Abstract
Lactic acid bacteria (LAB), a class of commonly existing microorganisms in nature, are important components of gut commensal microflora in humans and animals. Previous studies suggested that LAB exerted specific physiological and biochemical functions on the host such as improving intestinal microbial balance, immunomodulation, inhibiting tumor growth, lowering cholesterol levels, as well as regulating blood pressure and are therefore widely used in food manufacturing and functional food development. Due to the continuous development of modern molecular biology techniques, studies regarding exploiting LAB as expression hosts in addition to fermentation starter cultures and probiotics have received increasing attention from both academia and industry. In the 1980s, some researchers initiated molecular genetic research for LAB. They characterized lactose metabolism-related genes and proteins in LAB and established preliminary DNA delivery systems for LAB. Over the past decades, owing to the advances in modern DNA sequencing and gene characterization techniques, structures and functions of LAB genomes and plasmid-related genes have been further elucidated, which lays a solid theoretical foundation for the further development of LAB-based gene expression systems (Bolotin et al. 2001; Altermann et al. 2005).
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Lactic acid bacteria (LAB), a class of commonly existing microorganisms in nature, are important components of gut commensal microflora in humans and animals. Previous studies suggested that LAB exerted specific physiological and biochemical functions on the host such as improving intestinal microbial balance, immunomodulation, inhibiting tumor growth, lowering cholesterol levels, as well as regulating blood pressure and are therefore widely used in food manufacturing and functional food development. Due to the continuous development of modern molecular biology techniques, studies regarding exploiting LAB as expression hosts in addition to fermentation starter cultures and probiotics have received increasing attention from both academia and industry. In the 1980s, some researchers initiated molecular genetic research for LAB. They characterized lactose metabolism-related genes and proteins in LAB and established preliminary DNA delivery systems for LAB. Over the past decades, owing to the advances in modern DNA sequencing and gene characterization techniques, structures and functions of LAB genomes and plasmid-related genes have been further elucidated, which lays a solid theoretical foundation for the further development of LAB-based gene expression systems (Bolotin et al. 2001; Altermann et al. 2005).
2.1 LAB-Associated Gene Expression Systems
LAB expression systems were developed lately as compared to traditional Escherichia coli, Bacillus, and yeast expression systems. Generally, there’re still many limitations in LAB-based gene expression systems such as low gene expression efficiency, complicated procedures, and low transformation efficiency. Nevertheless, genetic engineering of LAB offers remarkable advantages over other traditional expression systems due to the intrinsic properties of LAB strains, thereby exhibiting great potential for applications in antigen screening, protease expression, and immunotherapy. The advantages of LAB-based expression systems are listed below: (1) LAB are safe and edible food-grade microorganisms granted by WHO and FAO and have been applied in food products for thousands of years; (2) LAB are inherent enteric microflora in humans and animals and play a key role in the establishment and maintenance of host immune system; (3) some LAB strains adhere tightly to the gut mucosa; and (4) LAB culture supernatants can be directly consumed without the necessity of purifying expressed heterologous proteins. LAB-based gene expression systems comprise host strains, expression vectors, and heterologous genes. This chapter will briefly describe host strains and cloning vectors in LAB-based gene expression systems in terms of their compositions and features.
2.1.1 Host Strains in Expression Systems for LAB
LAB strains of different genera or species and even different strains within the same species differed greatly in terms of their biochemical, ecological, and molecular immune properties (Meijerink et al. 2012; Ai et al. 2015). This strain diversity creates extensive host strain options in LAB but also causes difficulty of selecting proper strains as expression hosts since high-level production of heterologous proteins in a specific strain-based expression system cannot always be achieved in other strains (Rigaux et al. 2009).
Currently, several genera including Lactococcus, Enterococcus, and Lactobacillus have been most widely applied as host strains (Table 2.1) owing to their extraordinary features such as good resistance to the harsh conditions in the digestive tract, low immunogenicity, relatively high electro-transformation efficiency, and long retention time in the gastrointestinal tract. Due to the progress in molecular genetics research on LAB, LAB as food-grade delivery vehicles have received increasing attentions. In order to further facilitate the screening of recombinant LAB strains, host strains can be modified based on specific requirements such as lacF-deficient strains and strains with nisRK integrated into their chromosomes.
2.1.2 Vectors in LAB-Based Gene Expression System
2.1.2.1 Plasmids in LAB
Since Chassy and Flickinger (1987) detected plasmids in LAB, researchers have further studied plasmids that are present in LAB. The distribution of plasmids has been suggested to be highly uneven and strain-specific in LAB. More plasmids were found in LAB strains such as Lactobacillus reuteri, Lactobacillus helveticus, and Lactobacillus acidophilus when compared with strains belonging to other species. Moreover, the size (1–150 kb) and amounts of plasmids differed greatly in different LAB strains (Chassy et al. 1976; Vescovo et al. 1981; McKay and Baldwin 1990). Overall, plasmids in LAB are characterized by the following features: (1) Most plasmids in LAB are cryptic plasmids, and only a few of them are associated with the host’s specific phenotypes such as bacteriocin synthesis, sugar metabolism, and antibiotic resistance (Smiley and Fryder 1978; Fortina et al. 1993). (2) The copy number of the LAB plasmids is correlated with their size. Smaller plasmids are replicated at a higher number. (3) Plasmids from LAB, which have a broad host range, can replicate in a wide range of host bacteria.
Since most of plasmids from LAB are cryptic, their functions on gene transcription, translation, and protein secretion are still not completely clear, which might be one of causative factors for delayed development in LAB molecular biologics study. Nevertheless, owing to the advances in genetic engineering techniques, vectors for cloning, expression, and integration in LAB were successively developed through studying the regulatory elements within plasmids isolated from LAB. At present, cloning and expression vectors are the most extensively applied vectors in LAB.
2.1.2.2 Cloning Vectors for LAB
Genetic cloning techniques, one of the effective means of resolving the complexity of genetics, allow isolation of single desired genes and construction of new LAB starter strains carrying these isolated genes. Cloning vectors are used for amplification and propagation of foreign DNA inserts and have certain copy numbers within host cells. Cloning vectors have the following essential features: (1) suitable cloning sites, into which foreign DNA can be inserted, (2) autonomous replication or replication along with chromosomal DNA once plasmids are integrated into the host chromosome, and (3) selectable markers which facilitate the selection of transformed cells harboring DNA insert-containing plasmids. To date, various types of cloning vectors including plasmids, viral vectors/bacteriophages, and artificial vectors that incorporate segments from the plasmids, bacteriophage, or genomic DNA have been constructed. These artificial vectors might only have an origin of replication but not promoters for expression.
Even though Escherichia coli is the most frequently applied host in molecular cloning, other microorganisms sometimes are also used as hosts. Thus, shuttle vectors containing a second replication origin, which assures their replication in other types of microorganisms (e.g. LAB), are needed. The enterococcal plasmid pAMβ1, a θ-type-replicating plasmid with a broad host range, is the first plasmid used for constructing LAB cloning vectors. It is also one of the prototype cloning vectors for lactococci and lactobacilli. Several plasmids such as pIL252 and pIL253, which are based on pAMβ1, were established. Other common plasmid replicons such as pWV01 and pSH71 can propagate and replicate in various LAB strains. Moreover, there are other low-copy and high-copy derivative plasmids such as pGK1 and pGK12.
In order to screen the right transformants, one or multiple resistance genes are ligated into vectors as selectable markers. Since a large number of wild-type LAB strains are resistant to ampicillin, kanamycin, and tetracycline, erythromycin and chloramphenicol resistance genes serve as the common selection markers. Of note, since the transfer and dissemination of antibiotic resistance genes in the environment are potentially deleterious to the environmental ecosystem, resistance genes are not suitable selection markers in the food industry. Therefore, several food-grade expression systems for LAB based on the sugar utilization, sensitivity to PH and temperature, and bacteriocin resistance of host strains have been developed.
Apart from single-component cloning vectors, some researchers developed several two-component cloning systems. Emond et al. (2001) constructed a two-component food-grade cloning vector pVEC1, which is a pCD4 derivative and carries the functional pCD4 replicon. Another pCD4-derived plasmid pCOM1 was also constructed as a companion vector. Plasmid pCOM1, in which an erythromycin resistance gene serves as the dominant selection marker, is deficient of repB gene. After the selection of recombinant strains carrying both plasmids, recombinant bacterial cells will lose plasmid pCOM1 when grown under antibiotic-free conditions owing to the incompatibility between pVEC1 and pCOM1. By using this cloning system, Lactococcus lactis MG1363 and industrial strains were shown to exhibit stable phage-resistant phenotype.
2.1.2.3 Expression Vectors for LAB
To ensure effective expression of genes of interest, expression vectors must have elements for constitutive or inducible protein expression besides the basic elements in cloning vectors such as origins of replication and heterologous genes. In terms of the cellular location of expressed proteins, there’re three types of protein expression in LAB as described below:
-
1.
Cytoplasmic expression: it can effectively protect produced protein from the external environment. But cell lysis is required for intracellular protein release.
-
2.
Secreted expression: the secretion of recombinant protein into extracellular medium is directed by signal peptide.
-
3.
Cell surface displaying of target protein: it can be achieved by anchoring recombinant proteins to the cell wall.
In recent years, a large number of expression vectors in LAB have been constructed for different applications in the fields of food, medicine, and life sciences.
2.1.3 Intracellular Expression Systems for LAB
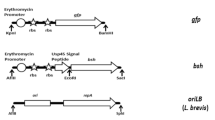
Intracellular expression of heterologous proteins in LAB is achieved by inserting foreign genes into LAB expression vectors without signal peptides and introducing recombined vectors into LAB hosts via electroporation. Plasmids for intracellular expression in LAB comprise promoters, multiple cloning sites for insertion of foreign genes, terminators, selection markers, and replicons. Thus far, most commonly applied promoters in LAB expression systems are lacA, lacR, lacF, T7, xylA, lacS, nisA/nisZ, and nisF (Kleerebezem et al. 1997). Based on the carried resistance gene, expression vectors in LAB can be divided into antibiotic resistance-based vectors and food-grade expression vectors. Traditional expression vectors for LAB carry one or multiple antibiotic resistance-encoding genes such as erythromycin and chloramphenicol resistance genes, and transformants are selected via antibiotic selection pressure. The plasmids pNZ8037 and pNZ8048 carrying chloramphenicol resistance genes were developed on the basis of the food-grade NICE system and are standard expression vectors for L. lactis. These two plasmids contain the pSH71 replicon and nisin-inducible promoter nisA and can be utilized as expression vectors in both Escherichia coli and L. lactis. To overcome plasmid pNZ8048-induced low-level expression in LAB, plasmids pNZ8148 and pNZ8150 carrying chloramphenicol resistance genes were constructed based on pNZ8048. These two upgraded plasmids are capable to drive highly efficient expressions in LAB.
2.1.4 Secreted Expression Systems for LAB
2.1.4.1 Features of Secretion Vectors in LAB
Exploiting LAB as expression hosts or cell factories to produce antigenic proteins, pharmaceutical molecules and other functional factors have become a new research filed on LAB. To achieve secretory expression of protein in LAB, heterologous genes are inserted expression vectors with signal peptides, which direct the secretion of intracellular recombinant proteins to extracellular environment after transforming LAB hosts with recombinant vectors via electroporation. Signal peptides, a vital determining factor for secretory expression, are peptides residing on recombinant precursor proteins, which can be targeted to the secretory pathway by signal peptides. Through the cleavage of signal peptidase, signal peptides are separated from the mature proteins, which are translocated to extracellular compartment afterward (Nielsen et al. 1997).
Compared with intracellular expression, secreted expression holds the advantage of synthesizing heterologous proteins in an active form without aggregation within the host cells, which effectively prevents the loss of proteins during protein recovery. Moreover, secreted recombinant proteins can be separated from other cellular proteins, thus simplifying the downstream purification procedures for target proteins. However, not all proteins are suitable for secretory expression, especially for those naturally nonsecreted proteins, whose secretion is determined by multiple factors such as their size, structure, charges, and signal peptide. For one specific protein, different signal peptides can result in differential secretion efficiencies (Meazza et al. 1997). Zhang et al. (2010) showed that modifying signal peptide structure pronouncedly improved the secretion efficiency of recombinant protein in LAB. Therefore, selection of proper signal peptides is of great importance for secretory expression system for LAB or other hosts.
2.1.4.2 Secretion Systems Based on Usp45 Signal Peptide
At present, the signal peptide of Usp45 protein isolated from L. lactis is the most extensively applied signal peptide for secretory expression in LAB and was identified in the genome of L. lactis MG1363 by van Asseldonk et al. (1990). LAB secretion systems can efficiently recognize Usp45 signal peptide, which resulted in enhanced secretion of heterologous proteins such as antigens (Ribeiro et al. 2002; Bermudez-Humaran et al. 2003b; Zhang et al. 2011) and antibodies (Bermudez-Humaran et al. 2003a; Zhang et al. 2010). To date, extensive studies have suggested that enhanced heterologous protein expressions via secretory expression are closely correlated with the abilities of signal peptides to stabilize variable proteins, inhibit the proteolysis by intracellular proteases, and improve protein secretion efficiency. In the study by Enouf et al. (2001), bovine rotavirus NSP4 protein was intracellularly and extracellularly expressed in L. lactis, but mature NSP4 protein could not be efficiently secreted. In addition to the Usp45-NSP4 precursor and mature NSP4 protein, two degradation products of mature NSP4 protein were also detected in the intracellular compartment of the NSP4-secreting LAB strain. This indicates that mature NSP4 protein might be partially degraded by intracellular proteases, which could be inhibited by infusion with the Usp45 signal peptide.
2.1.4.3 Secretion Systems Based on the Signal Peptide of S-Layer Protein
Signal peptide sequences exert crucial influences on the secretion efficiency of heterologous proteins in LAB. Due to the complex relation between protein secretion and signal peptides, it is not realistic to apply one specific signal peptide for the secretory expression of all different types of heterologous proteins in a specific LAB strain. Thus, searching for novel signal peptides to establish a diverse signal peptide library is an effective approach to elevate protein secretion expression levels in LAB. S-layer protein, whose relative molecular weights are in the range from 40,000 to 200,000, is a layer of bioactive macromolecules present on the cell wall surface of many bacteria and archaea. Most S-layer proteins consist of a single species of protein or glycoprotein. S-layer proteins account for 10–15% of the total bacterial proteins, and the gene expression machinery for their synthesis and secretion is quite strong.
