Abstract
In the last decades, many studies were addressed to focus the interplay between plant and microbial community into the soil and especially in the small soil zone in contact to plant root, called rhizosphere, which can be considered as a hotspot for interactions and therefore is a major target for improving nutrient use efficiency in crops. In this regard, unraveling the microbial activities that can be used to improve nutrient use efficiency may be the major challenge considering a sustainable agricultural contest. However, although using different approaches (metabolomics and transcriptomic) it has made it possible to characterize many interaction mechanisms, more remains largely unknown. Here, we summarize and discuss the abiotic and biotic factors that may manage plant-microbe interactions in the rhizosphere as well as in those parts of the soil furthest from the root, focusing on root architecture and nitrate as well.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
To meet the food needs of the population, an agricultural model based on the overuse of fertilizers has been adopted in recent decades. This model has created severe problems for both environmental quality and human health (Good et al. 2004). In addition, this approach has involuntarily led to the selection of genotypes with a low nutrient use efficiency. Given forecasts that the world population is increasing, around 9.2 billion on 2050 (FAO 2009), more crop production is needed using alternative strategies reducing the input of chemicals and improving food quality without negatively affecting the environment (Xu et al. 2012).
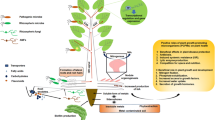
The rhizosphere is often defined as the area of soil around a root where the population of microorganisms depends on inputs from the plant. The rhizosphere is a soil microenvironment where plants and microorganisms can coexist in positive, negative, or neutral interactions (Lynch and Whipps 1990; Kardol et al. 2007) (Fig. 1). It is populated with numerous organisms, including fungi, bacteria, etc. and possibly exhibiting one of the highest levels of biological biodiversity of any environment in the world (Bender et al. 2016) (Fig. 1). Given the strong interaction between plants and microbes in the rhizosphere, it can be considered an extension of the plant’s genome (Berendsen et al. 2012; Verma et al. 2015). As crop yield is strongly dependent on water and nutrient uptake from the soil, the rhizosphere can be considered as a hotspot for interactions and therefore is a major target for improving nutrient use efficiency in crops.
Representative overview of different plant-rhizosphere microbe interactions and their activities (The figure has been adapted from Quiza et al. 2015 and modified)
Rhizosphere microbes may improve plant growth not only by making nutrients more available for uptake by the root (e.g., phosphate-solubilizing bacteria) but also through the production of phytohormones that can improve plant resistance to biotic and abiotic stresses (Berendsen et al. 2012). Plant roots secrete exudates derived from photosynthesis providing an important carbon supply for the growth of microorganisms (Brimecombe et al. 2007).
Therefore, engineering the rhizosphere may be a useful target for developing more sustainable agriculture (Zhang et al. 2017). Despite the importance of the rhizosphere and the innumerable publications in this field by using new technologies related to DNA sequencing and metabolomics, to date much remains to be discovered. More information is needed to understand the mechanisms involved in rhizosphere interactions and particularly how plants control their microbiome and vice versa how microorganisms influence crops.
The present review focuses on the recent knowledge of the rhizosphere, emphasizing particularly microbe and root interactions in relation to nitrogen supply.
Finally, this chapter reports some examples of how these interactions can be used to improve crops in a background scenario of more sustainable agriculture in the context of climate change.
2 Rhizosphere as an Active Network
The “rhizosphere” term was first conceived by Hiltner (1904) to delineate the plant root-soil interface. The rhizosphere was defined as the soil microenvironment where chemical, physical, and biological properties are affected by plant roots and their metabolic activities. Thus, the rhizosphere is one of the most dynamic networks of the terrestrial ecosystem, where direct plant-soil-microbe interaction takes place. Plant roots release up to 40% of photosynthetically fixed carbon directly into soil (Bais et al. 2006) mainly in the form of high and low molecular weight compounds (Newman 1985; McNear 2013). These compounds especially low molecular weight molecules such as amino and organic acids, phenolic compounds, sugar, etc. can be used by rhizosphere microbes as energy sources, which cause enrichment of the microbial biomass and activity (Doornbos et al. 2012). Moreover, exudation significantly influences soil physical and chemical properties (Nardi et al. 2000) such as soil pH (Javed et al. 2012), soil aggregation and erosion prevention (Naveed et al. 2017; Wang et al. 2017; Meena and Meena 2017), and water holding capacity (Young 1995). On the other side, root exudation helps the plants to gain essential nutrients from the soil through acidification and oxidation/reduction processes in the rhizosphere (McNear 2013). Nitrogen (N) and phosphorus (P) are key plant growth-limiting nutrients, and rhizosphere microbes play a key role through degradation and mineralization processes to deliver these essential nutrients to the plant. Much of the soil N and P are stored in complex organic forms that plants cannot easily access often requiring breakdown and solubilization by microbes before uptake by roots can occur. Thus, the rhizosphere is the apropos niche for plant and soil microbes, where key plant-soil-microbe interactions take place giving benefits to both organisms for plant mineral nutrient acquisition and substrate for microbial energy requirements. These interactions can either be beneficial (symbiotic) for the crop, for example, plant interactions with epiphytes, such as plant growth-promoting rhizobacteria (PGPR), nitrogen-fixing bacteria, and mycorrhiza fungi, or can be negative (parasitic) with plant pathogenic microorganisms (Singh et al. 2004; Raaijmakers et al. 2009; Walkers et al. 2003; Dadhich and Meena 2014). Hence, the rhizosphere is the hotspot where the plant and soil microbial community communicate with each other through the plant roots and mediated by root exudations.
3 Root Exudates Regulating Factors
Plant roots discharge immense amounts and ranges of organic compounds into the rhizosphere soil, known as root exudates or rhizodeposition, and through these exudates, plants can directly communicate with rhizosphere microbiota. Root exudates are an important nutrient for the microbial community in rhizosphere soil and therefore encourage root-microbe colonization (Bacilio et al. 2002). Plant roots can exude many different compounds such as acids, oxygen, and ions, but most are organic molecules (Uren 2000; Bais et al. 2006). These organic-based exudates can be categorized into high and low molecular weight (HMW and LMW, respectively) compounds. The majority of exudates are LMW, including amino and organic acids, sugars, phenolic compounds, and many other secondary metabolites, while considered HMW exudates are more complicated compounds such as mucilage and cellulose (Huang et al. 2014; Rovira 1969). Although root exudates are a key driver of plant-microbe communication in rhizosphere soils, less attention has been given to the mechanisms and regulatory processes that control root exudation. Plant age and genotype, as well as other external biotic such as nutrient availability and root architecture and abiotic factors like soil properties, temperature, stress, or toxic conditions, are known to regulate the quantity and quality of root exudates (Badri and Vivanco 2009; Varma et al. 2017).
3.1 Abiotic Factors
3.1.1 Soil Properties
Soil physical and chemical properties such as pH, texture, moisture, etc. significantly regulate the root exudation process; mainly soil moisture content is one of the key drivers of the root exudation, as high soil moisture leads to hypoxia condition due to the inadequate accessibility of O2 in the soil. Root mucilage exudation can improve the physical pathway for water and nutrient delivery to roots, and this is particularly important as soil dries (Carminati et al. 2016). Low soil moisture can cause temporary plant wilting which caused an increase in the release of amino acids from plant roots (Katznelson et al. 1954). Some bi-cropping studies suggested that high moisture content could help in the rapid transfer of maize root exudates to adjacent bean plants (Ivano 1962). Water stress, including both drought and flood conditions, can significantly impact on the quantity and quality of wheatgrass root exudates as the stress-induced exudation of organic acids such as malic, fumaric, malonic, succinic, and oxalic acids (Henry et al. 2007). Similarly, Song et al. (2012) also reported that exudation of various organic acids such as malic, lactic, acetic, succinic, citric, and maleic acid concentration was increased by osmotic stress in drought-tolerant and non-drought-tolerant maize in hydroponics. Furthermore, soil pH can have a significant effect on root exudation resulting in acidification and alkalinization. Stoltz and Greger (2002) found that the increased soil pH produced by Eriophorum angustifolium and E. scheuchzeri roots could decrease the leaching of toxic elements such as Cd, Cu, Pb, and Zn, while Wang et al. (2006) reported that a lower pH could lead to higher soil leaching of toxic elements such as Cd and Zn. Recent studies on E. angustifolium roots showed that changes in the rhizosphere to a more alkaline pH significantly influenced the exudation of organic acids (oxalic, succinic, and formic acids) in root mucilage under moderately toxic soil conditions (Javed et al. 2012). There are very few studies focused on how soil texture can influence the root exudation process. Soils with different textures have different chemical and physical properties which can cause direct or indirect induction or inhibition of LMW and HMW compound secretion by plant roots. Sandy substrates can induce root exudation as high secretion rates of amino acids were reported when plants were grown in quartz sand (Boulter et al. 1966). Since soil characteristics significantly affect root exudation, little attention has been given to these types of studies, which may be due to the plasticity and variability of root exudates. The study of root exudates is hindered by difficulties in the collection of representative samples, especially in the soil and under field conditions. Plant root exudates may be dissimilar in soil and hydroponics. However, recent molecular techniques combining with stable isotope probing could shed more light on the effect of soil physical characteristics on root exudation.
3.1.2 Temperature
Temperature can have major effects on root exudation. Rovira (1956) reported that higher temperature could cause the increase in the exudation of different amino acids, especially asparagine from Solanum lycopersicum (tomato) and Trifolium subterraneum (subterranean clover). Other studies showed that exudation of tannins and phenolic compounds was markedly inhibited at low temperature (4 °C) compared to the amount at 30 °C in fava beans (Vicia faba) (Bekkara et al. 1998). Contrariwise, Husain and Mckeen (1963) found that strawberry roots secrete higher amount of organic acids at low temperature (5–10 °C) than at higher temperature (20–30 °C), which also leads to root colonization with the pathogenic fungus Rhizoctonia fragariae (Husain and Mckeen 1963; Hale and Moore 1979). A generalized plant exudation response to temperature may not be found; it seems more likely that each species responds differently to temperature extremes.
