-His430, following to the kinase domain, presented an extended configuration and bound to the groove in the N-lobe of the neighbor MAP 2K7 molecule via the hydrogen bonds and hydrophobic interaction and likely worked as an intermolecular brake of MAP 2K7. The side chain of Leu426 in the middle of the C-terminal fragment intruded into the deep hydrophobic pocket involving Asn138, Trp151, and Val164, forming the side and bottom of the N-lobe groove. The intermolecular interaction likely induces the shift of the N-lobe toward the hinge region and subsequently the DFG-out conformation without forming the C-spine. Further structural inspections revealed that this N-lobe pocket of MAP 2K7 is unique among MAP 2Ks. The other MAP 2K could not accommodate the leucine residue with the region similar to the Leu426 binding pocket of MAP 2K7 because of the bulky or hydrophilic residue replaced by that of MAP 2K7 (Table. 3.2). Actually, the synthesized C-terminal peptide moderately but concentrate dependently inhibited the MAP 2K7 activity and thus could be defined as a seed compound of the Type-IV drug. The top of the N-lobe is used for the protein-protein interaction and autoregulation in several kinases such as MAP 2K4 and 3-phosphoinositide-dependent protein kinase (PDK1) and useful for the development of highly selective inhibitors. Inactive MAP 2K4 accommodates the substrate in the N-terminal groove as abovementioned [25]. PDK1 equips the PDK1-interacting fragment (PIF) pocket in the N-lobe, which primary recruits the substrate proteins. Rettenmaier et al. discovered the Type-IV inhibitors, which inhibit PDK1 in cells, based upon this structural insight [34]. Fortunately, these allosteric pockets of MAP 2K4, MAP 2K7, and PDK1 are nonoverlapping in the N-lobe and recognize the distinguished sequence.
Together all, we have the great chances to produce the near- and far-allosteric MAP 2K inhibitors (Type-III and Type-IV), which represent the tendency to be highly selective against the off-target kinases. However, we cannot quantitatively estimate the potential of the obtained inhibitor to date.
3.8 Far-Allosteric Inhibition of ERK2/Type-IV
Extracellular-regulated kinase 2 (ERK2) is a member of mitogen-activated protein kinases (MAPKs), regulated by MAP 2K1 via phosphorylation and defined as a drug discovery target for serious diseases such as cancer and type 2 diabetes. The ATP-competitive Type-I inhibitors for ERK2 have been discovered, and some of them are in clinical study [29, 42]. In addition to these, the far-allosteric inhibitors were discovered on the basis of the unique substrate recognition mechanism [15]. ERK2 anchors the substrate proteins via their consensus sequence of Lys(Arg)2-3-X1-6-Φa-X-Φb (Φ, hydrophobic amino acid; X, any amino acid) at the kinase interacting motif binding site (KIM site) which is distinct from the phosphorylation site recognizing the Ser(Thr)-Pro sequence of the substrate (Fig. 3.3b). The far-allosteric KIM site is located in the back side of the hinge region but inaccessible to the ATP binding site. Therefore, the KIM binder is classified as the Type-IV inhibitor. A synthesized peptide based on the consensus sequence in signal transducer and activator of transcription 3 (STAT3) inhibited the ERK2 activity with the IC50 value of 9.8 μM and had a significant effect on the model mice without the serious adverse effects [15]. The X-ray analysis conferred the separated sub-pockets in the KIM site. The two aliphatic residues of the peptide inhibitor are accommodated in the shallow pocket configured by the hydrophobic residues in the KIM region. The positive-charged residues of the peptide inhibitor were located in the negative-charged pool involving three aspartates and a glutamate residue in the KIM region of ERK2. The hydrophobic pocket and negative-charged pool are completely sequestered in the KIM region. The arbitrary sequence in the middle of the consensus peptide likely works as a sole linker of these characteristic regions. The KIM region forms a shallow groove and is disadvantageous for the gain of binding affinity compared with the ATP binding site. Nevertheless, the drug discovery studies indicated the potential to realize the far-allosteric inhibition of ERK2. The KIM binders discovered by the in silico screening displayed the moderate inhibitory activity and competitive behavior to the STAT3 peptide [19]. JNKs and p38 MAPKs, members of the MAP kinases involving ERK2, possess the common substrate recognition mechanism using the KIM region. MAPKs discriminate their own substrates with high fidelity through the recognition in the KIM region. Therefore, the KIM binding inhibitors are expected to possess high selectivity among MAPKs. However, the KIM inhibitors reported to date displayed the rather low activity. To solve this problem, the bivalent inhibitors were invented by merging the Type-I ATP competitive and KIM peptide inhibitors via the long linker segment [20]. The bivalent inhibitors exhibited high inhibitory potency against ERK2.