Many researchers consider abundant signal peptide options as one of the effective means to enhance the secretion efficiency in LAB. S-layer protein-encoding genes are highly efficiently expressed and secreted in LAB, which are closely linked to the high transcription efficiency of their promoters and high secretion efficiency of their signal peptides. Based on this characteristic of S-layer proteins in LAB, their one major application in LAB is to develop highly efficient (secretion) expression systems for LAB (Zhang et al. 2010). Kahala and Palva (1999) introduced the promoter of a S-layer protein SIpA in two different LAB hosts and found that this promoter significantly enhanced beta-glucuronidase (gusA) and aminopeptidase N (pepN) expression in L lactis and L. plantarum, respectively. Expression levels of gusA and pepN account for 15% and 28% of the total cellular proteins in L. lactis and L. plantarum, respectively. In addition, Sibakov et al. (1991) increased secretion of β-lactamase in a recombinant L. lactis strain by using the strong promoter and signal peptide of lactobacilli S-layer protein-encoding gene. Even though promoter and signal peptide have been suggested to be closely related to the expression and secretion efficiency of heterologous proteins in LAB, the characteristics of the expression hosts also affect the transcription efficiency of S-layer protein promoter. It was shown that using the same S-layer protein promoter yielded differential protein expression efficiencies in different LAB hosts (Kahala and Palva 1999), which further facilitates the research on signal peptide sequences of LAB expression systems.
2.1.4.4 Secretion Systems Based on Other Types of Signal Peptides
In addition to LAB-originated signal peptides, signal peptides derived from other bacteria have also been applied to construct secretion expression vectors for LAB. In a study by Le Loir et al. (2001), staphylococcal nuclease (nuc) was secreted by recombinant L. lactis with the native signal peptide of nuc gene, whose secretion efficiency was lower than when applying the Usp45 signal peptide. This might be attributed to the effects of signal peptides on protein conformation (Kajava et al. 2000). In addition to screening suitable native signal peptides, many researchers focused on modifying native signal peptide sequences to elevate the secretion efficiency of recombinant proteins in LAB. In L. lactis, insertion of a nine-residue synthetic propeptide (LEISSTCDA) after the Usp45 signal peptide sequence achieved a secretion efficiency of up to 80% for nuc, the yield of which was upregulated 2–4-folds (Le Loir et al. 2005).
2.1.5 The Surface Expression System of Lactic Acid Bacteria (LAB)
2.1.5.1 S-Layer Protein Expression System
In 1985, G. P. Smith took the advantage of major coat protein P3 of the filamentous bacteriophage to establish a molecular genetic system for phage, which plays various important roles in some fields, such as interactions between protein-protein and DNA-protein, analysis of antigen peptides, protein directed evolution, and signal transduction. However, the nature characteristics of the phage make it hard to express high molecular weight (HMW) protein, which limits the application of the phage expression system. To solve the above problem, some researchers did some efforts to develop new system by using bacteria that can fully express heterologous HMW protein. Based on the safety feature of LAB, its expression system received lots of attention and exhibited a high potential value in live vaccines, diagnosis, enzyme immobilization, and so on.
Recent studies indicate that some proteins that anchor on the cell surface of LAB play key roles in cell adhesion, immune responses, signal transduction, and other life activities. Based on the structural characteristics of surface protein, some were used to establish new expression system. It was apparent that the monomolecular crystalline array of proteinaceous subunit, which was termed as S-layer, was considered as one of the most common surface structures on bacteria. Sequence analysis shows that S-layer protein of LAB has two conserved domains (N-terminal secretion signal peptide and C-terminal anchoring peptide) and an intermediate variable region that participates in the protein refolding and crystallization process. Based on the above structure property of S-layer protein, the expression gene of heterologous protein can be inserted into S-layer protein coding gene, and then it was expressed on the cell surface of LAB following the S-layer protein expression. There is a growing body of evidence showing that bacterial S-layer protein expression system could exert great application potential in the field of microbiology, molecular biology, immunology, and biological catalysis. Due to its safety property, the expression system in LAB has received more attention from different study fields, such as recombinant vaccines, bacterial adhesion, and antibody.
2.1.5.2 Cell Wall Anchoring Expression System
To date, some enzymes that anchor surface proteins to the cell wall are proven to be used as cell wall anchoring domain of heterologous protein in the surface expression system based on LAB, especially cell wall hydrolases and the aggregation factor. Due to the difference in anchor position, the surface expression system can be divided into cell wall anchoring form, cell membrane anchoring form, and surface layer-associated proteins anchoring form. According to the bonding form between the anchoring protein and cell, the surface expression system is divided into covalent bond and non-covalent bond formation. On account of the binding site between heterologous protein and anchoring protein, it can be divided into two main forms: N-terminal and C-terminal formation. For the former, the expressed heterologous protein locates in the middle of the signal peptide and the anchoring region, such as the M6 protein from Streptococcus pyogenes, protein A of Staphylococcus aureus, and other surface protein in the Gram-positive bacteria. For the latter, the expressed protein locates the downstream of the anchoring region, such as proteinase PrtP from L. lactis, lysozymes from Bacillus phages, extracellular hydrolase from Lactobacillus strain, and peptidoglycan hydrolase of Enterococcus strain.
The LPXTG motif contained a hydrophobic domain and a positively charged tail, which was termed as a marker sequence of proteins which was anchored to the bacterial cell wall (Kuczkowska et al. 2015). After translocation, LPXTG motif would be firstly cleaved and then cross-linked at the threonine residue to a nucleophile, i.e., an active amino group of the peptidoglycan stems peptide or the lysine residue of the pilin motif (Kuczkowska et al. 2015). García-Mantrana et al. (2016) showed that two phytases from bifidobacteria could be cloned in L. casei under the control of a nisin-inducible promoter, and they were able to produce, export, and anchor to the cell wall. Kuczkowska et al. (2015) indicated that recombinant L. plantarum displaying CCL3 chemokine in fusion with HIV-1 Gag-derived antigen causes increased recruitment of T cells, in which the heterologous proteins were expressed in the cell surface.
LysM is widely distributed in more than 4000 proteins in both prokaryotes and eukaryotes, and this protein was firstly discovered in lysozyme of Bacillus phage φ29 acting as a C-terminal repeat comprising of 44 amino acids with seven amino acids inserted (Buist, Kok et al. 1995). The best-characterized LysM containing protein is the N-acetylglucosaminidase AcmA of L. lactis, which is required for cell separation and cell lysis during the stationary phase of L. lactis (García-Mantrana et al. 2016). The composition of Lysm domain family is highly abundant, which could be attributed to gene diversity, varied amount, and combinational diversity with other protein domain, and thus some of them were used as an anchor to display heterologous proteins on the surfaces of LAB. Hu et al. (2010) showed that a Novel LysM domain was isolated from L. fermentum bacteriophage endolysin and used as an anchor to display protein in the surfaces of LAB. To date, there is a growing body of evidence indicating that LAB could be used as a potential vehicle although some disadvantages need to be solved, such as thicker cell walls, less heterologous protein production, and so on.
2.1.6 Applications of LAB-Based Gene Expression System
2.1.6.1 LAB as Vaccine Delivery Vehicles
-
(1)
Applications of LAB vaccine delivery system
In recent years, due to the progress in developing LAB as mucosal vaccine delivery vectors, the application of recombinant LAB-based vaccines has been extensively expanded. Vaccines based on genetically engineered LAB strains have been exploited for disease management or as nutritional supplements. Herein, we will describe in detail the current status of preclinical laboratory research regarding diverse applications of recombinant LAB vaccines.
-
1.
Prevention and treatment of infectious diseases
At the early stage, the primary recombinant LAB-based mucosal vaccines against infectious diseases were genetically modified LAB strains expressing key antigen proteins (or fragments). Recombinant LAB vaccines are applied in two distinctive forms: immunoprophylaxis and immunotherapy. These two immunization forms differ in their timing of intervention. For immunoprophylaxis animals are administered with recombinant LAB strains prior to establishing experimental disease models, while for immunotherapy immunization animals administered with recombinant vaccines are carried out after animal models are developed. LAB vaccines against infections will be described below based on the types of pathogenic microorganisms involved.
-
(A)
Bacterial and fungal infections
Thus far, a large number of genetically modified LAB strains producing crucial antigens (fragments) of common human or animal pathogens have been successfully constructed and confirmed to confer immune protection against infections in various animal infection models (Wells et al. 1993; Norton et al. 1997; Maassen et al. 1999; Grangette and Muller-Alouf 2001; Robinson et al. 1997, 2004; Cheun et al. 2004; Corthésy et al. 2005; Hanniffy et al. 2007).
In order to optimize the immune efficacy of recombinant vaccines, researchers attempted to construct recombinant LAB strains expressing different antigen proteins or fragments derived from one specific pathogenic microorganism for immunization (Lee et al. 2001; Wu and Chung 2007; Wei et al. 2010; Hongying et al. 2014). Wu and Chung (2007) constructed a recombinant Lactobacillus reuteri strain producing the fusion protein of heat-stable enterotoxin and heat-labile enterotoxin B of enterotoxigenic Escherichia coli (ETEC) as a mucosal vaccine against ETEC infections. Wei et al. (2010) exploited Lactobacillus casei as a carrier for fimbriae protein of ETEC K99 in mucosal immunization.
Through certain evolved strategies, pathogens are capable to evade the host immune defense system so that they can continuously colonize, proliferate, and migrate in the hosts to cause infections. These strategies including specific cell surface components (e.g., surface capsules and pili) and synthesis of virulence factors (e.g., toxins and proteases) are unitized by pathogens to interfere or subvert host immune surveillance. Thus, in order to promote the recognition of specific pathogens by the host immune system and to trigger specific defensive immune responses, it is of vital importance to select essential virulence factor of pathogens as the target of vaccine development.
Besides, developing recombinant LAB strains as mucosal delivery vectors for antibody fragments to enhance host immunity against pathogens is another effective strategy against infections. Beninati et al. (2000) constructed recombinant Streptococcus gordonii strains expressing a microbicidal single-chain antibody (H6), which were found to inhibit Candida albicans-induced vaginal inflammation via vaginal immunization.
-
(B)
Viral infections
So far, researchers have successfully constructed genetically modified LAB strains against many common pathogenic viruses such as rotavirus, hepatitis B virus, and influenza virus and evaluated their efficacy in experimental animal models of viral infection (Xin et al. 2003; Ho et al. 2005; Perez et al. 2005; Lee et al. 2005; Poo et al. 2006; Lei et al. 2011; Liu et al. 2011; Zhang et al. 2011). It is promising that Xin et al. (2003) engineered a recombinant L. lactis strain to express and anchor the envelope protein of the human immunodeficiency virus (HIV) on its cell surface. They found that oral immunization of mice with this genetically modified LAB strain induced HIV-specific cellular (high level of IFN-γ-secreting lymphocytes in the spleen and intestinal lymph node) and humoral immune responses (high level of HIV-specific serum IgG and fecal IgA antibodies). Furthermore, expressed HIV antigen fragments were shown to be presented to T lymphocytes by dendritic cells, thereby mounting adaptive immune reactions (Xin et al. 2003).
-
(C)
Parasitic infections
Parasites, another major group of infectious organisms, can cause severe organ or tissue damage in the hosts. However, due to the difficulty in their in vitro cultivation, the development of anti-parasitic vaccines has been greatly hindered. Therefore, synthesis of recombinant parasite antigens using molecular biology techniques for immunization is of great significance for facilitating anti-parasitic vaccines development. Thus far, there have been some promising results with regard to applying LAB as vaccine vehicles against parasitic infections (Zhang et al. 2005; Ramasamy et al. 2006; Lee et al. 2009; Yam et al. 2011). Zhang et al. (2005) genetically modified Lactococcus lactis to express Plasmodium yoelii antigen fragment MSP-119 and orally immunized two strains of mice (BALB/c and C57BL/6) with this recombinant strain. They observed that this recombinant LAB strain effectively enhanced host immunity against malaria parasites in both strains of mice.
-
2.
Inflammatory bowel disease (IBD) management
IBD is group of chronic, recurrent gastrointestinal inflammatory disorders caused by intestinal dysfunction. IBD is often concomitant with parenteral complications and needs long-term therapy, thus leading to a marked decline in IBD patients’ quality of life. In spite of the similar clinical symptoms such as abdominal pain and diarrhea, IBD should be discriminated from pathogen-induced gastroenteritis. Ulcerative colitis (UC) and Crohn’s disease (CD) are two main forms of IBD. In recent years, the prevalence of IBD has been increasing worldwide; effective IBD prevention and treatment are desperately needed. So far, researchers have carried out a range of studies on mucosal vaccination with genetically engineered LAB against IBD based on differential IBD pathogenesis and therapeutic directions and obtained some promising results. The first attempt to apply genetically modified LAB strains against IBD was performed by Steidler and Hans (2000). They constructed a recombinant L. lactis strain expressing anti-inflammatory cytokine interleukin-10 (IL-10). Intragastric administration with this recombinant strain pronouncedly attenuated dextran sulfate sodium (DSS)-induced colitis in mice by 50% and also effectively suppressed the development of IL-10 gene deficiency-caused colitis in mice. More encouragingly, an “upgraded” recombinant IL-10-expressing L. lactis strain has been tested in a phase I clinical trial and demonstrated to be safe in CD patients and biologically contained (Braat et al. 2006). This is also the first clinical trial performed with genetically modified LAB (Braat et al. 2006), which greatly promotes research on recombinant LAB vaccines.
Apart from IL-10, some other anti-inflammatory molecules such as trefoil factor (TFF) and neuropeptide α-melanocyte-stimulating hormone (α-MSH) have also been expressed in genetically modified LAB strains for controlling IBD. TFF are small polypeptides with beneficial effects on mucosal protection and repair, while α-MSH is a neuroendocrine peptide with anti-inflammatory properties (Ren et al. 2005; Zhu et al. 2014). Vandenbroucke et al. (2004) observed pronounced alleviation of inducible acute colitis and spontaneous chronic colitis in mice after intragastric administration with TFF-expressing L. lactis. Yoon et al. (2008) found that oral immunization of mice with a α-MSH-secreting L. casei strain significantly ameliorated DSS-induced acute colitis in mice.