3.1.3 Light Intensity
A few studies have shown that the composition and quantity of root exudates were affected by light intensity, as it is directly linked to photosynthetic C fixation, the main C source for root exudates. For example, Rovira (1956) reported that trefoils grown under full daylight released higher amounts of serine, glutamic acid, and alanine but shadow inhibited the exudation of aspartic and glutamic acid, phenylalanine, and leucine in grown plants. Many authors reported the quantity of root exudates was significantly increased by the high light intensity and prolongation of the photoperiod, for example, phosphatidylserine in Zn-deficient wheat (Cakmak et al. 1998), catechin in spotted knapweed (Tharayil and Triebwasser 2010; Yadav et al. 2018), and citrate in white tulips (Cheng et al. 2014). Although fluctuations in light intensity combined with the altered photosynthetic spectrum (longer wavelengths) affected the synthesis of secondary metabolites in the leaves of birch and woody plants, the root exudates were not measured (Lavola et al. 1998). However, Yang (2016) reported that light intensity not only influences the quantity but also significantly impacts on the composition of root exudates in sugar beet. Recently Martin et al. (2018a, b) reported that fluctuations in light exposure increased the exudation of dissolved organic carbon and protein-like and humic-like dissolved organic matter, whereas constant low and continuous light significantly inhibit the exudation of total dissolved nitrogen. These findings suggest that light intensity and exposure time are two of the key factors that regulate synthesis and secretion of root exudates, and these should get more attention for future studies on root exudates and their regulating factors.
3.1.4 Nutrient Availability in the Rhizosphere: Nitrogen as Nutrient and Sensor
Although the rhizosphere is confined in a small area (about 2 mm) between root and soil, it is an important zone affecting nutrient dynamics influenced by a plethora of microbial activities useful for improving crop uptake efficiency (Dakora and Phillips 2002). Different factors combine to determine the availability of nutrients in this zone, such as soil chemistry, plant genotype, and nutrient supply (Jones et al. 2004). The availability of nutrients depends on the activity of microorganisms, which in turn are regulated by quantity and quality of the root exudates, thus creating different local micro-ecosystem compared to the bulk soil (Neumann et al. 2009; Meena et al. 2018). Various plant mechanisms have been developed to cope nutrient-limited conditions, but the exudation of organic acids and enzymes into the soil by roots can enhance soil organic matter (SOM) decomposition thus releasing nutrients making them available for plant uptake (Charholm et al. 2015). Some plant species exhibit specific rhizosphere effects on the availability of nutrients, such as the Solanaceae influencing phosphorus mobility in the soil (Safari and Rashidi 2012). Grasses can produce phytosiderophores to increase iron availability by chelation of the metal ion, particularly under Fe-limiting conditions (Ueno et al. (2007). From the bulk soil to the root surface, the microorganism populations increase in quantity but decrease in species diversity, thereby influencing the availability of nutrients. In the rhizosphere, there is an equilibrium between the plant, soil, and microorganisms which is usually characterized by low nutrient concentrations, where a constant turnover can ensure a steady supply for roots (Shen et al. 2013). Considering phosphorus, for example, it can be present in large amounts in the soil, but it is totally unavailable for the plant as both organic and inorganic forms are insoluble (Bhattacharyya and Jha 2012; Igual et al. 2001; Gyaneshwar et al. 2002). Applications of P fertilizers may not combat this limiting condition due to the phenomena of precipitation occurring in the soil (Mckenzie and Roberts 1990). However, in the rhizosphere, some microorganisms are defined as P-solubilizing, such as Azotobacter, Burkholderia, Erwinia, Flavobacterium, and Serratia (Bhattacharyya and Jha 2012), and these have the ability to make this element soluble by synthesizing the low molecular weight organic acids which dissolved the inorganic phosphorus converting it to soluble forms (Zaidi et al. 2009) and therefore available for plant uptake (Mehnaz and Lazarovits 2006).
N2 fixation is another important example of a microbe-plant interaction (discussed in Sect. 5.1), and its significance is dictated by the role that this element (N) has in plant physiology and is often limiting crop productivity. Among nutrients affecting plant growth and development, N plays a pivotal role, and nitrate, which is the main N-form in aerobic soils, can also be considered as a “signaling molecule.” Nitrate can act as a signal to regulate plant gene expression, metabolism, physiology, growth, and development (Krouk et al. 2011; Vidal and Gutiérrez 2008). Although the capacity of the plant to assimilate N depends from on carbon turnover, an increase in biomass is principally limited by nitrogen uptake from the rhizosphere. In plants grown using nitrate-free conditions, the presence of nitrate leads to the modulation of enzymes responsible for assimilation, such as nitrate reductase (NR), nitrite reductase (NiR), and glutamine synthetase (GS), which play fundamental roles in crop production (Crawford 1995; Stitt 1999). In the last decades, genomics, bioinformatics, and systematic have described a complex regulatory network at transcriptional and posttranscriptional levels for the plant’s responses to nitrate (Krouk et al. 2010). In Arabidopsis, nitrate and N metabolites regulate the expression of numerous genes involving a wide range of processes. Studies of the effect of nitrate supply on the quality and quantity of root exudates are missing, especially in space and time. According to Scheible et al. (2004), who provided an illustration of the signaling action of N in Arabidopsis grown in an N-free solution, nitrate supply determines a massive reprograming of genome expression. Within 30 min, the re-addition of nitrate induces gene expression involved in uptake, reduction of nitrate, and organic acid skeleton production. On the other hand, after longer times (about 3 h), the nitrate supply induces specific genes belonging to trehalose and hormone metabolism, protein kinases and phosphatases, receptor kinases, and transcription factors (Scheible et al. 2004), and some of these processes may be important in root-rhizosphere interactions, thereby increasing or reducing the crop nitrate uptake efficiency. As affirmed by Krouk et al. (2010), up to 10% of the transcriptome is responsive to nitrate, and many genes are considered as signals, as these genes are still nitrate-regulated in Arabidopsis NR-deficient mutants, where the first enzyme of the nitrate assimilation pathway is missing (Wang et al. 2004; Kumar et al. 2018).
Finally, the global nitrate signaling pathways show complex regulation either into the plants by transcriptions factor or external to the plants by nitrate availability, which in turn is strongly dependent on the rhizosphere activities. Thus, the rhizosphere is the hotspot where both the concentration of microorganisms is very high compared to bulk soil and the concomitant activity of roots and microorganisms allows the creation of a microenvironment favoring mineralization (Parkin et al. 2002) and denitrification (Qian et al. 1997). Finally, little is known about the mechanisms that microorganisms have on local nitrate signals in the rhizosphere.
3.2 Biotic Factors
3.2.1 Plant/Rhizosphere and Nutrient Use Efficiency
The current requirement in most agriculture is to reduce the inputs of fertilizers in formulating management practices for more sustainable production. New strategies for plant nutrition will require adequate consideration of rhizosphere processes for improving crop productivity (Shen et al. 2013; Zhang et al. 2017). Starting from a general point of view and focusing on specific topics, strategies to maximize the efficiency of rhizosphere processes related to plant nutrition can be assessed by manipulating root growth patterns and targeted fertilizer applications (Zhang et al. 2010; Chen et al. 2011).
Nutrient use efficiency (NUE) is determined by two components: uptake efficiency (UpE), or the root’s ability to take up nutrients from the soil, and utilization efficiency (UtE), as the plant capacity to assimilate or utilize the nutrients (Good et al. 2004; Xu et al. 2012; Lupini et al. 2017; Dadhich et al. 2015). In this context, as rhizosphere is strictly in contact with the roots, UpE is the component most influenced by the rhizosphere processes and on which it is possible to identify strategies for improving NUE.
In legume species Rhizobium spp. accommodated in specialized nodule structures can fix a considerable amount of N to supply the plant. Other microorganisms increase in the availability of nutrients in nonlegumes (Boddey et al. 2003). There are some microorganisms, such as Azospirillum spp., which can supply N to plants (Assmus et al. 1995) increasing the NUE more specifically the UpE. Relationships between the roots and microorganisms in the rhizosphere favor nutrient uptake: on the one hand, there are the root exudates that provide organic compounds to microorganisms (Parkin et al. 2002), and on the other side, they provide inorganic compounds more easily taken up by the plant (Marschner 1995). Some types of bacteria form a biofilm on the roots, thereby ensuring an improvement in nutrient uptake (Beauregard et al. 2013) by providing a robust water contact between soil particles and the root. Using PCR coupled with denaturing gradient gel electrophoresis and fluorescence approaches, Briones et al. (2003) demonstrated a strong correlation among ammonia-oxidizing bacteria and NUE of some rice cultivars also underlining that the presence of these bacteria improved nitrate uptake. Therefore, nutrient uptake may be improved through the selective production of root exudates, which favor the association with specific bacteria and decreasing the abundance of others. Nutrient availability in the rhizosphere is strongly regulated by exudation, but the manipulation of high-affinity nutrient transport systems at plasma membrane level is poorly studied, and increased chelating creates nutrient-limited zones around the roots (George et al. 2005; Liu et al. 2005). Furthermore, the selectivity of root exudates could be achieved through the molecular manipulation of the transporters to improve both the mineralization processes and the numbers of beneficial microorganisms in the rhizosphere. To date, few transporters involved in plant/microbe rhizosphere cross talk were completely functionally characterized (Kretzschmar et al. 2012; Rudrappa et al. 2008), and further studies are needed in crop species. Some authors have demonstrated that plant roots secrete organic compounds to inhibit nitrification (Subbarao et al. 2013) and denitrification (Cordero and Datta 2016), which may be used in agriculture to improve NUE. Manipulating these microbial processes in the rhizosphere can be used to develop crops with higher NUE by decreasing N leaching losses.