The FXFP region found in the MAP kinase insert region of MAPKs works as a second substrate anchoring site utilized by the transcription factor substrates (Fig. 3.3b) [1]. Structural studies revealed that the FXFP site was observed in the active ERK2 (doubly phosphorylated state) but inaccessible to the inactive state of ERK2 (non-phosphorylated state). In the inactive state, Phe181 and Leu182 in the A-loop occupied the FXFP site of ERK2. Double phosphorylation likely induces the conformation change in the A-loop and opens the FXFP site. The second anchoring system is also conserved in all MAPKs but divergent enough to discriminate their own substrates like the KIM site.
Upon activation, the dimer state is essential for the ERK2 extranuclear signaling. Herrero et al. reported that a small molecule inhibitor for ERK2 dimerization forestalls tumorigenesis [13]. Biochemical experiments revealed that this inhibitor interfered with the dimerization of ERK2 and probably bound to the dimerization interface of ERK2. Crystal structure indicated that the tip of the A-loop involving Asp175 intruded into the small pocket was located in the back side of the connecting loop between the β3-strand and αC-helix [13] (Fig. 3.3b).
To the best of our knowledge, three far-allosteric pockets involving the substrate recognition sites of KIM and FXFP and dimer interface are available for producing highly selective Type-IV inhibitors of ERK2.
3.9 Allosteric Inhibition of Receptor Tyrosine Kinases/Type-III
The receptor tyrosine kinase (RTK) family represents a typical auto-inhibition configuration in which accompanying the DFG-out transition, the phenylalanine residue in the DFG motif intrudes deeply into the ATP site (Fig. 3.5). In addition to that, the tyrosine residue as a phosphorylate acceptor in the A-loop is located at the substrate binding position in this auto-inhibition state (Fig. 3.5). The resulting allosteric pocket involving the DFG-out region is probably available for producing selective Type-III inhibitors (Fig. 3.5). FMS-like tyrosine kinase (FLT3), a member of the RTK family, is essential for the normal function of stem cells and the immune system and potential target for therapies of a variety of leukemia. The allosteric pocket in the typical RTK auto-inhibition structure of FLT3 is fully occupied by Tyr572 and its marginal residues in the juxtamembrane region (Fig. 3.5) [10] and is likely available for developing selective FLT3 inhibitors. Insulin-like growth factor-1 receptor (IGF-1R) is also a member of RTK family and associates to the signaling to tumor. Heinrich et al. showed that the indolealkylamine class inhibitors, which were discovered by a high-throughput screening, bound to the near-allosteric pocket involving the DFG-out region in the typical RTK auto-inhibition conformation of IGF-1R [12]. Thus, these inhibitors are defined as Type-III inhibitors and stabilize the RTK auto-inhibition conformation, concealing the phosphorylating tyrosine residue in the A-loop.