Neutralizing the inflammatory cytokine tumor necrosis factor (TNF)-α is another effective therapeutic strategy for IBD. In accordance with this strategy, several recombinant mucosal vaccines based on genetically modified LAB have been developed. A study by Vandenbroucke et al. (2009) confirmed the protective efficacy of recombinant L. lactis secreting anti-TNF-α nanobodies on both DSS- and IL-10 deficiency-induced colitis in mice after oral immunization. Furthermore, the affibody against TNF-α has been surface displayed in L. lactis and recombinant affibody was shown to exhibit TNF-α-binding capability. These results indicate that this affibody-expressing strain has a potential in binding intestinal TNF-α and might be used to mitigate gut inflammation in IBD (Ravnikar et al. 2010).
Since oxidative stress plays a key role in the pathogenesis of inflammatory disorders, antioxidant enzymes become another type of potential IBD therapeutic agents. Thus far, researchers have engineered LAB strains as mucosal delivery vectors for antioxidant enzymes such as superoxide dismutase (SOD) and catalase (CAT) to achieve the immunomodulation of IBD (Carroll et al. 2007; LeBlanc et al. 2011). LeBlanc et al. (2011) constructed recombinant L. casei expressing SOD or CAT and assessed their protective efficacy in mice with trinitrobenzenesulfonic acid (TNBS)-induced CD. It was observed that mice that received these SOD or CAT-producing LAB strains displayed less weight loss, less bacterial translocation to the liver, and alleviated colonic tissue damage.
Although the precise pathogenesis of IBD is still obscure, it has been proposed that insufficient innate antigenic stimulation can result in immune dysfunction, which triggers excessive immune reactions against innocuous intestinal antigens and thereby leads to inflammation and tissue damage. Therefore, utilization of anti-inflammatory agents to suppress excessive inflammatory responses in IBD patients is a major clinical treatment option for IBD. A study conducted by Foligne et al. (2007) offers new therapeutic targets for IBD management. It is known that some pathogens can effectively circumvent the host immune surveillance, stimulate anti-inflammatory cytokine production, and resist the host immune responses against them. Based on this theory, the low-calcium response V (LcrV) protein, one virulence factor of enteropathogenic Yersinia pseudotuberculosis was expressed in genetically engineered L. lactis (Foligne et al. 2007). The protective properties of this recombinant strain were demonstrated in TNBS- and DSS-induced murine colitis models. It was shown that this strain effectively induced IL-10 secretion and dampened colonic inflammation. Intriguingly, the protective effects of this recombinant strain in mice with TNBS-induced colitis were IL-10-dependent. Moreover, similar protective efficacy in TNBS-induced colitis model was observed by immunization with this LcrV-expressing L. lactis strain or an IL-10-expressing L. lactis strain (Foligne et al. 2007).
-
3.
Management of allergic disorders
Type I allergy, also known as immediate hypersensitivity reactions, is an IgE-mediated, immune disorder characterized by Th2-skewed immune responses. Clinical symptoms of type I allergy occur immediately upon allergen exposure. Food allergy, dust mite allergy, and pollen allergy are all immediate hypersensitivity disorders. In recent years, there has been accelerating incidence of type I allergy worldwide. However, there are still great difficulties in its effective clinical prevention or treatment. Traditional type I hypersensitivity treatment is allergen-specific immunotherapy, in which immune tolerance to specific allergens is developed. In the traditional desensitization therapy, patients are exposed to or injected with allergen extracts via non-mucosal routes for administration such as subcutaneous injection. Due to the limitation in the availability of highly pure allergen extracts, severe adverse reactions often occur in allergic patients receiving traditional desensitization therapy. Therefore, genetic engineering of LAB strains for producing and mucosal delivering recombinant allergens can effectively circumvent the aforementioned problems. The generally recognized safe status and immunomodulatory properties of LAB render them ideal vectors for presenting allergens to the host mucosal surface. So far, a number of allergen-expressing recombinant LAB strains have been successfully applied for type I allergy management (Adel-Patient et al. 2005; Charng et al. 2006; Cortes-Perez et al. 2007; Huibregtse et al. 2007; Rigaux et al. 2009; Schwarzer et al. 2011).
Allergic reactions are triggered upon the binding of allergen-specific IgE antibodies to the IgE binding epitopes on allergens. Allergen T cell epitopes is the molecular basis for the recognition of allergens by T cells, thereby conferring immunomodulatory effects. The progress in identifying allergen epitopes in recent years greatly facilitates the development of recombinant hypoallergenic allergens or T cell epitope peptide-based allergy therapies. T cell epitope peptides have been successfully applied to modulate cat and insect venom (Bohle 2006). To summarize, it is promising to apply recombinant LAB strains expressing novel allergen mutants as mucosal vaccines for the prophylaxis and alleviation of allergic diseases.
In recent years, some researchers have applied cytokine-expressing genetically engineered LAB against allergy, indicating a new research direction for managing allergic diseases (Frossard et al. 2007). Frossard et al. (2007) found that oral administration of mice with an IL-10 expressing recombinant L. lactis strain effectively inhibited allergic reactions and reduced serum Th2-type antibody (IgE and IgG1) production. Cytokine-expressing recombinant LAB strains were also used to boost the immunomodulatory effects of allergen (or allergen fragments)-expressing LAB in allergy management. Cortes-Perez and Ah-Leung (2007) showed that an IL-12-producing recombinant L. lactis strain enhanced the protective efficacy of a genetically engineered LAB strain expressing cow’s milk allergen in a cow’s milk allergy mouse model.
Cross-linking of allergen-specific IgE antibodies with cell surface receptors on basophils or mast cells is a prerequisite for triggering the release of inflammatory mediators such as leukotriene and histamine and mounting allergic reactions. Therefore, using humanized anti-human IgE antibodies to bind free IgE to prevent the cross-linking of IgE with mast cells is an effective approach for allergy prevention or alleviation. Based on this, Scheppler et al. (2005) genetically modified L. johnsonii to express and anchor either an IgE mimotopes or an anti-idiotypic single-chain fragment variable (scFv) mimicking an IgE epitope on the cell surface. This recombinant stain was shown to be recognized by anti-human IgE monoclonal antibody and induce systemic IgG antibodies against human IgE through both systemic and mucosal immunization. These results indicate that these recombinant strains can potentially induce anti-IgE response, preventing or relieving allergic symptoms.
-
(2)
Clinical applications of recombinant LAB as vaccines
As stated above, preclinical studies on recombinant LAB-based mucosal vaccines have achieved remarkable results, promoting their clinical research. Nevertheless, there are still some concerns regarding the clinical effectiveness and safety of recombinant LAB-based vaccines, which hinders the launch of their clinical trials. Steidler and Hans (2000) confirmed the remarkable protective efficacy of mucosal immunization of an IL-10-expressing genetically engineered L. lactis strain in two different mouse models of colitis. This research group developed a thymidine-deficient expression system (ActoBiotics™) for L. lactis to facilitate its clinical research. The thymidylate synthase-encoding gene thyA in L. lactis chromosome was replaced with the human IL-10 gene so that this modified recombinant strain cannot survive under thymidine- or thymine-free conditions. This modification prevents the release of this genetically engineered strain into the environment, thus eliminating the biosafety concerns toward this strain raised by its potential replication and pervasion (Bahey-El-Din 2012). Based on this thyA-deficient expression system, Braat et al. (2006) launched the first phase I clinical trial on recombinant LAB-based mucosal vaccines. After obtaining promising results in CD patients during its phase I clinical trial, its phase IIA clinical trial has also been conducted. Although the phase IIA clinical trial confirmed its biosafety status, it was not effective on mucosal repair (Bermúdez-Humarán et al. 2011).
Clinical studies have also been performed with another genetically engineered LAB strain (AG013), which was constructed to express human TFF1 based on ActoBiotics™ expression system. Its phase Ib trial demonstrated its safety and tolerability in participants as well as its efficacy in improving chemotherapy-induced ulcerative oral mucositis in patients with locally advanced head and neck cancer (Limaye et al. 2013).
2.1.6.2 Recombinant LAB in Enzyme Preparations
In addition to being developed as mucosal vaccines, genetically engineered LAB also exhibit great potential in the production of catalytically active enzymes. Relevant studies have also made some progress. We will describe their applications below based on their specific purposes.
Some researchers constructed enzyme-expressing LAB strains to regulate relevant biochemical reactions in food manufacturing, thereby improving food attributes such as flavor and texture. Yao et al. (2010) developed a recombinant L. lactis NZ9000 strain, which carries a secreted expression plasmid inserted with the bovine trypsin gene. Recombinant precursor protein with signal peptide was detected in the protoplast fraction of recombinant bacteria. The recombinant bovine trypsin was also shown to be biologically active. It avoids the biosafety risk from extracting bovine trypsin from cow pancreas, strengthens the proteolytic systems in LAB, and enhances bioactive peptide production during the manufacturing of fermented dairy products, thereby boosting the potential health benefits of fermented dairy products (Yao et al. 2010). Alpha-amylase has also been expressed in different cellular locations of genetically engineered LAB. Native α-amylase gene was introduced and expressed in two L. plantarum host strains (strain WCFS1 and a food-grade strain TGL02) with a secretion efficacy of 90% (Kanpiengjai et al. 2015). Notably, the properties of native wild-type α-amylase such as broad pH tolerability and maltose-producing activity were also found in secreted α-amylase by the recombinant TGL02 strain (Kanpiengjai et al. 2015). Furthermore, alpha-amylase was also effectively expressed on the cell surface of L. casei by using the Bacillus subtilis anchor protein PgsA (Narita et al. 2006). This α-amylase-displaying strain was shown to exhibit strong hydrolytic ability on soluble starch and to hydrolyze 36.3 g/L of soluble starch, yielding 21.8 g/L of lactic acid within 24 h (Narita et al. 2006).
Another application of enzyme-expressing recombinant LAB is to deliver recombinant enzymes with biocatalytic activities to host mucosa in order to modulate the host immune functions. Furthermore, significant progress has been made in related research in this field, which has been drawing great attention.
Oxidative stress is known to cause damage in cells or tissues, thereby triggering inflammatory diseases and accelerating host aging. Antioxidants have been a popular research topic in multiple fields in recent years owing to their capabilities of eliminating oxygen free radicals. Thus far, in the field of recombinant LAB-based mucosal vaccines, LAB have been genetically engineered to express antioxidant enzymes such as SOD and CAT, and their potential in managing gut inflammatory disorders has also been demonstrated in animal studies. Carroll et al. (2007) observed that genetically modified L. gasseri expressing Streptococcus thermophilus-derived SOD significantly alleviated intestinal inflammation in IL-10-deficient mice. Moreover, genetically engineered LAB (e.g., L. plantarum and L. casei) as delivery vectors for SOD or CAT were shown to prevent or mitigate TNBS- or DSS-induced gut inflammation in mice or rats (Rochat et al. 2007; Watterlot et al. 2010; LeBlanc et al. 2011).
In addition to antioxidant enzymes, some researchers also developed recombinant LAB as mucosal delivery vehicles for certain enzymes to improve clinical symptoms induced by the deficiency of these enzymes. Drouault et al. (2002) applied a Staphylococcus hyicus lipase-expressing L. lactis strain to improve pancreatic insufficiency-elicited defective lipid metabolism in pigs.
2.1.6.3 Recombinant LAB in Metabolic Regulation
In recent years, researchers have directionally modified or regulated certain metabolic pathways in LAB using genetic and biochemical engineering techniques, thereby altering the substrate utilization spectra of LAB, facilitating their substrate utilization, and directionally augmenting the production of desired metabolites. To a certain extent, these studies have greatly improved the production efficiency of LAB in the food industry.
Diacetyl, a natural by-product during dairy and alcoholic fermentations, has been widely exploited as a flavoring agent in the food industry due to its strong buttery flavor. LAB can utilize citric acid that is minorly present in milk to synthesize an intermediate α-acetolactate (α-AL), which is subsequently converted to diacetyl via oxidative decarboxylation. In order to enhance diacetyl production by LAB, researchers have attempted to regulate the production of crucial enzymes involved in diacetyl synthesis in LAB (Platteeuw and Hugenholtz 1995; Hugenholtz and Kleerebezem 2000). A lactate dehydrogenase (LDH)-deficient L. lactis NZ2700 strain was genetically modified to overexpress α-ALS, achieving a high production of the diacetyl analog acetoin with lactose as substrates. However, no high diacetyl production was found by this strain. Afterward, Hugenholtz and Kleerebezem (2000) constructed a genetically engineered α-AL decarboxylase (ALDB)-deficient L. lactis strain overexpressing NADH oxidase. This recombinant strain cannot convert α-AL to acetoin due to its α-ALDB deficiency. Moreover, high expression of NADH oxidase in this strain imparts it efficient synthesis of diacetyl.
Apart from diacetyl, regulating L-alanine synthesis is another classic case of utilizing genetic engineering tools for metabolic engineering in LAB. By genetically modifying a L-LDH- and alanine racemase-deficient L. lactis strain to express the alanine dehydrogenase (L-AlaDH) gene from B. sphaericus, Hols and Kleerebezem (1999) successfully altered the carbon flux in the sugar metabolism of wild-type L. lactis from homolactic fermentation to homoalanine fermentation. Besides, by using the LDH promoter from S. thermophilus, the B.subtilis AlaDH was overexpressed in recombinant L. lactis NZ9000, whose alanine production was upregulated 26 folds as compared to the untransformed L. lactis strain (Ye et al. 2010).
To date, a large number of studies have suggested the beneficial effects of LAB-produced exopolysaccharide (EPS) during milk fermentation on the hosts. EPS from LAB have been substantially applied as thickeners in the food industry because of its specific textural properties (Tong et al. 2015). However, limited production levels of EPS in many LAB strains hinder their industrial application. Therefore, some research teams have successfully applied genetic engineering tools to enhance EPS production by LAB (Levander and Svensson 2002; Boels et al. 2003; Svensson et al. 2005). By regulating the expression levels of key enzymes in the central pathways of carbohydrate metabolism, EPS expression levels were elevated in S. thermophilus (Levander and Svensson 2002; Svensson et al. 2005).