3.2.2 Plant Root as Main Trait to Improve NUE
The major limiting factors for the twenty-first-century crop production are the scarcity of water and nutrients in the soil. The plant root system is fundamental for acquiring nutrients and water which are primarily taken up from the soil. In this context, different approaches may be used to improve NUE: (i) changing root architecture to enhance nutrient acquisition especially in nutrient-limiting conditions and (ii) managing the rhizosphere processes starting from root modifications. In fact, it is now well established that root architecture plays a fundamental role in nutrient use efficiency (Lynch 2011) and influencing microorganisms in the soil too (Wang et al. 2011; Wu et al. 2012). Improving the capacity of the root to explore the soil by modifying root architecture is a pivotal task. Thus, understanding internal and/or external factors tuning the growth and development of this organ is an important target to improve water and nutrient use efficiency. Among macroelements, N (mostly nitrate) and phosphorus are the main limiting factors for plant growth and development (Gojon et al. 2009), and plants have adopted strategies to improve nutrient uptake efficiency by modifying root morphology and architecture. Nutrient dynamics in the rhizosphere can lead to substantial and specific changes in roots. Using varying N levels, it was possible to identify specific root adaptations regulated by local or systemic signaling (Bellegarde et al. 2017; Yadav et al. 2017). At the molecular level, genes, transcription factors, proteins, and miRNAs have all been identified to have their role in root architecture and nutrient use efficiency. Mlodzínga et al. (2015) showed the role of a gene (AHA2), a plasma membrane H+-ATPase in root morphology and architecture of rice under N-limiting conditions. Other molecular evidence was established in P-limiting conditions, as OsPHR2 gene and theOsMYB2P-1 transcription factor influenced root hair development and root architecture in rice (Wu and Wang 2008; Dai et al. 2012). QTLs were also associated with root architecture in maize (Li et al. 2015), rice (Li et al. 2009), and bean (Cichy et al. 2009). Of course, these results will be useful to use as molecular markers in breeding programs to improve root architecture and consequently nutrient use efficiency.
Root morphology is also a genetic factor modifying the rhizobiome. Diverse root types can contribute differently to microorganism selection in the rhizosphere. Recent publications point out how the microbe-community can change along roots and among root types. Edwards et al. (2015) showed how the dynamics of the root-associated microbes was strongly dependent on root niche in rice plants, whereas other authors have underlined the bacterial variation among plants, depending on age and genotypes (Lundberg et al. 2012) or different genotypes changing their ribotype profile (16S rRNA gene sequence) depending on host cells (Bulgarelli et al. 2012). Among different root zones, the highest number of bacteria is localized at the root tip, which may be able to select for specific microbes. In wheat seedlings, De Angelis et al. (2008), using high-density 16S rRNA microarray (PhyloChip), showed a differing number of microorganisms with the following hierarchy, bulk soil < mature root < root tips = root hairs, whereas specific colonization by Bacillus subtilis was observed in the root elongation zone of Arabidopsis, suggesting a possible protection mechanism against root pathogens (Massalha et al. 2017). The mature root zone included decomposers which may be associated with the degradation of cells from older root parts (Jones et al. 2009). Distinct communities were also confirmed in lateral roots as compared to tips and basal regions. In this respect, Brachypodium distachyon was employed as a monocotyledon model, and the communities assayed were dependent on root type (nodal or seminal) and root axis (Kawasaki et al. 2016a, b). Thus, as the plant genotype and root system regulates rhizosphere microbiota, more work in this field may be interesting for improving crop productivity. There is an emerging need to evaluate the differences among plant populations within the same species. Comparing wild and modern bean accessions, Pérez-Jaramillo et al. (2017) showed a difference in bacterial numbers that was highly correlated with root morphology. In conclusion, some authors affirm that root phenotype influences rhizosphere colonization by differences in cell wall structure, surface area, and exudate metabolic profiles (Pérez-Jaramillo et al. 2017; Saleem et al. 2016). Thus, it may be important to consider factors modifying root morphology such as nutrient and stresses to drive agronomic practices that optimize rhizosphere interactions.
4 Microbial Selection by Plants
The rhizosphere is where the plant and soil microbe communities are directly linked to each other based on their nutrient requirements. It is well documented that plants can shape their rhizobiome (rhizosphere microbial community), as plant genotype, growth stage, and soil type significantly affect the abundance, composition, and diversity of the rhizobiome (Berg and Smalla 2009; Philippot et al. 2013; Bulgarelli et al. 2013a, b). Selection of the rhizobiome is directly related to the plant root exudation pattern and other abiotic factors such as soil temperature, moisture, chemistry, and physical structure (Pugnaire et al. 2004; Hartmann et al. 2009; Bargett and van der Putten 2014). Recently, Burns et al. (2015) studied different predictors which can affect the rhizosphere microbial community using statistical methods, and they pointed out that plant species is the main determinant of the rhizosphere microbiome, which suggests there is an active microbial selection by plants. Plant-soil feedback strongly relies on plant species and rhizosphere microbe association (Kulmatiskietal. 2008) which implicates plant-microbe coexistence and community assembly (Bever et al. 2010). Furthermore, Burns et al. (2015) also reported that spatial location is also a key predictor which shapes the microbial community, and they also suggested that everything is not everywhere with localized hotspots on the root surface for specific populations.
Since Smalla et al. (2001) first studied the rhizosphere bacterial community using culture-independent techniques and suggested that each plant was colonized by their own bacterial community, numerous studies have focused on how different plant genotypes, plant growth period, and nutrient use efficiency have shaped or influenced abundance, composition, diversity, and functions of rhizosphere microbial communities. Due to recent advances in sequencing technology, there is a long list of studies on this topic, but here we will mainly focus on agriculturally important plants such as wheat, rice, and maize and how these plants and their genetic variation can influence the rhizosphere biome. Donn et al. (2014) studied the rhizobiome of two different wheat lines and suggested that the plant growth stage and level of adhesion of soil microbes to the root were main drivers of the bacterial community. The authors found a tenfold enrichment in the abundance of Pseudomonas, Actinobacteria, copiotrophs, and oligotrophs in the loosely bound rhizosphere soil during vegetative growth. Furthermore, Pseudomonas was highly abundant in the Janz wheat line, whereas copiotrophs were more abundant in the H45 line (Donn et al. 2014). Similar findings were observed by Kawasaki et al. (2016a, b) who showed that the binding of soil microbes to a root and root structure have a profound impact on the rhizobiome of Brachypodium distachyon (model plant for wheat) and its rhizosphere community was enriched by Burkholderiales. One recent study showed the intense effect of plant domestication and breeding on exudation which has directly influenced rhizosphere metabolites in wheat lines (Lannuci et al. 2017). Likewise, Liu et al. (2016) studied the effect of Jasmonic acid (JA) signaling on the wheat rhizosphere microbiome and showed plant organ-specific effects as JA signaling only had a significant impact on diversity and composition of root endophytes but not shoot endophytes or the rhizosphere community.
Lu et al. (2014) studied the rice rhizosphere microbiota using PLFA-based stable isotope labeling and found the rice plant had significantly influenced the microbial community through photosynthetic rhizodeposition. Breidenbach et al. (2016) studied the abundance and composition of the microbial community in the rice rhizosphere soil and showed that the abundance of archaea and bacteria was significantly higher (twofold) in the rhizosphere when compared with the bulk soil. Furthermore, these authors found profound fluctuations in Proteobacteria, Gemmatimonadetes, and Verrucomicrobia communities and showed their effect on functional groups such as iron reducers and fermenters. One recent study showed the autonomous behavior of rice plant in relation to the microbial community, as plants were grown in different types of soil, had the same effect on the composition of rhizosphere microbial community (Li et al. 2014). Similarly, the study of rice plants has showed that plant domestication and genotype play key roles in shaping the rhizosphere community (Lannuci et al. 2017; Shenton et al. 2016; Meena et al. 2015). Moreover, the enrichment of different bacterial phyla Anaerolineae and Methanotrophs in the wild and bred rice cultivars, respectively, were identified (Shenton et al. 2016).
Compared to other crops, there have been large numbers of studies on the maize rhizobiome. This may be due to its unique phenotypic and molecular diversity; we have discussed the more recent studies done on the maize rhizobiome. Cavaglieri et al. (2009) studied maize rhizobiome using microbial culturing-dependent techniques, and they reported that the plant growth stage has a significant influence on the rhizoplane and endo-rhizosphere community with noteworthy enrichment in some bacterial species such as Bacillus, Arthrobacter, Azotobacter, and Listeria and fungi such as Aspergillus and Fusarium. García-Salamanca et al. (2013) studied the bacterial community of the maize rhizosphere in carbonate-rich soil and reported a substantial enrichment of Gammaproteobacteria in rhizosphere soil which could be due to the high availability of carbonate. The rhizosphere bacterial diversity of 27 different maize inbred lines which were grown in field condition showed a remarkable influence of plant heredity on diversity and composition of rhizosphere bacterial community (Pieffer et al. 2013). Li et al. (2014) also studied the maize rhizobiome using a pyrosequencing approach, and they reported the preferential colonization of some bacterial groups such as Proteobacteria, Bacteroidetes, and Actinobacteria in rhizosphere. These authors also showed that the plant growth stage had a profound effect on the bacterial community composition in rhizosphere soil; fluctuations were mainly observed at the family, genus, and OTU level. Recently, few studies have focused on how maize plant N use efficiency (NUE) could shape the totals and functional diversity of rhizosphere soil microbes (Pathan et al. 2015a, b; Baraniya et al. 2016). These authors showed that the Lo5 maize line with higher NUE had stronger influence on the microbial community composition (bacteria and fungi). Both the composition and diversity of bacterial β-glucosidase and protease genes in rhizosphere soils were compared between Lo5 and T250 maize, a low NUE line. All these findings suggest the plant and its different characteristics such as plant heredity, growth stages, physiological traits, and plant nutrient requirements are key factors which drive the formation and structure of the rhizosphere microbial community. It should be noted that many of these studies have only been focused on bacteria and very few of them have focused on the fungal or archaeal communities, which suggests that future research should consider these other rhizosphere organisms in different plants.