An auto-inhibition configuration of FLT3 on behalf of the receptor tyrosine kinase. The phenylalanine residue in the DFG motif intrudes into the ATP binding site, resulting in the narrow ATP binding site. The tyrosine residue in the A-loop occupies the phosphorylation site and forms a hydrogen bond with the aspartate residue as a proton acceptor. In this conformation, the A-loop sequesters the DFG-out region from the ATP binding site
Tropomyosin receptor kinase A (TrkA) is a receptor for nerve growth factor (NGF) that mediates neuronal differentiation and cellular survival. TrkA belongs to the receptor tyrosine kinase family, consisting of an extracellular ligand-binding domain, a transmembrane helix, an intracellular juxtamembrane region (JM), and a tyrosine kinase domain. The TrkA signaling pathway plays a crucial role in the physiological function of chronic pain and is an attractive drug target for chronic pain. TrkA is a member of the Trk subfamily, which also includes TrkB and TrkC. Each Trk plays a distinct function. Thus, selective TrkA inhibitors are desired for the chronic pain therapy. Undesirable side effects have been reported for pan-Trk inhibitors. The several Types-I and Type-II inhibitors for TrkA have been developed based upon the crystal structures but disappointingly exerted the pan-activity. The crystal structures depicted that the inhibitor-binding environments in the ATP and DFG-out sites were completely conserved among the Trk family. These structural insights coincide with the low selectivity of the above inhibitors. However, some research organizations have reported the highly selective TrkA inhibitors, which were expected to be classified into Type-III, binding to the RTK allosteric pocket as observed in IGF-1R. The crystal structures of TrkA with the highly selective inhibitor unveiled that the inhibitor-binding pocket were assembled from the allosteric RTK pocket and JM region and completely sequestered from the ATP site [9, 38]. Therefore, these selective inhibitors were confirmed as the Type-III inhibitor. One-third of the pocket is composed of amino acid residues in the JM region. Owing to the low sequence homology of the JM region among Trks, these inhibitors exhibit high selectivity. Trks share significant sequence homology in the kinase domain involving the DFG-out region, whereas the JM region is highly divergent among the Trk family. The selective Type-III inhibitor binds to the JM region via hydrogen bonds, van der Waals interactions, and CH-π interactions at Leu486, His489, Ile490, etc. These residues of TrkA are not conserved in TrkB and TrkC and should contribute to the high binding affinity and selectivity.
Collectively, the receptor tyrosine kinases have a typical auto-inhibition structure and its close relatives useful for producing potent Type-III inhibitors, and the flexible region such as JM occasionally plays a definitive role for the high selectivity and potency of the inhibitors.
3.10 Molecular Switch Regulation of RSKs and MSKs/Type-IV
Dimethyl fumarate (DMF) has been used for the oral therapy of psoriasis and multiple sclerosis. More recently, Anderson et al. suggested that DMF covalently bound to the hydrophobic pocket in the C-lobe of p90 ribosomal S6 kinases (RSKs) and mitogen- and stress-activated kinases (MSKs) as its potential target proteins [2]. The crystal structure of the DMF-RSK2 complex showed that the Michael acceptor of DMF made a covalent bond with the Cys599 in the bottom of the C-lobe pocket formed by the hydrophobic residues Trp602, Val662, Ile633, and Leu710 [2]. The C-lobe pocket was estimated to be an allosteric regulator that bound the tip of the phosphorylated A-loop and stabilized the active conformation. The C-lobe pocket containing the cysteine residue is conserved among RSKs and MSKs. Therefore, the DMF-binding pocket is noteworthy for producing highly selective Type-IV inhibitors for RSKs and MSKs. Further structural dissections probably allow to discriminating the C-lobe pocket of these homologous kinases.