2.2 Food-Grade Expression System for Lactic Acid Bacteria
2.2.1 Basic Requirements of Food-Grade Expression System
The food-grade expression system is an expression system that maximizes the production of food and food-related products. Food-grade genetic expression system must contain the following characteristics: first, genetic expression vectors in the system are food-grade, consisting of DNA from known safe microbes and cannot contain non-food-grade DNA fragments. The lactic acid bacteria (LAB) vectors that are currently used generally carry one or more antibiotic (such as erythromycin, chloramphenicol) resistance genes to maintain a certain selection pressure. However, these resistance factors will drift, and if these carriers are used in daily life, they will have serious consequences for biosafety. Therefore, the use of food-grade selection markers that are harmless to the human body in place of antibiotic resistance markers is one of the effective means to solve this problem. Second, the expression host must be a safe, well-characterized, and stable food-grade microorganism. Lactobacillus, Lactococcus lactis, and Bifidobacteria are generally regarded as safe (GRAS) food-grade microorganisms. In addition, the host bacteria also need to be stable enough in food or in the body. Third, the inducer used in the expression system is also food-grade, such as sucrose, lactose, pyrimidine, nisin, etc.
2.2.2 Selective Marker of Food-Grade
Food-grade vectors of lactic acid bacteria require the vector is free of non-food-grade functional fragments. According to the difference of screening methods, selective marker of food-grade about LAB can be divided into two categories: complementary selection markers and dominant selection markers. A complementary selection marker requires a deletion mutation in the host chromosome, and then the vector’s selective marker is used to compensate for the deletion mutation, thereby restoring the host to a certain characteristic. Complementary markers often use genes that encode important proteins involved in metabolic transformation. The drawback of defective marker is that it can only be used for specific vectors – the host system. Dominant selection markers are mainly used to provide new phenotypic characteristics by taking advantage of the characteristics of the host bacteria themselves, and do not depend on the expression genes of the host. Therefore, such markers can be applied to other bacteria of the same genus or even to other lactobacillus. However, there are not enough food-grade dominant selection markers applied at present, mainly due to the following: (1) the selective process is complicated; and (2) sometimes the labeling system is too large because it needs to contain several genes.
Selective markers of food-grade that have been applied to LAB can be classified into four categories: saccharide utilization selective markers, auxotrophic complementary selection markers, bacteriocin resistance markers, and heavy metal resistance markers.
2.2.2.1 Saccharide Utilization Selective Markers
Sugar is an important raw material for industrial fermentation. Different LAB are different in using the types and efficiencies of sugar. Most LAB generally cannot use xylose, inulin, sucrose, and melibiose. Therefore, cloning genes associated with the utilization of these sugars into non-fermenting strains give them the ability to ferment a certain sugar.
The current research on lactose operons is the most in-depth. In the lactose operon, the lacF gene encodes the key enzyme IIA of the lactose phosphotransferase system in L. lactis. First, the complete lactose operon was integrated into the chromosome of L. lactis, and then the lacF deletion was made by double crossover homologous recombination to construct a lac− type receptor strain. At the same time, the LAB replicon pSH71, the promoter P32, and the lacF gene were used to construct the vector pFI846. When a plasmid containing the lacF gene is introduced into the lac− strain, the bacteria will restore the lac+ phenotype, and positive colonies can be screened on plate medium containing bromocresol purple (Platteeuw et al. 1996). Platteeuw et al. (1996) also constructed a food-grade vector by using the lacF gene as a selective marker, which includes the lacA promoter and the transcription terminator of the aminopeptidase N gene pep N. It enhances the stability of the vector, and the gusA gene was successfully expressed using this vector. Domestic Hesong et al. (2010) cloned the β-galactosidase gene from the L. acidophilus genome and expressed it in L.lactis. Positive clones were screened by 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal). The recombinant L. lactis was passaged for 60 generations by X-gal color development screening method, and the β-galactosidase enzyme activity and specific activity were measured. The results showed that positive clones could be successfully screened by expressing active β-galactosidase and X-gal color development. The β-galactosidase specific activity assay was performed on recombinant L. lactis after 60 generations by X-gal. There was no significant difference compared with the second generation of X-gal screening (P = 0.592 > 0.05) and no significant difference compared with the erythromycin screening (P = 0.882 > 0.05). Thus, the β-galactosidase gene has good activity and stability as a screening marker.
Posno and Heuvelmans (1991) cloned the xyl gene for xylose ferment of the L. pentosus MD353 into the E. coli-lactobacillus shuttle vector pLP3537 to obtain a recombinant pLP3537-xyl plasmid. Further, the plasmid was transferred to L. casei ATCC393 which could not utilize xylose. As a result, the transformant obtained the ability to utilize xylose, thereby obtaining xylose fermentation as a food-grade selective marker of Lactobacillus.
2.2.2.2 Auxotrophic Complementary Selection Markers
The auxotrophic complementary selection marker is a commonly used method for constructing food-grade expression systems. In bacteria, some products encoded by genes can catalyze the basic metabolic reactions of bacteria. When these genes are deleted or mutated, the bacteria will not be able to synthesize the corresponding products, thus causing the bacteria to fail to grow normally in the original growth environment. Only by supplementing the corresponding substrate will the bacteria return to its original phenotype, which is the auxotrophy of the bacteria. These genes are cloned into a plasmid and introduced into an auxotrophic strain, which is complementary to the host bacteria, and the bacteria can restore a certain characteristic. Therefore, you can choose according to this feature. Food-grade expression vectors have been successfully constructed with using the thyA, arl, thr, supB, and supD genes.
The thyA gene encodes a thymidylate synthase, which plays a key role in DNA synthesis. If the gene is missing, the strain cannot grow on the basic medium. The thyA gene is a safe food-grade selection marker that can be used to construct food-grade expression systems. At the same time, the content of thymine or thymidine in dairy products and the gut of animals (including humans) is very small, providing favorable conditions for the use of such carriers. Ross and O’Gara (1990) first constructed a food-grade vector with the thyA gene as a screening marker. However, since LAB lacking thy A gene were not constructed, the application of the gene in LAB was limited. Wang Chunfeng et al. (2001) of China Agricultural University obtained recombinant vectors by replacing the erythromycin resistance gene on plasmid pW425e with thyA. At the same time, thyA-deficient Lactobacillus acidophilus was screened as a recipient strain, and a food-grade carrier receptor system with thyA as a screening marker was obtained. The Eimeria tenella SO7 gene was introduced into the vector and expressed in LAB and E. coli. Xiong Yanwen et al. (2004) constructed a vector pSH91 applied to L. lactis. With a total length of 2337 bases, it is a food-grade vector composed of selective marker thyA gene, replicator of pWV01, and polyclonal site of plasmid pUC18. After transforming the vector into thyA-deficient L. lactis, the host bacteria returned to the wild type.
Alanine is a structural component of the Gram-positive bacteria wall, and D-alanine is an indispensable component of the bacterial cell wall peptidoglycan. When it is absent, the cell wall synthesis is blocked, causing the bacteria to die. D-Alanine is usually not contained in the raw materials of industrial fermentation, and L-alanine in the medium needs to be converted into D-alanine. Alanine racemase catalyzes the conversion of D-alanine and L-alanine. Therefore, the alr gene can serve as a food-grade complementary marker. Bron and Benchimol (2002) first used homologous recombination to mutate the alr gene on the chromosome to construct a mutant strain of the alr gene, providing a receptor strain. Bron and Benchimol (2002) obtained a mutant strain of L. plantarum and L. lactis arl gene as a recipient strain by using the deletion mutation. After introducing the vector of cloned alr gene into the host bacteria, the two alr gene mutant host bacteria can grow on the medium without D-alanine. Lu Wenwei (2014) established a selective marker for the alanine racemase gene (alr). First, the alanine racemase gene of L. lactis NZ9000 and L. casei BL23 was knocked out by the temperature-sensitive plasmid pG+host9 and the suicide integration vector pRV300, respectively. The host cell itself became D-Ala auxotrophy, and then the alanine racemase gene acts as a complementary selection marker to achieve functional complementation.
In the process of prokaryotic ruthenium synthesis, if the related genes are meteorite mutation, the strain will be purine auxotrophy. The product encoded by the nonsense suppressor gene supB can make up for the auxotrophy. Similarly, when the gene in the pyrimidine synthesis pathway undergoes an amber mutation, the nonsense suppressor gene supD gene product can compensate for this defect. Thus, food-grade vectors can be constructed using supB and supD as selective markers. Sorensen and Larsen (2000) constructed a new food-grade expression vector pFG200 based on the vector pFG1. The pFG200 expression vector uses supD as a selection marker and a pyrimidine auxotrophic strain as a host strain. Moreover, the pFG200 vector has no significant effect on the growth rate of the host bacteria and the acidification rate of the milk. However, a major drawback of such complementary systems is that specific mutations must be introduced into each mature host in a food-grade manner prior to application of the complementary plasmid.
2.2.2.3 Bacteriocin Resistance Markers
Since bacteriocin is a food-grade product, its resistance genes and immune genes can be used as food-grade selection markers. The bacteriocin resistance genes that have been utilized now include the nisin resistance gene (nsr), the immune gene (nis I), and the immunogenic gene of Lactobacillus F (laf I). A strain containing the nisin resistance gene (nsr) or the immunogenic gene Nis I can grow normally on a substrate containing a certain concentration of nisin. Therefore, the nisin resistance gene or immune gene is an ideal food-grade selection marker. The nsr gene contains an open reading frame of 957 bases that encodes a protein of 318 amino acids. Von Wright et al. (1990) inserted a fragment containing the nsr gene in the cloning vector pVS34 and eliminated the chloramphenicol resistance gene originally carried in the vector and constructed a food-grade vector with NSR as a selective marker. Hughes and Mc Kay (1992) constructed a food-grade cloning vector pFM011 with NSR as a selection marker and cloned a sequence encoding bacteriophage resistance in this vector to obtain a plasmid with both nisin resistance and phage resistance, named pFM012. The vector was introduced into L. lactis LM0230, which has phage resistance. This indicates that the nsr gene can be used to directly screen transformants, thereby replacing traditional antibiotic resistance markers.
The laf I gene is an immune gene of Lactobacillus F produced by L. johnsonii VPI 11088. Studies have shown that if laf I is destroyed, the strain is sensitive to Lactobacillus F. The vector pTRK434 carrying laf I was introduced into the Lactobacillus F-sensitive strain Lactobacillus johnsonii. The host bacteria restored the immunity of the bacteriocin and increased the immune tolerance by 64-fold compared with the non-transformant cells. Allison and Klaenhammer (1996) transformed the plasmid pTRK434 containing the laf I gene into the fermentative lactobacillus NCDO 1750. The transformant was selected using a medium containing Lactobacillus F to achieve selective labeling using laf I.
2.2.2.4 Selection Markers for Heavy Metal Resistance
Some plasmids of LAB contain resistance genes of metal ions such as cadmium (Cd) and copper (Cu), and food-grade carriers have been successfully constructed using these resistance genes. Liu et al. (1996) isolated a copper resistance gene (cuR) from L. lactis plasmid pND306 to construct a food-grade vector pND968. Wong et al. (2003) linked the cadmium ion resistance genes cadA and cadC to the S. thermophilus vector pND913 and removed the non-food-grade fragments to obtain the food-grade cloning vector pND919, which was successfully applied to the food-grade expression of S. thermophilus.
In addition, a dual plasmid selection marker system has great potential for use in food microbial applications and has been developed and utilized by researchers. The system comprises two plasmids, one is a vector carrying a functional replicon and the foreign gene to be expressed, but no selection marker, and the other is a concomitant plasmid, and the plasmid system with the antibiotic resistance selection marker is also applied to the food-grade selectable marker of LAB. Emond et al. (2001) successfully constructed a dual plasmid system in which two plasmids dissociate the major antibiotic markers of the vector plasmid. The vector consists entirely of L.lactis and complements the phenotypic characteristics of the associated plasmid. After transformation screening, the associated plasmid is readily removed in antibiotic-free medium and remains highly stable without selective pressure.
2.2.3 Food-Grade Inducer
In the process of cloning and expression of LAB, certain inducers are required. In food expression systems, it is required that the inducer must be food-grade and edible for humans, such as lactose, sucrose, nisin, and the like. Among them, nisin belongs to the wool sulfur bacteriocin, which is produced by L. lactis. The mature nisin molecule contains 34 amino acids with a molecular weight of 3510 Da. Nisin is a polypeptide substance, which is non-toxic and does not produce antigens in humans. After consumed, nisin can be inactivated by protease action in the digestive tract, so it will not change the intestinal flora structure. Nisin has been accepted as a food additive by the FAO/WHO expert committee in 1969 and is now used in more than 50 countries around the world. These properties of nisin determine that it can act as an inducer to induce the production of heterologous proteins in food-grade expression systems without any toxic effects on the human body and thus is a food-grade inducer. In addition to nisin, mutant derivatives of nisin and certain nisin analogs can also be used as inducers to induce the nisA promoter and can even be induced by the fermentation supernatant of nisin or its homologs or even nisin-producing bacteria. The using concentration of nisin is significantly lower than its minimum inhibitory concentration (MIC), ranging from 0.01 to 10 ng/mL. This concentration does not inhibit the growth of microorganisms, even if the host does not contain the NisI and NisFEG immune systems. In the process of inducing expression, the inducing agent needs to be added in the log phase, and the expression level can be controlled within a power range of 1000 times, and there is a linear dose-response relationship between the induced concentration and the protein production level in this range. The highest production rate arrives 2 h after induction, and the yield of the target protein can reach 60% of the soluble protein.