5 Plant-Microbe Interaction
5.1 N2-Fixing Bacteria
Nitrogen is a key nutrient for plant development, and it is usually a plant growth-limiting factor. Although more than two thirds of the global atmosphere comprise N2, plants are incapable of using this elemental form of N. On the other side, some soil prokaryotes known as diazotrophs have a dinitrogenase enzyme which enables these prokaryotes to convert N2 into ammonia followed by nitrite (ammonia oxidizers) and nitrate (nitrite reductase), N forms used by plants (Lam et al. 1996; Franche et al. 2009). Due to the plant’s large requirement for N, some have formed beneficial symbiotic associations with these diazotrophs (N2-fixing bacteria). This symbiotic association between plants and N2-fixing bacteria is a kind of mutualistic symbiosis where the host plant directly consumes inorganic N such as ammonia or nitrate from the soil which is fixed by the N2-fixing bacteria. In return the plant provides sheltered environment and fixed carbon and other nutrients to the diazotrophic prokaryotes. Legumes (Fabaceae family) are the foremost symbiotic plants that associate with rhizobia (gram-negative), a member of Alphaproteobacteria (Oldroyd and Downie 2008). Legume-rhizobia interaction instigates root nodule formation, which begins with the release of flavonoids by the plant under N starvation conditions (Oldroyd et al. 2011; Mcnear 2013). Flavonoid signals are responsible for activating nod genes in rhizobia which encoded lipochitooligosaccharides, known as nod factors (Mcnear 2013; Mus et al. 2016). Nod factors are pivotal symbiotic signals as they induce invasion of bacteria into the host plant and nodule formation where the bacteria ultimately are housed (Mcnear 2013; Mus et al. 2016). The structure of the nod factors such as the length of backbone, size, and saturation of fatty acyl chain depend on the rhizobia species (Mcnear 2013; Mus et al. 2016) which leads to host specificity during formation of the plant-rhizobia association (Oldroyd and Downie 2008). Even though two different species of rhizobium, etli and lati, carry identical nod factors, both species have distinct host specificity (Phaseolus spp. and Lotus spp., respectively) (Cárdenas et al. 1995). Host plant selection specificity is astounding as out of millions of microbes; only a tiny number are able to create efficacious symbiosis with host plants (Mcnear 2013). Only one nonlegume plant, called Parasponia species (Cannabaceae family), is capable to form a symbiotic association with rhizobia (Sytsma et al. 2002). Invasion of rhizobia occurs through crack entry into the host plant Parasponia (Lancelle and Torrey 1985), which is different from legume plants, where the rhizobia enter through root hair curling. Furthermore, the proliferation of rhizobia and N fixation takes place after the formation of fixation threads, and these threads branch all over the nodule cells; however, they still stay in contact with the apoplast (Behm et al. 2014; Mus et al. 2016; Ashoka et al. 2017). Somehow, development of the Parasponia-rhizobia association is quite young and is a primitive form of nodulation. Behm et al. (2014) suggested that the Parasponia-rhizobia symbiosis can be used as a model system to understand the control mechanisms which emerge during the early stages of N2-fixing mutualism evolution.
Apart from rhizobia, members of the Actinobacteria phylum, Frankia sp. (gram-positive), have been shown to nodulate (actinorhizas) with broad spectrum of woody plants, called actinorhizal plants. Actinorhizal plants are distributed in 8 different families, containing 17 genera and 150 species. Actinorhizal development processes are very similar to legume nodules, but they are much larger in size and longer lived compared to legume nodules. Actinorhizas have central vasculature and can fix the same amount of N2 as rhizobia nodules (Mus et al. 2016). Another group of bacteria called cyanobacteria, especially filamentous cyanobacteria (mainly Nosotc and Scytonema), are able to fix N2 with cells which are known as heterocysts. Cyanobacteria create this symbiotic association with a variety of higher and lower plants, algae, and fungi (Merks and Elhai 2002). Moreover, there are some other N-fixing bacteria, such as Azospirillum spp. and Azoarcus spp., that can colonize nonlegume plants without any nodule formation (Elmerich and Newton 2007), and these bacteria are known as N2-fixing endophytes (Döbereiner 1992; Baldani and Baldani 2005). Consequently, it may be important to create more symbiotic associations between N2-fixing bacteria and nonlegume plants, especially in different crops, thereby reducing the use of chemical N fertilizer and better manage greenhouse gases fluxes.
5.2 Plant Growth-Promoting Bacteria (PGPR)
Kloepper and Schroth (1978) were the first authors who coined the term “plant growth-promoting rhizobacteria” (PGPR) for bacteria inoculated on seeds that successfully colonized the plant root and promoted plant growth. Antoun and Kloepper (2001) reported that only a few bacteria (1–2%) have an aptitude to promote plant growth. PGPR are propitious free-living bacteria which can colonize the rhizosphere and rhizoplane and within the root itself (Gray and Smith 2005). Moreover, based on this colonization, PGPR can be divided into two subcategories: (i) bacteria which colonized the rhizosphere or rhizoplane are called extracellular PGPR (ePGPR) and (ii) those positioned inside root tissues are known as iPGPR (intracellular PGPR) (Viveros et al. 2010). To date, several different bacterial genera have been recognized as PGPR, many of them mainly belong to the Proteobacteria and Firmicutes phyla (Lugtenberg and Kamilova 2009; Drouge et al. 2012), though Bacillus spp. and Pseudomonas spp. are predominant (Podile and Kishore 2006).
PGPR can induce plant growth in two different ways, directly as biofertilizer or indirectly as biopesticide/biocontrol. Direct plant growth induction includes enhanced nutrient supply such as N, phosphorus, and potassium and increases in Fe through the release of siderophore, etc. or by modulation of phytohormones such as auxin and cytokinins (Arora et al. 2012; Bhardwaj et al. 2014). Indirect plant growth promotion entails control over the inhibitory effect of phytopathogens by producing antibiotics or trigger induced systemic resistance (Glick 1995; van Loon et al. 1998; Van Loon 2007). Colonization of a microbial community on the root surface was erratic, and interaction with roots occurred in various patches (McNear 2013). Nutrient availability and root surface physicochemical variations are key factors that induce changes in the abundance and structure of microbial communities (McNear 2013). Root exudation provides chemical compounds for microbes which leads to the formation of microcolonies on the root surface. Danhorn and Fuqua (2007) reported that root epidermal cell junctions, root hairs, axial grove, cap cells, etc. are common sites for the formation of bacterial colonies. Furthermore, microcolonies expand into larger bacterial biofilms which later become wrapped into an exopolymeric matrix (McNear 2013). Rudrappa et al. (2008) showed that bacterial abundance (PGPR) is one of the key factors controlling plant growth promotion. During the plant growth promotion time course, microbial biofilms act together performing quorum sensing and synchronize discharges of various compounds that directly or indirectly promote plant growth (Mcnear 2013). Each plant growth promotion mechanism and function (direct or indirect) have been discussed in detail (Gupta et al. 2015; Vecheron et al. 2013). Some recent findings also suggest that PGPR can also be used in the phytoremediation of contaminated soil (Zhuang et al. 2007; Shukla et al. 2011; Tak et al. 2013; Meena and Yadav 2015), as some PGPR enable improved plant resistance against abiotic factors, including heavy metal contamination, and in some way help plants to enhance resistance to heavy metals (Jing et al. 2007; Saharan and Nehra 2011; Tak et al. 2013).
5.3 Mycorrhizal Fungi
Among all microbial associations, fungi form symbiotic associations with plants, and between mycelial fungi and plants, the relationship is known as a mycorrhizal association. These words are derived from the Greek word mikos meaning fungi and rhiza meaning roots. In contrast to legume-rhizobia association, mycorrhizal association is pervasive and indiscriminative, resulting in colonization of nearly 80% of angiosperms and all gymnosperms. Mycorrhizal association is the earliest plant-microbe association, it first occurred approximately 450 million years ago, and this helps explain the pervasiveness of mycorrhiza overall the plant kingdom. Mainly the mycorrhizal association is a mutualistic association, where fungi provide phosphorus, water, and other micronutrient acquisitions by increasing the root surface; in return the fungi receive fixed carbon. Moreover, mycorrhizal fungi play a vital role in the fitness of natural plants (Allen et al. 1995).
Based on anatomical aspects, the mycorrhizal association is divided into two different subcategories, ectomycorrhizae and endomycorrhizae. In ectomycorrhizae (ECM), fungi grow and are colonized within root intercellular spaces by forming a hartig net around the root cortex (McNear 2013). In contrast, endomycorrhizae fungi are colonized within root cortical cells and form highly branched structures, called arbuscules (Harrison 2005). Moreover, endomycorrhizae fungi are subdivided into three subcategories, orchid, ericoid, and arbuscular mycorrhiza (AM), AM association being the most common occurring association. During association, fungal hyphae (hartig net or arbuscule) provide nutrients to the plant through the horizontal transfer and expanding the root surface area and exchanging photosynthetically fixed carbon from plants. While ECM prefers to associate with woody plants, AM fungi form a symbiotic association with various land plants, including many agricultural crops (Garcia et al. 2015). In stress conditions such as under extreme rainfall, the fungus can grow their hyphae outside soil nutrient-depleted zones, leading to expanded contact with the soil surface or particles which help to reach out into the soil for plant uptake (Barman et al. 2016; Datta et al. 2017). The mycorrhizal hyphal network also significantly affects soil quality by promoting soil aggregation and stability through various biochemical and biological mechanisms, which directly or indirectly increase plant productivity (Rillig and Mummey 2006).