3.11 The Pursuit of Unidentified Allosteric Pockets in Protein Kinases
In the near future, all the protein kinase structures would be available in Protein Data Bank and consequently accelerate the production of highly selective inhibitors. Bioinformatics technology and computational chemistry using artificial intelligence (AI) presumably play central roles in dissecting and characterizing the inhibitor-binding pockets of all protein kinases. The structural flexibility of protein kinases confers the innumerable conformations in solution, involving some energetically stable those such as the auto-inhibition state. The conformational selection by a protein kinase inhibitor results in the population shift in the protein kinase configuration. The DFG-out configuration was firstly identified as a result of the Type-II inhibitor binding but not observed as an apo state. Molecular dynamics studies facilitated the comprehensive understanding of the structural flexibility of protein kinases [7, 39]. With the upsurge in computational power, it would be possible to develop drug discovery techniques involving in silico screening against the conformational ensemble of a target protein kinase and the clarification of unprecedented conformations that could not been identified without crystal analyses. Meanwhile, several recent studies showed that the propensity of the slow dissociation kinetics of the protein kinase inhibitor was beneficial for drug efficacy and associated with conformational change. Ayaz et al. indicated that roniciclib, a Type-I inhibitor, exerted the kinetic selectivity for CDK2, indicating sustainable effect on retinoblastoma protein phosphorylation [4]. Furthermore, the derivatives of roniciclib prolonged the resident time by the inhibitor-induced conformational change in binding to the CDK2 [4]. Finally, the computation development involving molecular dynamics would dissect the conformational behavior and binding kinetics of protein kinases to produce highly selective protein kinase inhibitors.
3.12 Conclusion
It is necessary to resolve a large number of the significant challenges to develop inhibitors to be drugs. Protein kinase inhibitors with low selectivity suppress a variety of the off-target protein kinases as well as the target one and arguably exert the adverse effects on vital. To pursue the production of highly selective protein kinase inhibitors, a variety of the druggable allosteric sites involving the ATP back pocket (Sect. 3.3), the DFG-out region (Sect. 3.4), the αD-helix pocket and CK2β binding interface of CK2α (Sect. 3.6), the KIM and FXFP sites of ERK2 (Sect. 3.8), the N-lobe pockets (Sects. 3.6, 3.7 and 3.8), the C-lobe pocket (Sect. 3.10), the physiologically essential dimer interface (Sect. 3.8), and so on are available in the protein kinase domains (Table 3.3). These allosteric sites are primarily utilized for the diverse physiological function such as the activity regulation, substrate recognition, and subcellular localization of protein kinases. These allosteric molecular switches are quite unique among protein kinases. Therefore, the allosteric sites equipped in the protein kinase domain are noteworthy for gaining high selectivity. The approved kinase inhibitors gained the high selectivity by more or less using the allosteric site. Erlotinib, a Type-I EGFR inhibitor for the anticancer therapy, bound to the ATP back pocket (Sect. 3.3). Imatinib, a Type-II Bcr-Abl inhibitor for the chronic leukemia therapy, occupied the DFG-out region (Sect. 3.4). Trametinib, a Type-III MAP 2K1 inhibitor as anticancer drug, used only the near-allosteric region induced by the auto-inhibition (Sect. 3.7). The Type-IV kinase inhibitor binds to the far-allosteric region and thus is expected to be highly selective but displays the low inhibitory potency. The Type-V kinase inhibitor presents a considerable burden in gaining the high selectivity because of little or no use of the allosteric region.
The structural flexibility of protein kinases confers the innumerable conformations in solution, involving the unidentified allosteric pockets as discovered in the auto-inhibition state. Finally, the computation development involving molecular dynamics would dissect the conformational behavior of protein kinases and promote to produce highly selective protein kinase inhibitors.