2.2.4 Food-Grade Expression System of LAB and Its Application
2.2.4.1 NICE System
Eleven genes related to nisin biosynthesis are clustered into a DNA fragment about 14 kb in the order of nisA/Z, nisB, nisT, nisC, nisI, nisP, nisR, nisK, nisF, nisE, and nisG, in which nisR and nisK are two components of the regulatory system and the promoter nisA and nisF can be induced by nisin. In 1995, the Dutch Dairy Research Institute (now NIZO Food Research Institute) invented the food-grade expression system NICE (nisin-controlled gene expression system) by studying the self-regulating biosynthesis mechanism of nisin. Based on the promoter nisA of the nisin biosynthesis gene cluster and the two-component regulatory system gene nisRK, this system regulates gene expression through the induction of nisin. A valid nisin-induced NICE system consists of three parts: (1) Gram-positive bacteria containing the nisRK gene as host bacteria; (2) nisin, nisin analogs, or nisin mutants as inducers; (3) nisA or plasmid for the nisF promoter.
-
(1)
The working mechanism of the NICE system
In the process of nisin autonomic regulation in NICE system, the histidine kinase NisK acts as a sensor for nisin, and the NisR protein acts as a regulator of transcription, which activates the transcription of the target gene. Once the extracellular nisin exists, nisin binds to the receptor NisK. Subsequently, NisK transfers the phosphate group to NisR by autophosphorylation, and the activated NisR induces the nisin operon at the nisA promoter, which induces a regulatory process shown in Fig. 2.1.
-
(2)
Host
Gram-positive bacteria, including Lactobacillus, Streptococcus, Enterococcus, Bacillus, Leuconostoc, and particularly any of the NisR and NisK proteins that can express a certain level in Lactococcus can be used as a host for the NICE system. So far, researchers have built two series of host cells: (1) L. lactis that can produce nisin, such as L. lactis FI5876 and NZ9700; they are all modified from wild bacteria by plasmid elimination or phage elimination. Other examples include that L. lactis NZ9800 is obtained by reducing the four bases of the nisA gene; FI7332 integrates the erythromycin resistance gene into the nisA gene of FI5876. (2) Host cells that cannot produce nisin. These host cells integrate the nisRK gene into the genome, such as the commonly used host strains L. lactis NZ900 and L. lactis NZ3900. For nisin-producing bacteria, nisin not only induces its own production but also induces the expression of its target gene. For non-nisin-producing bacteria, nisin only induces the expression of the target gene. In addition to integrating nisK and nisR into the genome, the researchers designed multiple plasmids such as pNZ9520 and pNZ9530 to apply the NICE system to other hosts based on the plasmid pAMβ1 for wide host applications.
-
(3)
Plasmid vector
The plasmid vector requires a promoter comprising nisA or nisF for nisin-induced expression, and, to date, more than 20 related plasmid vectors have been constructed for expression of Gram-positive bacteria, even E. coli. Commonly used plasmid vectors are divided into five categories: One type is a transcriptional fusion vector, such as the common pNZ8020. The vector comprises a promoter of nisA, a NisR binding site, and the ribosome binding site. The polyclonal site is located behind the promoter region, and the gene expression needs its own initiation codon. The second type is a translational fusion vector, such as the commonly used pNZ8048. After the Nco I restriction endonuclease is inserted into the promoter region, since it contains ATG as a starting site, the inserted target gene can achieve high transcription efficiency and ultimately guarantee the expression level of target protein. Studies have shown that the use of this type of fusion vector to express β-glucosidase, its expression activity is six times higher than that of the transcriptional fusion vector. In the third mode, the promoter of Nisin A and the target gene were ligated in vitro and then cloned into the target vector together. The vector was used to successfully express germicidin PA-1 and Escherichia coli V. The fourth mode is a two-plasmid NICE system consisting of two compatible plasmids: one plasmid carries the nisRK regulatory gene and the other carries the target gene under the control of the nisA promoter. Its inducibility is the same as in L. lactis, but the time to reach the highest protein production level is significantly longer than L. lactis. The fifth mode is a vector for secretion expression, such as pNZ8110, which uses a signal peptide secreting the protein Usp45 for secretion expression of the protein of interest.
-
(4)
Application of NICE system
The NICE system has the characteristics of high-efficiency expression and the value of practical application. It can be applied to the high-efficiency expression of important proteins in biotechnology, such as bacteria, viruses and eukaryotic antigens, cytokines, bacteriocins, and membrane proteins in various LAB. In the cellular metabolic pathway, it is also possible to precisely modulate the activity of key enzymes and control the production of metabolites of interest.
-
1.
Overexpression of protein
Using the NICE system, proteins from different sources can be overexpressed to help people study the properties of proteins or enzymes. At the same time, because it is derived from Gram-positive bacteria, there is no inclusion body when expressing proteins with close relationship. For example, de Ruyter and Kuipers (1996) used the NICE system to homologously express the pepN gene, and its protein expression reached 50% of that of soluble cells, and no inclusion bodies were produced.
-
2.
Expression of membrane protein
Under normal physiological conditions, the natural expression level of membrane proteins is relatively low, and a large number of its heterologous expression has become a restrictive condition for the study of function and structure. Combined with the difficulty of purification and crystallization of membrane proteins, heterologous expression of membrane proteins has become a bottleneck in membrane protein-related research due to its hydrophobic properties and toxicity to host cells. The NICE system has certain advantages for overexpressing membrane proteins: (1) the proteolytic ability of L. lactis is weak; (2) the nisA promoter is a strong and rigorous promoter, which can make the expression of membrane proteins under reasonable control; and (3) the expressed membrane protein can be dissolved by a surfactant. The membrane proteins of many prokaryotic cells have been expressed by the NICE system, such as ABC transporter, MFS transporter, peptide transporter, etc., and the expression level can reach 10% of membrane protein. Bernaudat and Frelet-Barrand (2011) expressed several eukaryotic membrane proteins derived from Arabidopsis thaliana in L.lactis, including polycopper oxidase and naphthoquinone oxidoreductase. Some scholars have tried to promote the expression of eukaryotic membrane proteins through systematic transformation, such as the addition of fusion proteins, but the related research is generally relatively difficult and lagging.
-
3.
Secretory protein expression and surface display system
Food-grade lactobacillus is used in the production of protein in industrial fermentation but is also a suitable candidate for the delivery of heterologous proteins in food or the digestive tract. Antigens, vaccines, drugs, and the like can be expressed in LAB by secreting protein expression and surface display systems. Enouf et al. (2001) expressed the recombinant immunogen protein NSP4 using the NICE system. The results showed that the NICE system can produce rNSP4 protein with antigen and immunogenic properties and has the same immunogenic properties as viral proteins. Bermudez-Humaran and Langella (2002) used this system to produce IL-12. Experiments show that the activity of IL-12 produced by the system is similar to that of commercial IL-12, which can be used in mass production and in vivo application of IL-12.
-
(1)
Other bacteriocin-induced food-grade expression systems
In addition to the NICE system, some other LAB have a similar mechanism for bacteriocin production. But unlike the NICE system, the main function of peptide secretion is not the bacteriocins but the expression of the whole operating unit stimulated by external hormones. Sorvig et al. found a bacterial production operon in L. sakei and used it to construct a series of expression vectors. β-Glucuronidase and aminopeptidase were used as expression genes, respectively, and the expression effects of different vectors were compared. It was found that using pSH71 vector and sakacin P promoter, and using sakacin P as the inducer, the expression level of proglucuronidase was ten times that of the wild-type strain. However, the expression level of aminopeptidase accounted for half of the intracellular soluble protein (Sorvig and Gronqvist 2003; Sorvig and Mathiesen 2005). Nguyen and Nguyen (2012a, b) induced the expression of β-galactosidase (LacZ) of L. bulgaricus DSM 20081 in L. plantarum using the plasmids pSIP403 and pSIP409 constructed above. By adding the corresponding inducing peptide IP-673, the expression level of LacZ reached 53,000 U/L, accounting for 63% of intracellular soluble protein. In the same year, the author used these systems to express the chitosanase of B. licheniformis in L. plantarum with an expression level of 5 mg recombinant protein per liter of medium. Therefore, the above system is very valuable for the expression of LAB, particularly the expression of Lactobacillus proteins.
-
(2)
Sugar-induced food-grade expression system
Through genome-wide sequencing of a large number of LAB, it can be known that the LAB genome contains a plurality of sugar metabolism-related gene clusters, which can ensure that LAB grow on a medium containing various sugars as a carbon source. In the sugar-induced expression system of lactobacillus, the sugar selective marker is mainly used. As a food-grade inducer, sugar can be used as a marker for non-antibiotic resistance. Since the lactose operon is well studied, most expression systems use lactose as a selective marker to induce gene expression. Many lactose-induced food-grade expression systems were constructed by lacA promoter. Eaton et al. (1993) introduced the reported gene luxAB behind the promoter lacA and expressed L.lactis as the host bacteria. When lactose was used as the carbon source, the activity of the promoter was increased by seven times. Platteeuw et al. (1996) constructed a vector containing the lac A promoter with the lacF gene as a selection marker and inserted a transcription terminator of pep N into the vector to increase the stability of the vector and cloned DNA. The β-glucuronidase gene (gusA) was cloned downstream of the lac A promoter and introduced into the plasmid-free L. lactis NZ3000 receptor. The strain could be grown in lactose-containing medium, and the gus A gene was highly expressed with lactose induction. The food-grade plasmids pLEB590 and pLEB600 with L casei as host bacteria and pSH71 replicator and repA gene as the skeleton, respectively, carrying P45 and PpepR promoter, and screening with lactose selection markers. The proline iminopeptidase gene pep I was successfully expressed with these two plasmids (Takala and Saris 2002). Payne and MacCormick (1996) first inserted the lactose operon into the chromosome of L. lactis MG5267 and then integrated the lysin gene derived from Listeria phage LM-4 into the lac promoter, and its expression was controlled by the lac promoter. And ultimately the expression of the LM-4 lysin gene is controlled by lactose.
In addition, there are carbon sources such as xylose sucrose as selection markers. Using D-xylose as a selective marker, the e-lactobacillus lactobacillus shuttle vector pLP3537 was cloned and expressed the xylose reductase gene cluster xyl on the chromosome of L. pentosaceus, which enabled the recombinant lactobacillus to have the ability to utilize D-xylose. The possibility of using xylose as a selective marker was confirmed. Lokman et al. (1991) linked the gene of chloramphenicol acetyltransferase to the downstream of xylose promoter and introduced it into Lactobacillus pentosaceus, which were cultured in the medium containing xylose and glucose, respectively. The results showed that the activity of chloramphenicol acetyltransferase was 60~80 times that of glucose culture under the induction of xylose. Xylose-inducible expression system (XIES) is a sugar-dependent expression system. By using xylose-inducible promoter PxylT, intracellular or secreted proteins can be produced. The first use of this expression system was the heterologous expression of the S. aureus nuclease gene (intracellular and extracellular expression using the signal peptide of the Usp45 protein) in L. lactis NCDO2118. When the system is induced with an appropriate inducer, such as xylose or glucose, a large amount of the target protein can be expressed. The system can be a good complement to the NICE system, as its induction is tightly regulated. At the same time, when the system is used for protein secretion expression, the secreted protein is not observed to decompose (de Azevedo and Karczewski 2012).
Leenhouts and Bolhuis (1998) constructed a food-grade expression system with sucrose as the selective marker by the integration of lactobacillus. The gene scrA/scrB encoding sucrose and hydrolase was cloned into lactococcal plasmid pWV01. By using the copy of this plasmid, lactobacillus expression vector pINT124 pINT125 with sucrose as the screening marker was successfully constructed. Mahmoud and Sameh (2011) used this expression vector to express the bacteriocin gene pctA in L. lactis.
-
(3)
Zinc-induced expression system
Llull and Poquet (2004) designed a new expression system PZNZitR-driven heterogeneous expression system based on the promoter of zit operator. The zit operon is primarily involved in zinc regulation. When zinc is absent, such as when the cell is starving, the operator’s transcriptional repressor leaves its repressor site, and RNA polymerase can bind to the PZN promoter to induce subsequent protein expression. By expressing two target proteins, it was shown that this expression system is highly inducible, but the amount of protein obtained is lower than that of the NICE system.
Recently, a new zinc-induced expression system, Zirex, can induce protein expression in L. lactis. The promoter is induced by zinc with a concentration below toxicity, and the SczA repressor regulates the activation of PczcD and leads to the induced expression of subsequent fluorescent proteins. In addition, the NICE system and the Zilex system are combined to produce different proteins at different times in one microorganism.
-
(4)
Environmentally induced food-grade expression system
When LAB express proteins in vitro, they can induce protein expression by adding specific inducers. However, in vivo, the relevant inducers cannot be supplemented to the microorganisms at the appropriate concentration to achieve the induced concentration. To compensate for this deficiency, researchers have successively studied the stress-inducible expression system (SICE) and P170, which induce the expression of specific proteins in the gut.
Benbouziane and Ribelles (2013) found a stress-inducible expression system in L. lactis and studied it. The system is controlled by the pGroESL promoter and expresses the target protein only under specific stress conditions such as high salt, high temperature, and low pH. In addition, the system has great potential for the expression and transport of vaccine proteins at specific sites in the body for the treatment of inflammatory bowel disease (IBD) and human papillomavirus.
Another major feature of LAB is the production of lactic acid during growth. Based on this feature, a pH-induced expression system can be constructed. Madsen et al. (1999) found that the promoter P170 regulated by pH value was induced in the culture environment with a pH value of 6.5~6.0 and the bacteria entering the stable period. Through genetic manipulation, the deletion of 72 bases at the front end of P170 mRNA does not affect the promoter regulation, but increases the pH induction effect of the promoter by 150 times and the gene expression level accordingly.
Temperature is also an important factor influencing protein expression. Nauta et al. (1997) designed a thermo-unstable Rro repressor variant Rrol2 using lac Z as a reporter gene to construct a temperature-controlled expression system. When the temperature was increased from 24 to 42 °C, the amount of β-glucosidase expression was increased by 500 times.
2.3 Gene Knockout System in Lactic Acid Bacteria
2.3.1 The Mechanisms and Characteristics of Gene Knockout Vector
A gene knockout is a genetic technique in which foreign gene is inserted into site-specific position, and its purpose is to make the target gene have a directed change. This technique conquered the defect of random mutation (blindness and contingency) and is an ideal method for genetically modified organisms. Compared with other organisms, the development of gene knockout in LAB is relatively low speed owing to thicker cell wall, low conversion rate, and less vectors. The gene knockout technique can be used to modulate the metabolic responses, decrease ferment cost, and enhance the production and purity of the aimed product. Moreover, it could be used to study the structure and function of genes. So far, the most common ways used in gene knockout depend on a traditional homologous recombination, and a few studies adopt means of site-specific recombination, homologous recombination involving single-stranded DNA substrate, and CRISPR/Cas system.