Unlike the rhizobia symbiosis, the chemical signaling processes of mycorrhizae are less well understood. Much focus has been given to the ECM association since both partners can be grown easily, while endomycorrhizae is difficult to grow in vitro due to their obligate behavior (McNear 2013). Plant root exudates such as various flavonoids (mainly rutin) (Lagrange et al. 2001), abietic acid (Fries 1987), and strigolactones (only in AM fungus) (Akiyama et al. 2005) initiate the spore germination and hyphal growth and branching of the fungus by activation of genes in mycorrhiza fungus which produce lipochitooligosaccharides or short chitooligosaccharides, known as Myc factors (Maillet et al. 2011; Genre et al. 2013). There is some resemblance between Myc factors and nod factors (produced by rhizobia in the legume-rhizobia association) (McNear 2013), and this could explain the phenomena of “common symbiosis pathway” which originated in AM symbiosis, later adopted by legume-rhizobia association (Garcia et al. 2015). Due to these chemical dialogues, fungi interact with host plants and begin the hyphae proliferation into host plant roots, called hyphopodium (in AM), or growth between dermal cells (in ECM). Formation of arbuscules is the final step of the symbiotic process, and through these arbuscules, fungi enter into the root cell cytoplasm. Selosse et al. (2006) reported that two or more plants can share nutrients through the same mycorrhizae associations as fungal hyphae can create a “common mycorrhizal network” (CMN). Similar phenomena may occur in the ECM symbiosis, but more research is needed to understand the molecular mechanisms of ECM symbiotic associations.
5.4 Pathogenic Microorganisms
In contrast to beneficial (symbiotic/naturalistic) associations with the rhizosphere microbiota, plants can also interact with some soil microbes that inhibit plant growth and health, commonly referred as pathogenic microbes or phytopathogens. These can include fungi, bacteria, viruses, oomycetes, and nematodes. Phytopathogens are one of the major constraints to global food production and security. While many of soil-borne pathogens survive and grow into bulk soil, the rhizosphere is a key niche where pathogenic microbes form parasitic associations with plant roots. Soil-borne pathogens are more persistent when compared to pathogens associated with aboveground parts of the plant (Bruehl 1987). While information has been available on how root exudates modulate/regulate plant-symbiont association, the limited focus has been given on how rhizodeposition can attract and activate the phytopathogens. Agrios (2005) has divided plant pathogens into four main groups: virus, bacteria, fungi, and nematodes. Among these four groups, fungi are the main soil-borne pathogens compared to others. Bacteria and viruses need natural openings or wounds to enter into plant tissues. Furthermore, the soil is not a suitable habitat for nonspore-forming bacteria, while some filamentous bacteria such as Streptomyces spp. can grow easily in soil and able to cause infection to host plants. Ralstonia solanacearum (Pseudomonas) can colonize the xylem and cause wilt to a variety of plants, mainly tomato (Genin and Boucher 2004), pepper, and eggplants. While Agrobacterium tumefaciens is well known for causing crown gall (Nester et al. 2005), through the transmission of DNA (T-DNA), these two are the best-understood examples of soil-borne pathogenic bacteria. Only a few viruses can infect plant roots, as they required a vector for transmission and in soil nematodes (e.g., Nepoviruses; Brown et al. 1995), and some zoosporic fungi such as Olpidium (Campbell 1996) are the main vectors for viral transmission. Nematodes can be free-living worms that normally ingest other microbes such as bacteria or fungi or other nematodes. However some can form parasitic relationships with host plants. There are three main types of associations that nematodes form with host plants: (i) ectoparasitic, in which nematodes contact with only the outer body of roots; (ii) endoparasitic, in which nematodes are able to reach out the inner body of plant roots; and (iii) sedentary endoparasitic, in which nematodes persist in the inner body of roots for reproduction process (Raaijamakers et al. 2009).
Fungi and oomycetes can be soil-borne phytopathogens. While oomycetes are phylogenetically closer to blue algae, their morphological characteristics are identical to fungi, and their parasitism and diseases have similar symptoms to fungi (Raaijamakers et al. 2009). Most of the soil pathogenic fungi and oomycetes are necrotrophic, and only a few of them are biotrophic such as Plasmodiophora brassicae (cause of cabbage “clubroot”) and Plasmopara halstedii (“downy mildew” in sunflower, Friskop et al. 2009). Phytophthora sojae is semi-biotrophic and causes powdery mildew and rusts, especially in soybean. Due to their generalist behavior, necrotrophic pathogens have a wider selection of hosts and can easily infect many plants. Fungi penetrate the host plants through the germ tubes or zoospores and infect epidermal root cells (tips, hairs, etc.) or strike at emerging shoots or seed radicles. Pythium, fast-growing and important soil-borne pathogen, affects tree seedling production through attacking plant seeds even before they start to emerge; the disease is called “damping off” (Vaartaja 1975; Weiland et al. 2013). Fungi produce degrading enzymes or use hydrostatic pressure to enter into host root cells (young or juvenile roots) and further colonized into the root cortex. After cortex colonization, fungal mycelia still continue to spread to other parts of the plant and sometimes grow externally and can also cause disease in adjoining plants. Fusarium oxysporum and Verticillium dahlia and other fungi can spread into the plant vascular tissues through the root endodermis and blocking water flow that causes wilting. This fungal problem can infect many different plants including potato, cotton, tomato, and eggplants and some tree species such as olive (Beckman 1987). A number of different diseases are caused by a fungus such as decay, rot, damping off, and wilt; among all of them, root rot is often the first stage. By infecting or destroying plant roots, plant water and nutrient uptake capacity is decreased causing nutrient deficiency symptoms, reduced size, and drought stress (Raaijamakers et al. 2009).
6 Concluding Remarks and Future Applications
The rhizosphere represents a plethora of interactions between plant-microbes, plant-soil, microbe-soil, and microbe-microbe, and because this network is highly complex, we can affirm that the study of this environment is still in its infancy (Zheng et al. 2017; Berendesen et al. 2012; Sihag et al. 2015). A pivotal and strong link is highlighted among soil-microorganism-plant, thereby creating a hotspot in which to study and improve crop health and NUE. In the present work, we have discussed the dynamic processes in the rhizosphere and especially how root exudates, as well as the rhizobiota, are influenced by biotic and abiotic factors to understand and thus improve nutrient use efficiency in the context of more sustainable agriculture. Moreover, the emphasis was given to the role of the roots both in the selection of microbes and in the efficiency of use of resources focusing particularly on nitrate as nutrient and signal.
Rhizosphere interaction studies include measurements of the microbiota, root morphology, and rarely their interactions, especially in field condition. Our limited knowledge is presented on how the plants control the microbe communities present in the soil and how different root types influence the availability of nutrients and also their absorption by rhizobiota. Furthermore, new omic technologies will contribute to identifying new signaling compounds which allow us to set new strategies for promoting plant growth and health.