References
Akella R, Moon TM, Goldsmith EJ (2008) Unique MAP Kinase binding sites. Biochim Biophys Acta 1784(1):48–55
Andersen JL, Gesser B, Funder ED, Nielsen CJF, Gotfred-Rasmussen H, Rasmussen MK, Toth R, Gothelf KV, Arthur JSC, Iversen L, Nissen P (2018) Dimethyl fumarate is an allosteric covalent inhibitor of the p90 ribosomal S6 kinases. Nat Commun 9(1):4344
Atwell S, Adams JM, Badger J, Buchanan MD, Feil IK, Froning KJ, Gao X, Hendle J, Keegan K, Leon BC, Muller-Dieckmann HJ, Nienaber VL, Noland BW, Post K, Rajashankar KR, Ramos A, Russell M, Burley SK, Buchanan SG (2004) A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase. J Biol Chem 279(53):55827–55832
Ayaz P, Andres D, Kwiatkowski DA, Kolbe CC, Lienau P, Siemeister G, Lucking U, Stegmann CM (2016) Conformational adaption may explain the slow dissociation kinetics of roniciclib (BAY 1000394), a type I CDK inhibitor with kinetic selectivity for CDK2 and CDK9. ACS Chem Biol 11(6):1710–1719
Betzi S, Alam R, Martin M, Lubbers DJ, Han H, Jakkaraj SR, Georg GI, Schonbrunn E (2011) Discovery of a potential allosteric ligand binding site in CDK2. ACS Chem Biol 6(5):492–501
Brear P, North A, Iegre J, Hadje Georgiou K, Lubin A, Carro L, Green W, Sore HF, Hyvonen M, Spring DR (2018) Novel non-ATP competitive small molecules targeting the CK2 alpha/beta interface. Bioorg Med Chem 26(11):3016–3020
De Vivo M, Masetti M, Bottegoni G, Cavalli A (2016) Role of molecular dynamics and related methods in drug discovery. J Med Chem 59(9):4035–4061
Fischmann TO, Smith CK, Mayhood TW, Myers JE, Reichert P, Mannarino A, Carr D, Zhu H, Wong J, Yang RS, Le HV, Madison VS (2009) Crystal structures of MEK1 binary and ternary complexes with nucleotides and inhibitors. Biochemistry 48(12):2661–2674
Furuya N, Momose T, Katsuno K, Fushimi N, Muranaka H, Handa C, Ozawa T, Kinoshita T (2017) The juxtamembrane region of TrkA kinase is critical for inhibitor selectivity. Bioorg Med Chem Lett 27(5):1233–1236
Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, Lippke J, Saxena K (2004) The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell 13(2):169–178
Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9(8):576–596
Heinrich T, Gradler U, Bottcher H, Blaukat A, Shutes A (2010) Allosteric IGF-1R inhibitors. ACS Med Chem Lett 1(5):199–203
Herrero A, Pinto A, Colon-Bolea P, Casar B, Jones M, Agudo-Ibanez L, Vidal R, Tenbaum SP, Nuciforo P, Valdizan EM, Horvath Z, Orfi L, Pineda-Lucena A, Bony E, Keri G, Rivas G, Pazos A, Gozalbes R, Palmer HG, Hurlstone A, Crespo P (2015) Small molecule inhibition of ERK dimerization prevents tumorigenesis by RAS-ERK pathway oncogenes. Cancer Cell 28(2):170–182
Iegre J, Brear P, De Fusco C, Yoshida M, Mitchell SL, Rossmann M, Carro L, Sore HF, Hyvonen M, Spring DR (2018) Second-generation CK2alpha inhibitors targeting the alphaD pocket. Chem Sci 9(11):3041–3049
Kinoshita T, Doi K, Sugiyama H, Kinoshita S, Wada M, Naruto S, Tomonaga A (2011a) Knowledge-based identification of the ERK2/STAT3 signal pathway as a therapeutic target for type 2 diabetes and drug discovery. Chem Biol Drug Des 78(3):471–476
Kinoshita T, Hashimoto T, Sogabe Y, Fukada H, Matsumoto T, Sawa M (2017) High-resolution structure discloses the potential for allosteric regulation of mitogen-activated protein kinase kinase 7. Biochem Biophys Res Commun 493(1):313–317