2.3.1.1 Gene Knockout Based on Homologous Recombination
The theoretical basis of classic gene knockout technique is based on homologous recombination. Homologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. Based on the above theory, the targeted gene deletion, insertion, and missense mutation could be reached via homologous recombination between a recombinant vector and host genome in the host cell.
Considering the difference in the number of homologous recombination, the gene knockout can be simply divided into two types: one-time homologous recombination and twice homologous recombination. Leenhouts and Bolhuis (1998) showed that the targeted gene of L. lactis was inactivated via insertion of foreign gene by the single-crossover integration system. In addition, prolyl dipeptidyl aminopeptidase gene was inactivated in the chromosomal of L. helveticus via recombination between the pepXP-derived repeat (Bhowmik and Steele 1993). Compared with one-time homologous recombination, two recombination reactions were observed in the latter, and it caused two different results, in which the targeted gene was completely deleted in one result and the gene was disrupted in other. Ferain et al. (1996) demonstrated that two lactate dehydrogenases played a major impact on peptidoglycan precursor synthesis in L. plantarum via two recombination reactions. L. citreum is an important lactic acid bacterium in fermented foods, but dextran production often causes undesired ropiness. To prevent this side effect, a dextransucrase knockout mutant was constructed by Jin and Li (2014), and it indicated that L. citreum dextransucrase not only synthesizes dextran for cell protection but also provides fructose as an important carbon source for cell growth.
2.3.2 Gene Knockout Based on Improved Homologous Recombination
2.3.2.1 Homologous Recombination with a Counterselectable Selection Marker
Homologous recombination can be used for gene disruption, but gene targeting is inefficient because of low homologous recombination frequency in the secondary reaction, which leads to some difficulty in screening the mutants. To solve the above problem, the use of a reasonable counterselectable selection marker is helpful in screening the correct mutant (Fig. 2.2). It means that the mutant containing this counterselectable selection marker can be died in an exact selection condition, and the mutant without this selection marker is able to survive in the same condition. The use of selection marker could reach the aim of rapid screening of exact mutant without introducing new gene.
The fourth intermediate in the pathway was identified to be orotate, which was also confirmed that it could not be utilized as a pyrimidine source by most L. lactic strains due to lack of the special transporter (Kilstrup, Hammer et al. 2005). Furthermore, the gene responsible for utilizing orotate as a sole pyrimidine source by an auxotrophic mutant has been already identified to be oroP, which also lead to its sensitivity to its toxic analog 5-fluoroorotate. Based on the above property, a new selection vector pCS1966 was constructed by Solem and Defoor (2008), considered as an efficient tool for special strain construction, which was proper for sequence-specific integration based on homologous recombination and also that in a bacteriophage attachment site.
In addition, another counterselectable selection marker gene that was always used in LAB is the upp gene that encodes uracil phosphoribosyltransferase results in resistance to 5-fluorouracil (Martinussen and Hammer 1994). The main drawbacks of using the upp is that this gene is found in almost every organism and that 5-fluorouracil may be toxic even in a upp mutant (Martinussen and Hammer 1995). Previous study showed that the upp gene deletion did not affect pyrimidine metabolism and biological characteristics. In 2009, a 3.0-kb pOR-based counterselectable integration vector, which beared a upp expression cassette, called pTRK935, was constructed by Goh et al., and this tool was identified to be very valuable to determine how L. acidophilus performed its probiotic function (Goh and Azcarate-Peril 2009). Furthermore, a temperature-sensitive replicon called pWV01 was also adopted by Song and Cui (2014) in order to construct two integration plasmids with chloramphenicol-resistant using upp gene as a counterselective marker for L. casei ATCC393 and L. lactis MG1363. Many results have showed that no significant difference existed between the wild-type and mutant lactic acid bacteria except for 5-FU resistance through determining the genetic stability, growth curve, carbon utilization, and also scanning electronic microscopy.
Although the upp gene was widely used in the screening of resistant strains, some disadvantages still exist in the gene knockout process. Because the upp gene widely exists in some bacteria and may participate in some important metabolic responses, the upp gene deletion in the host cell could cause some influence in the growth.
2.3.2.2 Homologous Recombination with a Temperature-Sensitive Type System
The replicon of a thermosensitive plasmid is a thermosensitive replicon, which can make the plasmid replicate at lower temperature and shut off at elevated temperature. This genetic system for homologous recombination allows the thermosensitive plasmid massive replication at lower temperature and increases the transformation frequency. Maguin et al. (1992) isolated a replication-thermosensitive mutant pVE6002 and used it for the construction of a thermosensitive plasmid. The result showed that its transposition frequencies were about 1%, which were higher than wild type. Biswas et al. (1993) constructed a thermosensitive broad host range rolling-circle plasmid, pG+ host5, which contains a pBR322 replicon for propagation in E. coli at 37 °C, and developed a protocol for gene replacement. Cultures were first maintained at 37 °C to select for a bacterial population enriched for plasmid integrants; activation of the integrated rolling-circle plasmid by a temperature shift to 28 °C resulted in efficient plasmid excision by homologous recombination and replacement of a chromosomal gene by the plasmid-carried modified copy. These results show that gene replacement can be obtained at an extremely high efficiency by making use of the thermosensitive rolling-circle nature of the delivery vector. Atiles et al. (2000) disrupted IlvE activity in L. lactis LM0230 via a thermosensitive plasmid, indicating that IlvE is the only enzyme capable of synthesis of Ile and Val from their biosynthetic precursors. To study the role of propanediol dehydrogenases pdh30 during glycerol metabolism, the gene disruption mutant L. brevis INIA ESI38::pORI28-pdh30 was constructed by Langa and Arques (2015) by site-specific integration of plasmid pORI28 and the temperature-sensitive plasmid pVE6007. HPLC analysis of the glycerol fermentation products showed an involvement of the pdh30 in the 3-hydroxypropionic acid (3-HP) biosynthesis. The temperature-sensitive plasmid is a valuable tool in the gene knockout system, and it could be used in some Gram-positive bacteria. However, this genetic system is unstable, and its high-temperature condition also limits its application in LAB.
2.3.3 Gene Knockout System Based on Site-Specific Recombination
It is known for us that site-specific recombination possesses significant conservative characteristics, and this recombination would occur in these DNA strands with segments possessing at least a certain degree of sequence homology. A recombinase enzyme and also the corresponding recombination sites are enough for some certain systems to carry out all the reactions; however, many accessory proteins and/or the corresponding sites would be necessary. Cre/loxp and TP901–1/att family are considered as the two most common site-specific systems according to the amino acid sequence homology and mechanistic relatedness.
2.3.3.1 cre/loxp Recombination System
cre/loxp recombination system just included a single enzyme, called cre recombinase, recombining a pair of short sequences, also known as lox sequence, in which the enzyme and the lox site are separated from bacteriophage P1. There are 4 subunits and 2 domains (C-terminal and N-terminal domains) among Cre protein with 343 amino acids in total. The structure of C-terminal domain is similar to that of integrase family protein in lambda phage (Sternberg and Hamilton 1981). LoxP is a special site on bacteriophage P1, which consisted 34 bp, including two sets of 13-bp symmetric sequences with asymmetric 8-bp sequence in the middle of the sequence, in which the symmetric 13-bp structure is palindromic and the middle 8-bp sequence is not. This special structure provided a certain direction for loxP. Generally, the loxP site existed in pairs when genetic manipulation was performed. The behavior of floxed sequence depended on the orientation of loxP sites, while it will be excised with loxP site in the same direction and it will be inverted with loxP sites in the opposite orientation. The Cre recombinase can not only recognize wild LoxP sequence but also recognize the mutant LoxP sequence in which a set of symmetric 13-bp sequences or asymmetric 8-bp sequences have slight changes, which further expand its application range. Lambert and Bongers (2007) constructed an effective mutagenesis vector that contains Cre/Lox cassette and disrupted bile salt hydrolase bsh gene and α-galactosidase melA gene in L. plantarum WCFS1. Remus and Kranenburg (2012) studied the effect of four clusters of genes on surface polysaccharide production by using the Cre/LoxP system.
The role of selection marker gene is easy to screen the exact mutants, but its existence could also cause some unnecessary changes in the expression of its upstream and downstream genes in the genome. Considering the characteristics of the Cre/LoxP system, it is always used to remove the market gene in the genome. Zhu and Zhao (2014) constructed the thymidylate synthase gene (thyA)-deficient strain derived from L. lactis NZ9000 using the Cre-loxP recombination system and used it as a food-grade selection marker for L. lactis in food and industry applications.
2.3.3.2 TP901-1/att Recombination System
TP901-1/att recombination system can stably replicate, and site-specific integrate their DNA into the host chromosome, which is constructed by integrating TP901-1, lactococcal temperate bacteriophage, into the chromosome of Lactococcus subspecies. The phage attachment site (attP) and the chromosomal attachment site (attB) can be integrated into new hybrid sites: attL and attR. Zhu and Zhao (2014) used the integration elements encoded by the temperate lactococcal bacteriophage TP901-1 to obtain chromosomal single-copy transcriptional fusions in L. lactis. A genetic tool special for repetitive, marker-free, and site-specific integration was constructed by Petersen and Martinussen (2013) in L. lactis. In this tool, a vector with nonreplicating plasmid, called pKV6, included a phage attP, useful to be integrated into a bacterial attB. i pKV6 would be integrated into the chromosome of the host with being flanked by attL and attR hybrid attachment sites in high frequency just when the corresponding vector was transformed into L. lactis with the ability to express the phage TP901-1. loxP recombinants would be selected based on the 5-fluoroorotic acid just when a plasmid responsible for expressing cre recombinase lacks the replicating ability in L. lactis. The gene involved in controlling xylose utilization was usefully adopted to be integrated into the chromosome of L. lactis strain MG 1363 in two steps in order to determine whether the constructed system would perform its function.
2.3.3.3 Linear Transformation Knockout System
Linear DNA, containing the mutated or deleted gene flanked by homologous regions of the chromosome, is transformed into recombination-proficient strains, which is one of the hot spots on study of gene knockout method. It’ s reported that a recombinant E. coli strain was constructed through replacing the RecBCD function with phageλ’s Red function via recombining the chromosome with short linear DNA fragments at a greatly elevated rate by Murphy (1998), with at least 70-fold higher than that by arecBC, sbcBC, or recD strain. The rate is at least 70-fold higher than that exhibited by arecBC sbcBC or recD strain. Compared with double crossover homologous recombination, the homologous arm of the above method is about 35~50 bp, and it has a high recombination efficiency. However, this method is always used in Gram-negative bacteria, and few studies were found in LAB.
Recently, single-stranded DNA recombination is always used in the gene knockout system in LAB. The single-stranded DNA binding proteins produced by redβ and redT gene are always used as recombinases that can promote annealing of complementary DNA strands. Van Pijkeren and Britton (2012) constructed the plasmid pJP042 and pJP005 to assess the ability of the L. reuteri RecT protein to support recombineering. It showed that the intrinsic vancomycin resistance of L. reuteri was significantly lower in the mutations, and the minimum inhibitory concentration of vancomycin was reduced from >256 to 1.5 mu g/mL by creating a single AA change in the d-Ala-d-Ala ligase enzyme. Compared with classic double crossover homologous recombination, ssDNA recombineering exhibits more advantages, such as easy operation, high recombination efficiency, not restricted by restriction enzyme cutting site, shorter homologous arms, and acquisition by direct PCR.
2.3.4 New Gene Knockout Method
With the development of molecular biology, some new gene knockout methods have attracted lots of attention, such as RNA interference, transcription activator-like effector nuclease technique, zinc finger nuclease gene targeting technique, CRISPR/Cas system, and so on. Among them, CRISPR/Cas system could be used in the gene knockout of lactic acid bacteria in the future. The exact mechanism of CRISPR/Cas system is that CRISPR-derived RNA form a compound of tracrRNA/crRNA with trans-activating RNA, which guides the Cas 9 nuclease cut the DNA target specified by the guide RNA. By designing the above two RNA, it can form a leading indicator short guide RNA that guide Cas nuclease cut the targeted DNA. As a RNA-oriented dsDNA binding protein, Cas9 nuclease is one of the first known unifying factors that collectively recognize RNA, DNA, and protein. The compounds of protein and Cas9 nuclease-null can specifically bind any DNA sequence together with moderate amount of sgRNA. The terminal of sgRNA can bind with the target DNA without affecting the combination between Cas9 and the target DNA. Therefore, Cas9 can deliver any compounds of protein and RNA to any DNA sequence (Liu and Xu 2015).
CRISPR-Cas9 selection system has been identified to be the most ideal system used to edit some genes in lactic acid bacterium for its high efficiency (Oh and van Pijkeren 2014), in which the efficiency in recovering subtle changes in the genome would range from 90% to 100%. CRISPR-Cas9 has been successfully used in recombineering, for example, the codon saturation mutagenesis in L. reuteri chromosome and also identification of such low-efficiency events as oligonucleotide-mediated chromosome deletion. CRISPR-Cas 9 would be also useful in identifying the recombinant bacterial cells with low recombineering efficiency and also eliminating the need for ssDNA recombineering optimization procedures.
2.3.5 The Common Vectors and Their Application
The gene knockout can be used to modulate the metabolic flux, interfere with the production of unnecessary secondary metabolism, reduce ferment cost, and raise the production and purity of target product. In addition, it can also be used to study the structure and function of genome of LAB and its beneficial mechanisms.