Abbreviations
- AM:
-
Arbuscular mycorrhizal
- ECM:
-
Ectomycorrhizae
- GS:
-
Glutamine synthetase
- HMW:
-
High molecular weight
- JA:
-
Jasmonic acid
- LMW:
-
Low molecular weight
- N:
-
Nitrogen
- NiR:
-
Nitrite reductase
- NR:
-
Nitrate reductase
- NUE:
-
Nutrient use efficiency
- P:
-
Phosphorus
- PGPR:
-
Plant growth-promoting rhizobacteria
- PLFA:
-
Phospholipid fatty acid analysis
- SOM:
-
Soil organic matter
- T-DNA:
-
Transmission of DNA
- UpE:
-
Uptake efficiency
- UtE:
-
Utilization efficiency
References
Agrios GN (2005) Plant pathology, 5th edn. Elsevier Academic Press Inc., New York
Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824–827
Allen MF, Morris SJ, Edwards F, Allen EB (1995) Microbe– plant interactions in Mediterranean-type habitats: shifts in fungal symbiotic and saprophytic functioning in response to global change. In: Moreno JM, Oechel WC (eds) Global change and Mediterranean-type ecosystems. Springer, New York, pp 287–305
Antoun H, Kloepper JW (2001) Plant growth promoting rhizobacteria. In: Brenner S, Miller JH (eds) Encyclopedia of genetics. Academic, New York, pp 1477–1480
Arora NK, Tewari S, Singh S, Lal N, Maheshwari DK (2012) PGPR for protection of plant health under saline conditions. In: Maheshwari DK (ed) Bacteria in agrobiology: stress management. Springer, Heidelberg, pp 239–258
Ashoka P, Meena RS, Kumar S, Yadav GS, Layek J (2017) Green nanotechnology is a key for eco-friendly agriculture. J Clean Prod 142:4440–4441
Assmus B, Hutzler P, Kirchhof G et al (1995) In situ localization of Azospirillum brasiliense in the rhizosphere of wheat with fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol 61:1013–1019
Bacilio JM, Aguilar FS, Ventura ZE, Perez CE, Bouquelet S, Zenteno E (2002) Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 249:271–277
Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32:666–681
Bais HP, Wier TL, Perry LG, Vivanco JM (2006) The role of root exudates in rhizosphere interaction with plants and other microorganisms. Annu Rev Plant Biol 57:233–266
Baldani JI, Baldani VLD (2005) History on the biological fixation research in graminaceous plants: special reference on the Brazilian experience. An Acad Bras Cienc 77:549–579
Baraniya D, Puglisi E, Ceccherini MT, Pietramellara G, Giagnoni L, Arenella MR, Nannipieri P, Renella G (2016) Protease encoding microbial communities and protease activity of the rhizosphere and bulk soils of two maize lines with different N uptake efficiency. Soil Biol Biochem 96:176–179
Bardgett RD, van der Putten WH (2014) Belowground biodiversity and ecosystem functioning. Nature 515:505–511
Barman J, Samanta A, Saha B, Datta S (2016) Mycorrhiza: the oldest association between plant and fungi. Resonance, General article 21: 1093–1103
Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R (2013) Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA 110:E1621–E1630
Beckman CH (1987) The nature of wilt diseases of plants. American Phytopathological Society Press, St. Paul
Behm JE, Geurts R, Kiers ET (2014) Parasponia: a novel system for studying mutualism stability. Trends in Plant Sci 19:757–763
Bekkara F, Jay M, Viricel MR, Rome S (1998) Distribution of phenolic compounds within seed and seedlings of two Vicia faba varieties differing in their seed tannin content and study of their seed and root phenolic exudations. Plant Soil 203:27–36
Bellegarde F, Gojon A, Martin A (2017) Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J Exp Bot 68:2553–2565
Bender SF, Wagg C, van der Heijden MGA (2016) An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31:440–452
Berendsen RL, Pieterse CM, Bakker PA (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Bever JD, Dickie IA, Facelli E, Facelli JM, Klironomos J, Moora M, Rillig MC, Stock WD, Tibbett M, Zobel M (2010) Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol 25:468–478
Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N (2014) Biofertilizer function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Fact 13:1–10
Bhattacharyya PN, Jha DK (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28:1327–1350
Boddey RM, Uruquiaga S, Alves BJR et al (2003) Endophytic nitrogen fixation in sugarcane: present knowledge and future applications. Plant Soil 252:139–149
Boulter D, Jeremy JJ, Wilding M (1966) Amino acids liberated into the culture medium by pea seedling roots. Plant Soil 24:121–127
Breidenbach B, Pump J, Dumont MG (2016) Microbial community structure in the rhizosphere of rice plants. Front Microbiol 6:1537
Brimecombe MJ, De Leij FAAM, Lynch JM (2007) Rhizodeposition and microbial populations. In: Pinton R et al (eds) The rhizosphere biochemistry and organic substances at the soil–plant interface. CRC Press, Boca Raton, pp 73–109
Briones AM, Okabe S, Umemiya Y et al (2003) Ammonia – oxidizing bacteria on root biofilms and their possible contribution to N use efficiency of different rice cultivars. Plant Soil 250:335–248
Brown DJF, Robertson WM, Trudgill DL (1995) Transmission of viruses by plant nematodes. Annu Rev Phytopathol 33:223–249
Bruehl GW (1987) Soilborne Plant Pathogens. Macmillan Publishing Company, New York
Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488(7409):91–95
Bulgarelli D et al (2013a) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P (2013b) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Burns JH, Anacker BL, Strauss SY, Burke DJ (2015) Soil microbial community variation correlates most strongly with plant species identity, followed by soil chemistry, spatial location and plant genus. AoB Plants 7:plv030
Cakmak I, Erenoglu B, Gülüt KY, Derici R, Römheld V (1998) Light-mediated release of phytosiderophores in wheat and barley under iron or zinc deficiency. Plant Soil 202:309–315
Campbell RN (1996) Fungal transmission of plant viruses. Annu Rev Phytopathol 34:87–108
Cárdenas L, Domínguez J, Quinto C, López-Lara IM, Lugtenberg BJJ, Spaink HP, Rademaker GJ, Haverkamp J, Thomas-Oates JE (1995) Isolation, chemical structures and biological activity of the lipo-chitin oligosaccharide nodulation signals from Rhizobium etli. Plant Mol Biol 29(453):464
Carminati A, Zarebanadkouki M, Kroener E, Ahmed MA, Holz M (2016) Biophysical rhizosphere processes affecting root water uptake. Ann Bot 118:561–571. https://doi.org/10.1093/aob/mcw113
Cavaglieri L, Orlando J, Etcheverry M (2009) Rhizosphere microbial community structure at different stages and root locations. Microbial Res 164:391–399
Chen XP, Cui ZL, Vitousek PM et al (2011) Integrated soil–crop system management for food security. PNAS 108:6399–6404
Cheng L, Tang X, Vance CP, White PJ, Zhang F, Shen J (2014) Interactions between light intensity and phosphorus nutrition affect the phosphate-mining capacity of white lupin (Lupinus albus L.). J Exp Bot 65:2995–3003
Cichy KA, Blair MW, Mendoza CHG, Snapp SS, Kelly JD (2009) QTL analysis of root architecture traits and low phosphorus tolerance in an Andean bean population. Crop Sci 49:59–68
Clarholm M, Skyllberg U, Rosling A (2015) Organic acid induced release of nutrients from metal-stabilized soil organic matter. The unbutton model. Soil Biol Biochem 84:168–176
Cordero OX, Datta MS (2016) Microbial interactions and community assembly at microscales. Curr Opin Microbiol 31:227–234
Crawford NM (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7:859–868
Dadhich RK, Meena RS (2014) Performance of Indian mustard (Brassica juncea L.) in Response to foliar spray of thiourea and thioglycollic acid under different irrigation levels. Indian J Ecol 41(2):376–378
Dadhich RK, Meena RS, Reager ML, Kansotia BC (2015) Response of bio-regulators to yield and quality of Indian mustard (Brassica juncea L. Czernj. and Cosson) under different irrigation environments. J Appl Nat Sci 7(1):52–57
Dai X, Wang Y, Yang A, Zhang WH (2012) OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphatestarvation responses and root architecture in rice. Plant Physiol 159:169–183
Dakora FD, Phillips DA (2002) Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245:35–47
Datta R, Kelkar A, Baraniya D, Molaei A, Moulick A, Meena RS, Formanek P (2017) Enzymatic degradation of lignin in soil: a review. Sustain MDPI 9(7):1163; 1–18
De Angelis KM, Lindow SE, Firestone MK (2008) Bacterial quorum sensing and nitrogen cycling in rhizosphere soil. FEMS Microbial Ecol 66:197–207
Danhorn T, Fuqua C (2007) Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61(1):401–422
Döbereiner J (1992) History and new perspective of diazotrophs in association with non-leguminous plants. Symbiosis 13:1–13
Donn S, Kirkegaard JA, Perera G, Richardson AE, Watt M (2014) Evolution of bacterial communities in wheat crop rhizosphere. Environ Microbiol 17:610–621
Doornbos RF, van Loon LC, Bakker PAHM (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron Sustain Dev 32:227–243
Drogue B, Dore H, Borland S, Wisniewski-Dyé F, Prigent-Combaret C (2012) Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res Microbiol 163:500–510
Edwards J et al (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. PNAS 112:E911–E920
Elmerich C, Newton WE (2007) Associative and endophytic Nitrogen fixing Bacteria and Cyanobacterial Associations – introduction. Springer, Dordrecht
FAO (2009) The state of food and agriculture, Rome
Franche C, Lindström K, Elmerich C (2009) Nitrogen fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 321:35–59
Fries N (1987) Ecological and evolutionary aspects of spore germination in the higher Basidiomycetes. Trans Br Mycol Soc 88:1–7
Friskop A, Marker S, Gulya T (2009) Downy mildew of sunflower. State Agricultural Experiment Station, New York
Garcia K, Delaux P, Cope KR, Ané J (2015) Molecular signals require for the establishment and maintenance of ectomycorrhizal symbiosis. New Phytol 208:79–87
Genin S, Boucher C (2004) Lessons learned from the genomeanalysis of Ralstonia solanacearum. Annu Rev Phytopathol 42:107–134
Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, Barker DG (2013) Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198:190–202
George TS, Simpson RJ, Hadobas PA, Richardson AE (2005) Expression of a fungal phytase gene in Nicotiana tabacum improves phosphorus nutrition of plants grown in amended soils. Plant Biotechnol J 3:129–140