Kinoshita T, Nakaniwa T, Sekiguchi Y, Sogabe Y, Sakurai A, Nakamura S, Nakanishi I (2013) Crystal structure of human CK2alpha at 1.06 A resolution. J Synchrotron Radiat 20(Pt 6):974–979
Kinoshita T, Sekiguchi Y, Fukada H, Nakaniwa T, Tada T, Nakamura S, Kitaura K, Ohno H, Suzuki Y, Hirasawa A, Nakanishi I, Tsujimoto G (2011b) A detailed thermodynamic profile of cyclopentyl and isopropyl derivatives binding to CK2 kinase. Mol Cell Biochem 356(1-2):97–105
Kinoshita T, Sugiyama H, Mori Y, Takahashi N, Tomonaga A (2016) Identification of allosteric ERK2 inhibitors through in silico biased screening and competitive binding assay. Bioorg Med Chem Lett 26(3):955–958
Lechtenberg BC, Mace PD, Sessions EH, Williamson R, Stalder R, Wallez Y, Roth GP, Riedl SJ, Pasquale EB (2017) Structure-guided strategy for the development of potent bivalent ERK inhibitors. ACS Med Chem Lett 8(7):726–731
Liao JJ (2007) Molecular recognition of protein kinase binding pockets for design of potent and selective kinase inhibitors. J Med Chem 50(3):409–424
Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS (2013) Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol 20(2):146–159
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298(5600):1912–1934
Marcie AG, Gregory DC, Min L, Brittany D, Kenneth A, Ross LS, Kenneth SK (2007) New approaches to the discovery of cdk5 inhibitors. Current Alzheimer Research 4(5):547–549
Matsumoto T, Kinoshita T, Kirii Y, Yokota K, Hamada K, Tada T (2010) Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state. Biochem Biophys Res Commun 400(3):369–373
Matsumoto T, Kinoshita T, Matsuzaka H, Nakai R, Kirii Y, Yokota K, Tada T (2012) Crystal structure of non-phosphorylated MAP 2K6 in a putative auto-inhibition state. J Biochem 151(5):541–549
Niefind K, Putter M, Guerra B, Issinger OG, Schomburg D (1999) GTP plus water mimic ATP in the active site of protein kinase CK2. Nat Struct Biol 6(12):1100–1103
Nishiyama A, Yamada T, Kita K, Wang R, Arai S, Fukuda K, Tanimoto A, Takeuchi S, Tange S, Tajima A, Furuya N, Kinoshita T, Yano S (2018) Foretinib overcomes entrectinib resistance associated with the NTRK1 G667C mutation in NTRK1 fusion-positive tumor cells in a brain metastasis model. Clin Cancer Res 24(10):2357–2369
Ohori M, Kinoshita T, Okubo M, Sato K, Yamazaki A, Arakawa H, Nishimura S, Inamura N, Nakajima H, Neya M, Miyake H, Fujii T (2005) Identification of a selective ERK inhibitor and structural determination of the inhibitor-ERK2 complex. Biochem Biophys Res Commun 336(1):357–363
Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, Mueller WT, Delaney A, Omer C, Sebolt-Leopold J, Dudley DT, Leung IK, Flamme C, Warmus J, Kaufman M, Barrett S, Tecle H, Hasemann CA (2004) Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol 11(12):1192–1197
Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA (2001) Mechanism-based design of a protein kinase inhibitor. Nat Struct Biol 8(1):37–41
Park JH, Liu Y, Lemmon MA, Radhakrishnan R (2012) Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochem J 448(3):417–423
Raaf J, Brunstein E, Issinger OG, Niefind K (2008) The CK2 alpha/CK2 beta interface of human protein kinase CK2 harbors a binding pocket for small molecules. Chem Biol 15(2):111–117
Rettenmaier TJ, Sadowsky JD, Thomsen ND, Chen SC, Doak AK, Arkin MR, Wells JA (2014) A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1. Proc Natl Acad Sci U S A 111(52):18590–18595
Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J (2000) Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289(5486):1938–1942
Sogabe Y, Hashimoto T, Matsumoto T, Kirii Y, Sawa M, Kinoshita T (2016) A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP 2K7. Biochem Biophys Res Commun 473(2):476–481
Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, Kraemer O, Himmelsbach F, Haaksma E, Adolf GR (2012) Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharmacol Exp Ther 343(2):342–350
Su HP, Rickert K, Burlein C, Narayan K, Bukhtiyarova M, Hurzy DM, Stump CA, Zhang X, Reid J, Krasowska-Zoladek A, Tummala S, Shipman JM, Kornienko M, Lemaire PA, Krosky D, Heller A, Achab A, Chamberlin C, Saradjian P, Sauvagnat B, Yang X, Ziebell MR, Nickbarg E, Sanders JM, Bilodeau MT, Carroll SS, Lumb KJ, Soisson SM, Henze DA, Cooke AJ (2017) Structural characterization of nonactive site, TrkA-selective kinase inhibitors. Proc Natl Acad Sci U S A 114(3):E297–E306
Tong M, Seeliger MA (2015) Targeting conformational plasticity of protein kinases. ACS Chem Biol 10(1):190–200
Uitdehaag JC, de Roos JA, van Doornmalen AM, Prinsen MB, de Man J, Tanizawa Y, Kawase Y, Yoshino K, Buijsman RC, Zaman GJ (2014) Comparison of the cancer gene targeting and biochemical selectivities of all targeted kinase inhibitors approved for clinical use. PLoS One 9(3):e92146
Walter NM, Wentsch HK, Buhrmann M, Bauer SM, Doring E, Mayer-Wrangowski S, Sievers-Engler A, Willemsen-Seegers N, Zaman G, Buijsman R, Lammerhofer M, Rauh D, Laufer SA (2017) Design, synthesis, and biological evaluation of novel type I(1)/2 p38alpha MAP kinase inhibitors with excellent selectivity, high potency, and prolonged target residence time by interfering with the R-Spine. J Med Chem 60(19):8027–8054
Ward RA, Colclough N, Challinor M, Debreczeni JE, Eckersley K, Fairley G, Feron L, Flemington V, Graham MA, Greenwood R, Hopcroft P, Howard TD, James M, Jones CD, Jones CR, Renshaw J, Roberts K, Snow L, Tonge M, Yeung K (2015) Structure-guided design of highly selective and potent covalent inhibitors of ERK1/2. J Med Chem 58(11):4790–4801
Wu P, Nielsen TE, Clausen MH (2016) Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov Today 21(1):5–10
Yamada K, Zhang JH, Xie X, Reinhardt J, Xie AQ, LaSala D, Kohls D, Yowe D, Burdick D, Yoshisue H, Wakai H, Schmidt I, Gunawan J, Yasoshima K, Yue QK, Kato M, Mogi M, Idamakanti N, Kreder N, Drueckes P, Pandey P, Kawanami T, Huang W, Yagi YI, Deng Z, Park HM (2016) Discovery and characterization of allosteric WNK kinase inhibitors. ACS Chem Biol 11(12):3338–3346
Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Janne PA (2009) Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 462(7276):1070–1074
Zuccotto F, Ardini E, Casale E, Angiolini M (2010) Through the “Gatekeeper Door”: exploiting the active kinase conformation. J Med Chem 53(7):2681–2694
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kinoshita, T. (2019). Protein Allostery in Rational Drug Design. In: Zhang, J., Nussinov, R. (eds) Protein Allostery in Drug Discovery. Advances in Experimental Medicine and Biology, vol 1163. Springer, Singapore. https://doi.org/10.1007/978-981-13-8719-7_3
Download citation
DOI: https://doi.org/10.1007/978-981-13-8719-7_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8718-0
Online ISBN: 978-981-13-8719-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)