Increasing reports have focused on the peptidoglycan-degrading enzyme involved in a range of bacterial processes and also the interaction between the host and the microbe; however, the function of this enzyme in lactobacilli is still unknown. Systematic gene deletion system has been adopted by Rolain and Bernard (2012) in exploring the functional role of the peptidoglycan hydrolase (PGH) complement located in the genome of L. plantarum. The role of N-acetylglucosaminidase Acm2 and NplC/P60 D, L-endopeptidase LytA, as key determinants in the morphology of L. plantarum has been shown in that study. eps gene cluster has been reported in L. johnsonii requiring for the biosynthesis of homopolymeric exopolysaccharides (EPS)-1 and heteropolymeric EPS-2 which was useful in constructing a capsular layer. epsA is the first gene of the cluster with putative function as transcriptional regulator, and the function has been identified by the result that deletion of epsA gene would result in complete loss of the ability of growing the EPS-1 and EPS-2 on the cell surface, and this ability would be fully restored just when this gene was complemented. All these results showed that epsA gene could regulate the EPS production positively. Reutericyclin is a unique antimicrobial tetramic acid produced by some strains of L. reuteri, but its synthesized mechanism is still unclear. Lin and Lohans (2015) showed that deletions of rtcNRPS or rtcPhlABC in L. reuteri TMW1.656 abrogated reutericyclin production but did not affect reutericyclin resistance. To raise the production of diacetyl in LAB, Platteeuw and Hugenholtz (1995) cloned the als gene for alpha-acetolactate synthase of L. lactis MG1363 in a multicopy plasmid under the control of the inducible L. lactis lacA promoter. Furthermore, the effect of alpha-acetolactate synthase overproduction on the formation of end products in various L. lactis strains was also determined under different fermentation conditions. These metabolic engineering studies suggest that more than 80% of the lactose can be converted via the activity of the overproduced alpha-acetolactate synthase in L. lactis.
Change history
06 October 2022
The original version of the book was inadvertently published with errors. The following corrections have been made after the original publication.
References
Adel-Patient K, Ah-Leung S, Creminon C et al (2005) Oral administration of recombinant Lactococcus lactis expressing bovine β-lactoglobulin partially prevents mice from sensitization. Clin Exp Allergy 35(4):539–546
Ai CQ, Zhang QX et al (2015) Protective effect of Streptococcus thermophilus CCFM218 against house dust mite allergy in a mouse model. Food Control 50:283–290
Allison GE, Klaenhammer TR (1996) Functional analysis of the gene encoding immunity to lactacin F, laf I, and its use as a Lactobacillus-specific food-grade genetic marker. Appl Environ Microbiol 62:4450–4460
Altermann EWM, Russell MA, Azcarate-Peril R et al (2005) Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci U S A 102(11):3906–3912
Atiles MW, Dudley EG, Steele JL (2000) Gene cloning, sequencing, and inactivation of the branched-chain aminotransferase of Lactococcus lactis LM0230. Appl Environ Microbiol 66(6):2325–2329
Bahey-El-Din M (2012) Lactococcus lactis-based vaccines from laboratory bench to human use: an overview. Vaccine 30(4):685–690
Benbouziane B, Ribelles P (2013) Development of a stress-inducible controlled expression SICE system in Lactococcus lactis for the production and delivery of therapeutic molecules at mucosal surfaces. J Biotechnol 1682:120–129
Beninati C, Oggioni MR, Boccanera M et al (2000) Therapy of mucosal candidiasis by expression of an anti-idiotype in human commensal bacteria. Nat Biotechnol 18(10):1060–1064
Bermudez-Humaran LG, Langella P (2002) Production of human papillomavirus type 16 E7 protein in Lactococcus lactis. Appl Environ Microbiol 682:917–922
Bermudez-Humaran LG, Cortes-Perez NG, Le Loir Y et al (2003a) Fusion to a carrier protein and a synthetic propeptide enhances E7 HPV-16 production and secretion in Lactococcus lactis. Biotechnol Prog 19(3):1101–1104
Bermudez-Humaran LG, Langella P, Commissaire J et al (2003b) Controlled intra- or extracellular production of staphylococcal nuclease and ovine omega interferon in Lactococcus lactis. FEMS Microbiol Lett 224(2):307–313
Bermúdez-Humarán LG, Kharrat P, Chatel JM et al (2011) Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb Cell Factories 10(1):1–10
Bernaudat F, Frelet-Barrand A (2011) Heterologous expression of membrane proteins: choosing the appropriate host. PLoS One 612:e29191
Bhowmik T, Steele JL (1993) Development of an electroporation procedure for gene disruption in Lactobacillus helveticus CNRZ 32. J Gen Microbiol 139(7):1433–1439
Biswas I, Gruss A, Ehrlich SD et al (1993) High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol 175(11):3628–3635
Boels IC, Van Kranenburg R, Kanning MW et al (2003) Increased exopoly- saccharide production in Lactococcus lactis due to increased levels of expression of the NIZO B40 eps gene cluster. Appl Environ Microbiol 69(8):5029–5031
Bohle B (2006) T-cell epitopes of food allergens. Clin Rev Allergy Immunol 30(2):97–108
Bolotin A, Wincker P, Mauger S (2001) The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11(5):731–753
Braat H, Rottiers P, Hommes DW et al (2006) A Phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol 4(6):754–759
Bron PA, Benchimol MG (2002) Use of the alr gene as a food-grade selection marker in lactic acid bacteria. Appl Environ Microbiol 6811:5663–5670
Carroll IM, Andrus JM, Bruno-Bárcena JM et al (2007) Anti-inflammatory properties of Lactobacillus gasseri expressing manganese superoxide dismutase using the interleukin 10-deficient mouse model of colitis. Am J Physiol Gastrointest Liver Physiol 293(4):G729–G738
Charng YC, Lin CC, Hsu CH (2006) Inhibition of allergen-induced airway inflammation and hyperreactivity by recombinant lactic-acid bacteria. Vaccine 24(33–34):5931–5936
Chassy BM, Flickinger JL (1987) Transformation of Lactobacillus casei by electroporation. FEMS Microbiol Lett 44(2):173–177
Chassy BM, Gibson EV, Giuffrida AL (1976) Evidence for extrachromosomal elements in Lactobacillus. J Bacteriol 127(3):1576–1578
Cheun HI, Kawamoto K, Hiramatsu M et al (2004) Protective immunity of SpaA-antigen producing Lactococcus lactis against Erysipelothrix rhusiopathiae infection. J Appl Microbiol 96(6):1347–1353
Cortes-Perez NG, Ah-Leung S (2007) Intranasal coadministration of live lactococci producing interleukin-12 and a major cow’s milk allergen inhibits allergic reaction in mice. Clin Vaccine Immunol 143:226–233
Cortes-Perez NG, Lefèvre F, Corthier G et al (2007) Influence of the route of immunization and the nature of the bacterial vector on immunogenicity of mucosal vaccines based on lactic acid bacteria. Vaccine 25(36):6581–6588
Corthesy B, Boris S, Isler P et al (2005) Oral immunization of mice with lactic acid bacteria producing Helicobacter pylori Urease B subunit partially protects against challenge with Helicobacter felis. J Infect Dis 192(8):1441–1449
de Azevedo M, Karczewski J (2012) In vitro and in vivo characterization of DNA delivery using recombinant Lactococcus lactis expressing a mutated form of Listeria monocytogenes Internalin A. BMC Microbiol 12:299
de Ruyter PG, Kuipers OP (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 6210:3662–3667
Drouault S, Juste C, Marteau P et al (2002) Oral treatment with Lactococcus lactis expressing Staphylococcus hyicus lipase enhances lipid digestion in pigs with induced pancreatic insufficiency. Appl Environ Microbiol 68(6):3166–3168
Eaton TJ, Shearman CA, Gasson MJ (1993) The use of bacterial luciferase genes as reporter genes in Lactococcus: regulation of the Lactococcus lactis subsp. lactis lactose genes. J Gen Microbiol 139:1495–1501
Emond E, Lavallée R, Drolet G, Moineau S, LaPointe G (2001) Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl Environ Microbiol 67(4):1700–1709
Enouf V, Langella P, Commissaire J et al (2001) Bovine rotavirus nonstructural protein 4 produced by Lactococcus lactis is antigenic and immunogenic. Appl Environ Microbiol 67(4):1423–1428
Ferain T, Hobbs JN Jr et al (1996) Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J Bacteriol 178(18):5431–5437
Foligne B, Dessein R, Marceau M et al (2007) Prevention and treatment of colitis with Lactococcus lactis secreting the immunomodulatory Yersinia LcrV protein. Gastroenterology 133(3):862–874
Fortina MG, Parini C, Rossi P et al (1993) Mapping of three plasmids from Lactobacillus helveticus ATCC 15009. Lett Appl Microbiol 17(6):303–306
Frossard CP, Steidler L, Eigenmann PA (2007) Oral administration of an IL-10–secreting Lactococcus lactis strain prevents food-induced IgE sensitization. J Allergy Clin Immunol 119(4):952–959
García-Mantrana I, Yebra MJ, Haros M et al (2016) Expression of bifidobacterial phytases in Lactobacillus casei and their application in a food model of whole-grain sourdough bread. Int J Food Microbiol 216:18
Goh YJ, Azcarate-Peril MA (2009) Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl Environ Microbiol 7510:3093–3105
Grangette C, Muller-Alouf H (2001) Mucosal immune responses and protection against tetanus toxin after intranasal immunization with recombinant Lactobacillus plantarum. Infect Immun 693:1547–1553
Hanniffy SB, Carter AT, Hitchin E et al (2007) Mucosal delivery of a pneumococcal vaccine using Lactococcus lactis affords protection against respiratory infection. J Infect Dis 195(2):185–193
Ho PS, Kwang J, Lee YK (2005) Intragastric administration of Lactobacillus casei expressing transmissible gastroentritis coronavirus spike glycoprotein induced specific antibody production. Vaccine 23(11):1335–1342
Hols P, Kleerebezem M (1999) Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat Biotechnol 176:588–592
Hongying F, Xianbo W, Fang Y et al (2014) Oral Immunization with recombinant Lactobacillus acidophilus expressing the adhesin Hp0410 of Helicobacter pylori induces mucosal and systemic immune responses. Clin Vaccine Immunol 21(2):126–132
Hu S, Kong J, Kong W, Guo T et al (2010) Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl Environ Microbiol 76(8):2410–2418
Hugenholtz J, Kleerebezem M (2000) Lactococcus lactis as a cell factory for high-level diacetyl production. Appl Environ Microbiol 669:4112–4114
Hughes BF, Mc Kay LL (1992) Deriving phage-insensitive Lactococci using a food-grade vector encoding phage and nisin resistance. J Dairy Sci 75:914–923
Huibregtse IL, Snoeck V, de Creus A et al (2007) Induction of ovalbumin-specific tolerance by oral administration of Lactococcus lactis secreting ovalbumin. Gastroenterology 133(2):517–528
Jimenez JJ, Diep DB, Borrero J et al (2015) Cloning strategies for heterologous expression of the bacteriocin enterocin A by Lactobacillus sakei Lb790, Lb. plantarum NC8 and Lb. casei CECT475. Microb Cell Fact 14:166
Jin Q, Li L (2014) Construction of a dextran-free Leuconostoc citreum mutant by targeted disruption of the dextransucrase gene. J Appl Microbiol 1174:1104–1112
Kahala M, Palva A (1999) The expression signals of the Lactobacillus brevis slpA gene direct efficient heterologous protein production in lactic acid bacteria. Appl Microbiol Biotechnol 51(1):71–78
Kajava AV, Zolov SN, Kalinin AE et al (2000) The net charge of the first 18 residues of the mature sequence affects protein translocation across the cytoplasmic membrane of gram-negative bacteria. J Bacteriol 182(8):2163–2169
Kandaswamy K, Liew TH, Wang CY et al (2013) Focal targeting by human beta-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci U S A 110(50):20230–20235
Kanpiengjai A, Lumyong S, Wongputtisin P et al (2015) Efficient secretory expression of gene encoding a broad pH-stable maltose-forming amylase from Lactobacillus plantarum S21 in food-grade lactobacilli host. J Korean Soc Appl Biol Chem 58(6):901–908
Kleerebezem M, Beerthuyzen MM, Vaughan EE et al (1997) Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol 63(11):4581–4584
Kuczkowska K, Mathiesen G, Eijsink VG et al (2015) Lactobacillus plantarum displaying CCL3 chemokine in fusion with HIV-1 Gag derived antigen causes increased recruitment of T cells. Microb Cell Factories 14(1):1
Lambert JM, Bongers RS (2007) Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol 734:1126–1135
Langa S, Arqués JL (2015) Glycerol and cobalamin metabolism in lactobacilli: relevance of the propanediol dehydrogenase pdh30. Eur Food Res Technol 2412:173–184
Le Loir Y, Nouaille S, Commissaire J et al (2001) Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl Environ Microbiol 67(9):4119–4127
Le Loir Y, Azevedo V, Oliveira SC et al (2005) Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb Cell Factories 4(1):2
LeBlanc JG, del Carmen S, Miyoshi A et al (2011) Use of superoxide dismutase and catalase producing lactic acid bacteria in TNBS induced Crohn’s disease in mice. J Biotechnol 151(3):287–293
Lee MH, Roussel Y, Wilks M et al (2001) Expression of Helicobacter pylori urease subunit B gene in Lactococcus lactis MG1363 and its use as a vaccine delivery system against H-pylori infection in mice. Vaccine 19(28–29):3927–3935
Lee JS, Poo H, Han DP et al (2005) Mucosal immunization with surface-displayed severe acute respiratory syndrome coronavirus spike protein on Lactobacillus casei induces neutralizing antibodies in mice. J Virol 80(8):4079–4087
Lee P, Abdul-Wahid A, Faubert GM (2009) Comparison of the local immune response against Giardia lamblia cyst wall protein 2 induced by recombinant Lactococcus lactis and Streptococcus gordonii. Microbes Infect 11(1):20–28
Leenhouts K, Bolhuis A (1998) Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Appl Microbiol Biotechnol 494:417–423