Giles E.D. Oldroyd, J. Allan Downie, (2008) Coordinating Nodule Morphogenesis with Rhizobial Infection in Legumes. Annual Review of Plant Biology 59 (1):519-546
Giles E.D. Oldroyd, Jeremy D. Murray, Philip S. Poole, J. Allan Downie, (2011) The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annual Review of Genetics 45 (1):119-144
Glick BR (1995) The enhancement of plant growth by free-living bacteria. Can J Microbiol 41:109–117
Gojon A, Nacry P, Davidian JC (2009) Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol 12:328–338
Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9:597–605
Gray EJ, Smith DL (2005) Intracellular and extracellular PGPR: commonalities and distinctions in the plant-bac-terium signaling processes. Soil Biol Biochem 37:395–412
García-Salamanca A, Molina-Henares MA, van Dillewijn P, Solano J, Pizarro-Tobías P, Roca A, Duque E, Ramos JL (2013) Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb Biotechnol 6(1):36–44
Gupta G, Parihar SS, Ahirwar NK, Snehi SK, Singh S (2015) Plant growth promoting rhizobacteria (PGPR): current and future prospective for development of sustainable agriculture. J Microbial Biochem Technol 7:2
Gyaneshwar P, Naresh Kumar G, Parekh LJ, Poole PS (2002) Role of soil microorganisms in improving P nutrition of plants. Plant Soil 245:83–93
Hale MG, Moore LD (1979) Factors affecting root exudation. Adv Agron 31:93–124
Harrison MJ (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol 59:19–24
Hartmann A, Schmid M, Van Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant Soil 321:235–257
Hennery A, Doucette W, Norton J, Bugbee B (2007) Changes in crested wheat grass root exudation caused by flood, drought and nutrient stress. J Environ Qual 36:904–912
Hiltner L (1904) Ueber neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie und unter besonderer BerUcksichtigung der Grundungung und Brache. Arb Deut Landw Gesell 98:59–78
Huang XF, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM (2014) Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92:267–275
Husain SS, Mckeen WE (1963) Interactions between strawberry roots and Rhizoctonia fragariae. Phytopathology 53:541–545
Igual JM, Valverde A, Cervantes E, Velázquez E (2001) Phosphatesolubilizing bacteria as inoculants for agriculture: use of updated molecular techniques in their study. Agronomie 21:561–568
Ivanov VP (1962) Mutual effect through the root system of a mixed crop of corn and broad beans. Fiziol Rast 9:179–188
Javed MT, Stoltz E, Lindberg S, Greger M (2012) Changes in pH and organic acids in mucilage of Eriophorum angustifolium roots after exposure to elevated concentrations of toxic elements. Environ Sci Pollut Res Int. https://doi.org/10.1007/s11356-012-1413-z
Jing Y, He Z, Yang X (2007) Role of soil rhizobacteria in phytoremediation of heavy metal contaminated soils. J Zhejiang Univ Sci B 8:192–207
Jones DL, Hodge A, Kuzyakov Y (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163:459–480
Jones DL et al (2009) Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321:5–33
Kardol P, Cornips NJ, van Kempen MML, Bakx-Schotman JMT, van der Putten WH (2007) Microbe-mediated plant–soil feedback causes historical contingency effects in plant community assembly. Ecol Monogr 77:147–162
Katznelson H, Rouatt JW, Payne TMB (1954) Liberation of amino acids by plant roots in relation to Desiccation. Nature 174:1110–1111
Kawasaki A et al (2016a) Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLoS One 11:e0164533
Kawasaki A, Donn S, Ryan PR, Mathesius U, Devilla R, Jones A et al (2016b) Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for Wheat. PLoS One 11(10):e0164533
Kloepper JW, Schroth MN (1978) In Proceedings of the 4th international conference on plant pathogenic bacteria vol. 2 (ed. Station de Pathologie Végétale et Phytobactériologie) 879–882 (Gibert-Clarey, Tours)
Kretzschmar T et al (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signaling and branching. Nature 483:341–344
Krouk G, Crawford NM, Coruzzi GM, Tsay Y-F (2010) Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13(3):265–272
Krouk G, Ruffel S, Gutierrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16:178–182
Kulmatiski A, Beard KH, Stevens JR, Cobbold SM (2008) Plant-soil feedbacks: a meta-analytical review. Ecol Lett 11:980–992
Kumar S, Meena RS, Bohra JS (2018) Interactive effect of sowing dates and nutrient sources on dry matter accumulation of Indian mustard (Brassica juncea L.). J Oilseed Brassica 9(1):72–76
Lagrange H, Jay-Allgmand C, Lapeyrie F (2001) Rutin, the phenolglycoside from eucalyptus root exudates, stimulates Pisolithus hyphal growth at picomolar concentrations. New Phytol 149:349–355
Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular genetics of nitrogen assimilation into amino acids in high plants. Annu Rev Plant Physiol Plant Mol Biol 47:569–593
Lancelle SA, Torrey JG (1985) Early development of rhizobium induced root nodules of Parasponia rigida. I infection and early nodulation. Protoplasm 123:26–37
Lannucci A, Fragasso M, Beleggia R, Nigro F, Papa R (2017) Evolution of crop rhizosphere: impact of domestication on root exudates in Tetraploid wheat (Triticum turdigum L.). Front Plant Sci 8:2124
Lavola A, Julkunen-Tiitto R, Roininen H, Aphalo P (1998) Host-plant preference of an insect herbivore mediated by UV-B and CO2 in relation to plant secondary metabolites. Biochem Syst Ecol 26:1e12
Li JZ, Xie Y, Dai AY, Liu LF, Li ZC (2009) Root and shoot traits responses to phosphorus deficiency and QTL analysis at seedling stage using introgression lines of rice. J Genet Genomics 36:173–183
Li X, Rui J, Xiong J, Li J, He Z, Zhou J, Yannarell AC, Mackie RI (2014) Functional Potential of Soil Microbial Communities in the Maize Rhizosphere. PLoS One 9(11):e112609
Li PC, Chen FJ, Cai HG, Liu JC, Pan QC, Liu ZG, Gu RL, Mi GH, Zhang FS, Yuan LX (2015) A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J Exp Bot 66:3175–3188
Liu JQ, Samac DA, Bucciarelli B, Allan DL, Vance CP (2005) Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J 41:257–268
Liu H, Carvalhais LC, Schenk PM, Dennis PG (2016) Effect of Jasmonic acid signalling on wheat micorbiome differ between body sites. Sci Rep 7:41766
Lu H, Murase J, Watanabe A, Sugimoto A, Kimura M (2014) Linking microbial community dynamics to rhizosphere carbon flow in a wetland rice soil. FEMS Microbiol Ecol 48:179–186
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Lundberg DS et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Lupini A, Princi MP, Araniti F, Miller AJ, Sunseri F, Abenavoli MR (2017) Physiological and molecular responses in tomato under different forms of N nutrition. J Plant Physiol 216:17–25
Lynch JP (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol 156:1041–1049
Lynch JM, Whipps JM (1990) Substrate flow in the rhizosphere. Plant Soil 129:1–10
Maillet F, Poinsot V, Andre O, Puech-Pages V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Andres Martinez E, Driguez H, Cecard G, Denarie J (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhizal. Nature 469:58–64
Marschner H (1995) Mineral Nutrition of Higher Plants. Academic, London
Martin BC, Statton J, Siebers AR, Grierson PF, Ryan MH, Kendrick GA (2018a) Colonizing tropical seagrasses increase root exudation under fluctuating and continuous low light. Limnol Oceanogr 63:S381–S391
Martin BC, Glesson D, Statton J, Siebers AR, Grierson PF, Ryan MH, Kendrick GA (2018b) Low light availability alters root exudation and reduces putative beneficial microorganisms in seagrass roots. Front Microbiol 8:2667
Massalha H et al (2017) Live imaging of root–bacteria inter-actions in a microfluidics setup. PNAS 114:4549–4554
McKenzie RH, Roberts TL (1990) Soil, and fertilizers phosphorus update. In: Proceedings of Alberta soil science workshop proceedings, February, 20–22, Edmonton, pp 84–104
McNear DH Jr (2013) The Rhizosphere- roots, soil and everything in between. Nat Educ Knowl 4:1
Meeks JC, Elhai J (2002) Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol Mol Biol Rev 66:94–121
Meena H, Meena RS (2017) Assessment of sowing environments and bio-regulators as adaptation choice for clusterbean productivity in response to current climatic scenario. Bangladesh J Bot 46(1):241–244
Meena RS, Yadav RS (2015) Yield and profitability of groundnut (Arachis hypogaea L) as influenced by sowing dates and nutrient levels with different varieties. Legum Res 38(6):791–797
Meena RS, Dhakal Y, Bohra JS, Singh SP, Singh MK, Sanodiya P (2015) Influence of bioinorganic combinations on yield, quality and economics of Mungbean. Am J Exp Agric 8(3):159–166
Meena RS, Kumar V, Yadav GS, Mitran T (2018) Response and interaction of Bradyrhizobium japonicum and Arbuscular mycorrhizal fungi in the soybean rhizosphere: a review. Plant Growth Regul 84:207–223
Mehnaz S, Lazarovits G (2006) Inoculation effects of Pseudomonas putida, Gluconacetobacter azotocaptans, and Azospirillum lipoferum on corn plant growth under greenhouse conditions. Microbial Ecol 51:326–335
Mlodzinska E, Klobus G, Christensen MD, Fuglsang AT (2015) The plasma membrane H+-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Plant Physiol 154:270–282
Mus F, Crook MB, Garcia A, Garcia-Costas MK, Geddes BA, Kouri ED, Paramasivan P, Ryu MH, Oldroyd GE, Poole PS, Udvardi MK, Voigt CA, Ané JM, Peters JW (2016) Symbiotic nitrogen fixation and challenges to extending it to non-legumes. Appl Environ Microbiol 82:3698–3710
Nardi S, Concheri G, Pizzeghello D, Sturaro A, Rella R, Parvoli G (2000) Soil organic matter mobilization by root exudates. Chemosphere 41:653–658
Naveed M, Brown LK, Raffan AC, George TS, Bengough AG, Rosse T, Sinclair I, Koebernick N, Cooper L, Hackett CA, Hallett PD (2017) Plant exudates may stabilize or weaken soil depending on species, origin and time. Eur J Soil Sci 68:806–816
Nester EW, Gordon MP, Kerr A (2005) Agrobacterium Tumefaciens: from plant pathology to biotechnology. APS Press, St. Paul
Neumann G, George TS, Plassard C (2009) Strategies and methods for studying the rhizosphere-the plant science toolbox. Plant Soil 321:431–456