Lei H, Sheng Z, Ding Q et al (2011) Evaluation of oral immunization with recombinant avian influenza virus HA1 displayed on the Lactococcus lactis surface and combined with the mucosal adjuvant cholera toxin subunit B. Clin Vaccine Immunol 18(7):1046–1051
Levander F, Svensson M (2002) Enhanced exopolysaccharide production by metabolic engineering of Streptococcus thermophilus. Appl Environ Microbiol 682:784–790
Limaye SA, Haddad RI, Cilli F et al (2013) Phase 1b, multicenter, single blinded, placebo-controlled, sequential dose escalation study to assess the safety and tolerability of topically applied AG013 in subjects with locally advanced head and neck cancer receiving induction chemotherapy. Cancer 119(24):4268–4276
Lin XB, Lohans CT (2015) Genetic determinants of reutericyclin biosynthesis in Lactobacillus reuteri. Appl Environ Microbiol 816:2032–2041
Liu B, Xu H (2015) CRISPR/Cas: a faster and more efficient gene editing system. J Nanosci Nanotechnol 153:1946–1959
Liu CQ, Leelawatcharamas V, Harvey ML et al (1996) Cloning vectors for lactococcus based on a plasmid encoding resistance to cadmium. Curr Microbiol 33:35–39
Liu DQ, Qiao XY, Ge JW et al (2011) Construction and characterization of Lactobacillus pentosus expressing the D antigenic site of the spike protein of transmissible gastroenteritis virus. Can J Microbiol 57(5):392–397
Llull D, Poquet I (2004) New expression system tightly controlled by zinc availability in Lactococcus lactis. Appl Environ Microbiol 70(9):5398–5406
Lokman BC, Santen PV, Verdoes JC et al (1991) Organization and characterization of three genes involved in d -xylose catabolism in Lactobacillus pentosus. Mol Gen Genomics 230(1):161–169
Maassen CB, Laman JD, den Bak-Glashouwer MJ et al (1999) Instruments for oral disease-intervention strategies: recombinant Lactobacillus casei expressing tetanus toxin fragment C for vaccination or myelin proteins for oral tolerance induction in multiple sclerosis. Vaccine 17(17):2117–2128
Madsen SM, Arnau J, Vrang A et al (1999) Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis. Mol Microbiol 32(1):75–87
Maguin E, Duwat P, Hege T et al (1992) New thermosensitive plasmid for gram-positive bacteria. J Bacteriol 174(17):5633–5638
Mahmoud KT, Sameh EM (2011) Heterologous expression of pctA gene expressing propionicin T1 by some lactic acid bacterial strains using pINT125. Alexandria University, Netherlands
Mannam P, Jones KF, Geller BL (2004) Mucosal vaccine made from live, recombinant Lactococcus lactis protects mice against pharyngeal infection with Streptococcus pyogenes. Infect Immun 72(6):3444–3450
McKay LL, Baldwin KA (1990) Applications for biotechnology: present and future improvements in lactic acid bacteria. FEMS Microbiol Rev 7(1–2):3–14
Meazza R, Gaggero A, Neglia F et al (1997) Expression of two interleukin-15 mRNA isoforms in human tumors does not correlate with secretion: role of different signal peptides. Eur J Immunol 27(5):1049–1054
Meijerink M, Wells JM, Taverne N et al (2012) Immunomodulatory effects of potential probiotics in a mouse peanut sensitization model. FEMS Immunol Med Microbiol 65(3):488–496
Murphy KC (1998) Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol 180(8):2063–2071
Narita J, Okano K, Kitao T et al (2006) Display of alpha-amylase on the surface of Lactobacillus casei cells by use of the PgsA anchor protein, and production of lactic acid from starch. Appl Environ Microbiol 72(1):269–275
Nauta A, van den Burg B, Karsens H et al (1997) Design of thermolabile bacteriophage repressor mutants by comparative molecular modeling. Nat Biotechnol 15(10):980–983
Nguyen HA, Nguyen TH (2012a) Chitinase from Bacillus licheniformis DSM13: expression in Lactobacillus plantarum WCFS1 and biochemical characterisation. Protein Expr Purif 812:166–174
Nguyen TT, Nguyen HA (2012b) Homodimeric beta-galactosidase from Lactobacillus delbrueckii subsp. bulgaricus DSM 20081: expression in Lactobacillus plantarum and biochemical characterization. J Agric Food Chem 607:1713–1721
Nielsen H, Engelbrecht J, Brunak S et al (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10(1):1–6
Norton PM, Wells JM, Brown HW et al (1997) Protection against tetanus toxin in mice nasally immunized with recombinant Lactococcus lactis expressing tetanus toxin fragment C. Vaccine 15(6–7):616–619
Oh JH, van Pijkeren JP (2014) CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 4217:e131
Oliveira MLS, Areas APM, Campos IB et al (2006) Induction of systemic and mucosal immune response and decrease in Streptococcus pneumoniae colonization by nasal inoculation of mice with recombinant lactic acid bacteria expressing pneumococcal surface antigen A. Microbes Infect 8(4):1016–1024
Payne J, MacCormick CA (1996) Exploitation of a chromosomally integrated lactose operon for controlled gene expression in Lactococcus lactis. FEMS Microbiol Lett 1361:19–24
Perez CA, Eichwald C, Burrone O et al (2005) Rotavirus vp7 antigen produced by Lactococcus lactis induces neutralizing antibodies in mice. J Appl Microbiol 99(5):1158–1164
Petersen KV, Martinussen J (2013) Repetitive, marker-free, site-specific integration as a novel tool for multiple chromosomal integration of DNA. Appl Environ Microbiol 7912:3563–3569
Platteeuw C, Hugenholtz J (1995) Metabolic engineering of Lactococcus lactis: influence of the overproduction of alpha-acetolactate synthase in strains deficient in lactate dehydrogenase as a function of culture conditions. Appl Environ Microbiol 6111:3967–3971
Platteeuw C, van Alen-Boerrigter I et al (1996) Food-grade cloning and expression system for Lactococcus lactis. Appl Environ Microbiol 623:1008–1013
Poo H, Pyo HM, Lee TY et al (2006) Oral administration of human papillomavirus type 16 E7 displayed on Lactobacillus casei induces E7-specific antitumor effects in C57/BL6 mice. Int J Cancer 119(7):1702–1709
Posno M, Heuvelmans PT (1991) Complementation of the inability of lactobacillus strains to utilize D-xylose with D-xylose catabolism-encoding genes of Lactobacillus pentosus. Appl Environ Microbiol 579:2764–2766
Ramasamy R, Yasawardena S, Zomer A et al (2006) Immunogenicity of a malaria parasite antigen displayed by Lactococcus lactis in oral immunisations. Vaccine 24(18):3900–3908
Ravnikar M, Štrukelj B, Obermajer N et al (2010) Engineered lactic acid bacterium Lactococcus lactis capable of binding antibodies and tumor necrosis factor alpha. Appl Environ Microbiol 76(20):6928–6932
Remus DM, Kranenburg R (2012) Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb Cell Factories 111:149
Ribeiro LA, Azevedo V, Le Loir Y et al (2002) Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: a first step towards food-grade live vaccines against brucellosis. Appl Environ Microbiol 68(2):910–916
Rigaux P, Daniel C, Isbergues M et al (2009) Immunomodulatory properties of Lactobacillus plantarum and its use as a recombinant vaccine against mite allergy. Allergy 64(3):406–414
Robinson K, Chamberlain LM, Schofield KM et al (1997) Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat Biotechnol 15(7):653–657
Robinson K, Chamberlain LM, Lopez MC et al (2004) Mucosal and cellular immune responses elicited by recombinant Lactococcus lactis strains expressing tetanus toxin fragment C. Infect Immun 72(5):2753–2761
Rochat T, Bermúdez-Humarán L, Gratadoux JJ et al (2007) Anti-inflammatory effects of Lactobacillus casei BL23 producing or not a manganese-dependant catalase on DSS-induced colitis in mice. Microb Cell Factories 6(1):1–10
Rolain T, Bernard E (2012) Identification of key peptidoglycan hydrolases for morphogenesis, autolysis, and peptidoglycan composition of Lactobacillus plantarum WCFS1. Microb Cell Factories 111:137
Ross P, O’Gara F (1990) Thymidylate synthase gene from Lactococcus lactis as a genetic marker: an alternative to antibiotic resistance genes. Appl Environ Microbiol 567:2164–2169
Scheppler L, Vogel M, Zuercher AW et al (2002) Recombinant Lactobacillus johnsonii as a mucosal vaccine delivery vehicle. Vaccine 20(23–24):2913–2920
Scheppler L, Vogel M, Marti P et al (2005) Intranasal immunisation using recombinant Lactobacillus johnsonii as a new strategy to prevent allergic disease. Vaccine 23(9):1126–1134
Schwarzer M, Repa A, Daniel C et al (2011) Neonatal colonization of mice with Lactobacillus plantarum producing the aeroallergen Bet v 1 biases towards Th1 and T-regulatory responses upon systemic sensitization. Allergy 66(3):368–375
Sibakov M, Koivula T, von Wright A, Palva I (1991) Secretion of TEM beta-lactamase with signal sequence isolated from the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol 57(2):341–348
Smiley MB, Fryder V (1978) Plasmids, lactic acid production, and N-acetyl-D-glucosamine fermentation in Lactobacillus helveticus subsp. Appl Environ Microbiol 35(4):777–781
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228(4705):1315–1317
Solem C, Defoor E (2008) Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl Environ Microbiol 7415:4772–4775
Song L, Cui H (2014) Construction of upp deletion mutant strains of Lactobacillus casei and Lactococcus lactis based on counterselective system using temperature-sensitive plasmid. J Microbiol Methods 102:37–44
Sorensen KI, Larsen R (2000) A food-grade cloning system for industrial strains of Lactococcus lactis. Appl Environ Microbiol 664:1253–1258
Sorvig E, Gronqvist S (2003) Construction of vectors for inducible gene expression in Lactobacillus sakei and L plantarum. FEMS Microbiol Lett 2291:119–126
Sorvig E, Mathiesen G (2005) High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 151(Pt 7):2439–2449
Steidler L, Hans W (2000) Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2895483:1352–1355
Svensson M, Waak E, Svensson U et al (2005) Metabolically improved exopoly- saccharide production by Streptococcus thermophilus and its influence on the rheological properties of fermented milk. Appl Environ Microbiol 71(10):6398–6400
Takala TM, Saris PE (2002) A food-grade cloning vector for lactic acid bacteria based on the nisin immunity gene nisI. Appl Microbiol Biotechnol 594–5:467–471
van Asseldonk M, Rutten G, Oteman M et al (1990) Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95(1):155–160
van Pijkeren JP, Britton RA (2012) High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 4010:e76
Vandenbroucke K, Hans W, Van Huysse J et al (2004) Active delivery of trefoil factors by genetically modified Lactococcus lactis prevents and heals acute colitis in mice. Gastroenterology 127(2):502–513
Vandenbroucke K, de Haard H, Beirnaert E et al (2009) Orally administered L. lactis secreting an anti-TNF nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3(1):49–56
Vescovo M, Bottazzi V, Sarra PG et al (1981) Evidence of plasmid deoxyribonucleic acid in Lactobacillus. Microbiologica (Bologna) 4(4):413–419
von Wright A, Wessels S, Tynkkynen S et al (1990) Isolation of a replication region of a large lactococcal plasmid and use in cloning of a nisin resistance determinant. Appl Environ Microbiol 56:2029–2035
Watterlot L, Rochat T, Sokol H et al (2010) Intragastric administration of a superoxide dismutase-producing recombinant Lactobacillus casei BL23 strain attenuates DSS colitis in mice. Int J Food Microbiol 144(1):35–41
Wei CH, Liu JK, Hou XL et al (2010) Immunogenicity and protective efficacy of orally or intranasally administered recombinant Lactobacillus casei expressing ETEC K99. Vaccine 28(24):4113–4118
Wells JM, Wilson PW, Norton PM et al (1993) Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol 8(6):1155–1162
Wong WY, Su P, Allison GE et al (2003) A potential food-grade cloning vector for Streptococcus thermophilus that uses cadmium resistance as the selectable marker. Appl Environ Microbiol 69(69):5767–5771
Wu CM, Chung TC (2007) Mice protected by oral immunization with Lactobacillus reuteri secreting fusion protein of Escherichia coli enterotoxin subunit protein. FEMS Immunol Med Microbiol 50(3):354–365
Xin KQ, Hoshino Y, Toda Y et al (2003) Immunogenicity and protective efficacy of orally administered recombinant Lactococcus lactis expressing surface-bound HIV Env. Blood 102(1):223–228
Yam KK, Hugentobler F, Pouliot P et al (2011) Generation and evaluation of A2-expressing Lactococcus lactis live vaccines against Leishmania donovani in BALB/c mice. J Med Microbiol 60(9):1248–1260
Yao LY, Man CX, Zhao F et al (2010) Expression of bovine trypsin in Lactococcus lactis. Int Dairy J 20(11):806–809
Ye W, Huo G, Chen J et al (2010) Heterologous expression of the Bacillus subtilis (natto) alanine dehydrogenase in Escherichia coli and Lactococcus lactis. Microbiol Res 165(4):268–275
Yoon SW, Lee CH, Kim JY et al (2008) Lactobacillus casei secreting alpha-MSH induces the therapeutic effect on DSS-induced acute colitis in Balb/c Mice. J Microbiol Biotechnol 18(12):1975–1983
Zhang ZH, Jiang PH, Li NJ et al (2005) Oral vaccination of mice against rodent malaria with recombinant Lactococcus lactis expressing MSP-119. World J Gastroenterol 11(44):6975–6980
Zhang Q, Zhong J, Liang X et al (2010) Improvement of human interferon alpha secretion by Lactococcus lactis. Biotechnol Lett 32(9):1271–1277
Zhang Q, Zhong J, Huan L (2011) Expression of hepatitis B virus surface antigen determinants in Lactococcus lactis for oral vaccination. Microbiol Res 166(2):111–120
Zhu D, Zhao K (2014) Construction of thyA deficient Lactococcus lactis using the cre-loxp recombination system. Ann Microbiol 653:1–7
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd. and Science Press
About this chapter
Cite this chapter
Chen, H., Chen, C., Ai, C., Ren, C., Gao, H. (2019). Genetic Operation System of Lactic Acid Bacteria and Its Applications. In: Chen, W. (eds) Lactic Acid Bacteria. Springer, Singapore. https://doi.org/10.1007/978-981-13-7832-4_2
Download citation
DOI: https://doi.org/10.1007/978-981-13-7832-4_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-7831-7
Online ISBN: 978-981-13-7832-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)