Newman EI (1985) The rhizosphere: carbon sources and microbial populations. In: Fitter AH (ed) Ecological interactions in soil. Blackwell Scientific Publications, Oxford, p 107
Oldroyd GED, Downie JA (2008) Coordinating nodule morphogenesis with Rhizobial infection in legumes. Annu Rev Plant Biol 59(1):519–546
Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-Rhizobial Symbiosis. Annu Rev Genet 45(1):119–144
Parkin TB, Kaspar TC, Cambardella C (2002) Oat plant effects on net nitrogen mineralization. Plant Soil 243:187–195
Pathan SI, Ceccherini MT, Pietramellara G, Puschenreiter M, Giagnoni L, Arenella M, Varanini Z, Nannipieri P, Renella G (2015a) Enzyme activity and microbial community structure in the rhizosphere of two maize lines differing in N use efficiency. Plant Soil 387:413–424
Pathan SI, Ceccherini MT, Hansen MA, Giagnoni L, Ascher J, Arenella M, Sørensen SJ, Pietramellara G, Nannipieri P, Renella G (2015b) Maize lines with different Nitrogen Use Efficiency (NUE) also differ for molecular diversity of bacterial β-glucosidase gene and glucosidase activity in their rhizosphere. Biol Fertil Soils 51:995–1004
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL et al (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. PNAS 110:6548–6553
Pérez-Jaramillo JE, Carrión VJ, Bosse M, Ferrão LFV, de Hollander M, Garcia AAF, Ramírez CA, Mendes R, Raaijmakers JM (2017) Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J 10:2244–2257
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799
Podile AR, Kishore GK (2006) Plant growth-promoting rhizobacteria. In: Gnanamanickam SS (ed) Plant-associated Bacteria. Springer, Dordrecht, pp 195–230
Pugnaire FI, Armas C, Valladares F (2004) Soil as a mediator in plant-plant interactions in a semi-arid community. J Veg Sci 15:85–92
Qian JH, Doran JW, Walters DT (1997) Maize plant contributions to the root zone available carbon and microbial transformations of nitrogen. Soil Biol Biochem 29:1451–1462
Quiza L, St-Arnaud M, Yergeau E (2015) Harnessing phyto-microbiome signaling for rhizosphere microbiome engineering. Front Plant Sci 6:507
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009) The rhizosphere: a playground and battel filed for soil borne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Rillig MC, Mummey DL (2006) Mycorrhizas and soil structure. New Phytol 171:41–53
Rovira AD (1956) Plant root excretions in relation to the rhizosphere effect. I. The nature of root exudate from oats and peas. Plant Soil 7:178–194
Rovira A (1969) Plant root exudates. Bot Rev 35:35–57
Rudrappa T et al (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148:1547–1556
Safari SAA, Rashidi T (2012) The relationship between available P and selected biological properties in the rhizosphere of ten crop species under glasshouse conditions. Span J Soil Sci 2:74–89
Saharan BS, Nehra V (2011) Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res 21:1–30
Saleem M, Law AD, Moe LA (2016) Nicotiana roots recruit rare rhizosphere taxa as major root-inhabiting microbes. Microbial Ecol 71:469–472
Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136:2483–2499
Selosse MA, Richard F, He X, Simard S (2006) Mycorrhizal networks: les liaisons dangerousness. Trends Ecol Evol 11:621–628
Shen J, Li C, Mi G, Ki L, Yuan L, Jiang R, Zhang F (2013) Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. J Exp Bot 64:1181–1192
Shenton M, Iwamato C, Kureta N, Ikeo K (2016) Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition. Rice 9:42
Shukla KP, Sharma S, Singh NK, Singh V, Tiwari K, Singh S (2011) Nature and role of root exudates: efficacy in bioremediation. Afr J Biotechnol 10:9717–9724
Sihag SK, Singh MK, Meena RS, Naga S, Bahadur SR, Gaurav YRS (2015) Influences of spacing on growth and yield potential of dry direct seeded rice (Oryza sativa L.) cultivars. Ecoscan 9(1–2):517–519
Singh BK, Millard P, Whiteley AS, Murrell JC (2004) Unravelling rhizosphere– microbial interactions: opportunities and limitations. Trends Microbiol 12:386–393
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot R, Heuer H, Berg G (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751
Song F, Han X, Zhu X, Herbert S (2012) Response to water stress of soil enzymes and root exudates from drought and non-drought tolerant corn hybrids at different growth stages. Can J Soil Sci 92:501–507
Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2:178–186
Stoltz E, Greger M (2002) Cotton grass effects of trace elements in submersed mine tailings. J Environ Qual 31:1477–1483
Subbarao GV, Sahrawat KL, Nakahara K, Rao IM, Ishitani M, Hash CT, Kishii M, Bonnett DG, Berry WL, Lata JC (2013) A paradigm shifts towards low-nitrifying production systems: the role of biological nitrification inhibition (BNI). Ann Bot 112:297–316
Sytsma KJ, Morawetz J, Pires JC, Nepokroeff M, Conti E, Zjhra M, Hall JC, Chase MW (2002) Urticalean rosids: circumscription, rosid ancestry, and phylogenetics based on rbcL, trnL-F, and ndhF sequences. Am J Bot 89:1531–1546
Tak HI, Ahmad F, Babalola OO (2013) Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. In: Whitacre DM (ed) Reviews of environmental contamination and toxicology, vol 223. Springer, New York, pp 33–52
Thomas Danhorn, Clay Fuqua, (2007) Biofilm Formation by Plant-Associated Bacteria. Annual Review of Microbiology 61 (1):401-422
Tharayil N, Triebwasser DJ (2010) Elucidation of a diurnal pattern of catechin exudation by Centaurea stoebe. J Chem Ecol 36:200–204
Ueno D, Rombola AD, Iwashita T, Nomoto K, Ma JF (2007) Identification of two novel phytosiderophores secreted by perennial grasses. New Phytol 174:304–310
Uren NC (2000) Types, amounts, and possible functions of compounds released into the rhizosphere by soil-grown plants. In: Pinto R, Varanini Z, Nannipieri P (eds) The rhizosphere: biochemistry and organic substances at the soil-plant interface. Marcel Dekker, New York, pp 19–40
Vaartaja O (1975) Pythium sylvaticum in Canadian forest nurseries. Can Plant Dis Surv 55:101–102
Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moënne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dyé F, Prigent-Combaret C (2013) Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci 4:356
Van Loon LC (2007) Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol 119:243–254
Van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Varma D, Meena RS, Kumar S (2017) Response of mungbean to fertility and lime levels under soil acidity in an alley cropping system in Vindhyan Region, India. Int J Chem Stud 5(2):384–389
Verma JP, Meena VS, Kumar A, Meena RS (2015) Issues and challenges about sustainable agriculture production for management of natural resources to sustain soil fertility and health: a book review. J Clean Prod 107:793–794
Vidal EA, Gutiérrez RA (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11:521–529
Viveros OM, Jorquera MA, Crowley DE, Gajardo G, Mora ML (2010) Mechanism and practical considerations involved in plant growth promotion by rhizobacteria. J Soil Sci Plant Nutr 10:293–319
Walker TS, Bais HP, Grotewold E, Vivanco JM (2003) Root exudation and rhizosphere biology. Plant Physiol 132:44–51
Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi GM, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136:2512–2522
Wang AS, Angle JS, Chaney RL, Delorme TA, Reeves RD (2006) Soil pH effects on uptake of Cd and Zn by Thlaspi caerulescens. Plant Soil 281:325–337
Wang XR, Pan QA, Chen FX, Yan XL, Liao H (2011) Effects of coinoculation with arbuscular mycorrhizal fungi and rhizobia on soybean growth as related to root architecture and availability of N and P. Mycorrhiza 21:173–181
Wang ZH, Fang H, Chen M (2017) Effects of root exudates of woody species on the soil anti-erodibility in the rhizosphere in a karst region. China Peer J 5:e3029
Weiland J, Littke W, Haase D (2013) Forest nurseries face critical choices with the loss of methyl bromide fumigation. Calif Agric 67:153–161
Wu P, Wang X (2008) Role of OsPHR2 on phosphorus homeostasis and root hairs development in rice (Oryza sativa L.). Plant Signal Behav 3:674–675
Wu QS, Zou YN, Liu CY, Lu T (2012) Interacted effect of arbuscular mycorrhizal fungi and polyamines on root system architecture of citrus seedlings. J Integr Agric 11:1675–1681
Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182
Yadav GS, Lal R, Meena RS, Datta M, Babu S, Das LJ, Saha P (2017) Energy budgeting for designing sustainable and environmentally clean/safer cropping systems for rainfed rice fallow lands in India. J Clean Prod 158:29–37
Yadav GS, Das A, Lal R, Babu S, Meena RS, Saha P, Singh R, Datta M (2018) Energy budget and carbon footprint in a no-till and mulch based rice–mustard cropping system. J Clean Prod 191:144–157
Yang L (2016) Root exudation pattern of sugar beet (Beta vulgaris L.) as influenced by light intensity and P deficiency. PhD dissertation, University Göttingen
Young IM (1995) Variation in moisture contents between bulk soil and the rhizosheath of Triticum aestivum L. cv. New Phytol 130:135–139
Zaidi A, Khan MS, Ahemad M, Oves M (2009) Plant growth promotion by phosphate solubilizing bacteria. Acta Microbiol Immunol Hung 56:263–284
Zhang FS, Shen JB, Zhang JL, Zuo YM, Li L, Chen XP (2010) Rhizosphere processes and management for improving nutrient use efficiency and crop productivity: implications for China. In: Sparks DL (ed) Advances in agronomy, vol 107. Academic, San Diego, pp 1–32
Zhang R, Vivanco JM, Shen Q (2017) The unseen rhizosphere root-soil-microbe interaction for crop production. Curr Opin Microbiol 37:8–14
Zhuang X, Chen J, Shim H, Bai Z (2007) New advances in plant growth promoting rhizobacteria for bioremediation. Environ Int 33:406–413
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Pathan, S.I., Ceccherini, M.T., Sunseri, F., Lupini, A. (2020). Rhizosphere as Hotspot for Plant-Soil-Microbe Interaction. In: Datta, R., Meena, R., Pathan, S., Ceccherini, M. (eds) Carbon and Nitrogen Cycling in Soil. Springer, Singapore. https://doi.org/10.1007/978-981-13-7264-3_2
Download citation
DOI: https://doi.org/10.1007/978-981-13-7264-3_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-7263-6
Online ISBN: 978-981-13-7264-3
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)