Abstract
A myriad of small RNAs (18–25 nt in length) undergo heterogeneous modifications to inflect RNA stability and other complex physiological processes like stress responses, metabolism, immunity, and epigenetic inheritance of environmentally acquired traits. Such small RNAs include microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), and tRNA-derived small RNAs (tsRNAs). Worldwide crop production and human health are affected when plants are attacked by pathogens and pests. Therefore, a large collection of genes get up- or down regulated to mediate the defense responses in plants against pathogens (bacteria, fungi, oomycetes, and viruses). Host endogenous small RNAs, thus, come into play to counter biotic stress where RNA silencing machinery is utilized to facilitate pathogen-associated molecular pattern-triggered immunity and effector-triggered immunity. RNA interference (RNAi) pathways trigger gene silencing in interacting species from even different kingdoms (cross-kingdom RNAi). Diverse pathways are involved in regulating the defense mechanism including Dicer-like proteins (DCLs), double-stranded RNA (dsRNA) binding protein, RNA-dependent RNA polymerases (RDRs), RNA polymerase IV and V, small RNA methyltransferase HEN1, and Argonaute (AGO) proteins showcasing their functional specificities as well as verbosity. Transgenic plants are newly emerging players that help in solving the problem of pathogen attack in fields. In this chapter, the recent breakthrough on the function of sRNAs in response to biotic stress, mainly in plant-pathogen interaction, and its application in disease control is discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
8.1 Introduction
8.1.1 Zigzag Model
World population is increasing at a constant rate leading to agricultural land loss. This problem caters for diverse means to improve global food production. Another problem accounted for is the loss in crop productivity and grain quality due to bacteria, fungi, oomycetes, viruses, and insects. Therefore, it is required to unleash the biotic stress responses in plants and develop innovative tools using traditional and modern breeding approaches for crop protection against pathogens and pests (Bebber and Gurr 2015). On the contrary, pathogens have acquired the ability to counter such barriers to access nutrients and flourish inside plants thereby provoking their immune system. Nevertheless, plants have derived a defense mechanism to overcome pathogen infection by activating or suppressing a large array of genes (Jones and Dangl 2006). The “zigzag model” is proposed which explains in an easier way the different layers of innate immunity when plants are infected with pathogens (Jones-Rhoades et al. 2006). To avoid spreading infection by pathogen, the very first means of defense against them is pattern recognition receptors (PRRs). These receptors are cell surface-localized, transmembrane proteins and can detect conserved pathogenic patterns known as microbe−/pathogen−/host danger-associated molecular patterns (MAMPs or PAMPs or DAMPs) and hence shoot up the MAMP-/PAMP-triggered immunity (MTI/PTI) to limit the spread of pathogen (Jones and Dangl 2006). Flagellin peptide, elongation factor Tu protein (EF-Tu), and chitin are the best-studied MAMPs that form a major component of fungal cell walls and lipopolysaccharides (LPS). The perception of MAMPs relies on PRRs where FLS2 and EFR recognizing flagellin and EF-Tu possess to have a same structural construction formed by extracellular leucine-rich repeats (LRR) and a cytoplasmic kinase domain. On the contrary, CERK1, an Arabidopsis PRR, recognizes chitin containing three extracellular LysM domains and a cytoplasmic kinase domain. This recognition helps in inducing callose deposition, producing reactive oxygen species, accumulating salicylic acid (SA), and expressing pathogenesis-related (PR) genes (Yang and Huang 2014). Pathogens, on the other hand, have developed schemes to outpower MTI by sending effector proteins inside plant cells that abolish early recognition and downstream signaling events of MTI, therefore, resulting in effector-triggered susceptibility (ETS) (Feng and Zhou 2012). But plants too have emerged to protect themselves from this infection by using their resistance (R) proteins that recognize the specific effectors and activate effector-triggered immunity (ETI). This immune response is more sturdy and speedy (Chisholm et al. 2006). There is another hypersensitive response (HR), which causes cell death at the site of infection to restrain the growth of the pathogen. The effector proteins that are produced are called Avr factors. In the latter case, the R proteins [nucleotide binding site (NBS) and an LRR domain] guard the Avr factors and detect their modification caused by the effector proteins (Mackey et al. 2002). MAP kinase gets activated when pathogen’s molecules are perceived by PRRs or R proteins leading to a reprogramming in host’s gene expression along with the activation of genes with antimicrobial function (PR, pathogenesis related) (Tsuda and Katagiri 2010).
The war of defense and counter-defense between pathogens and plants has resulted in distinct collection of pathogen effectors and resistance genes.
8.2 Role of RNA
Posttranscriptional modifications are found extensively in stable and structured RNAs (tRNA and rRNA, mRNAs, and an expanding catalog of small and large noncoding RNAs) (Li and Mason 2014). Recent discovery of reversible 6-methyladenosine (m6A) modifications in mRNAs (Dominissini et al. 2012) as well as key enzymes for their dynamic regulation is observed. Other studies have documented pseudouridine (Li et al. 2015), 5-methylcytidine (m5C) (Hussain et al. 2013), and most recently, 1-methyladenosine (m1A) (Dominissini et al. 2016) in mRNAs. RNA modifications are also observed in small RNAs to perform various cellular functions that include development in plants, metabolic study, maintenance of genome integrity, immunity against pathogens, and abiotic stress responses. Regulation of gene expression is performed by small RNA in a sequence-specific manner either transcriptionally or posttranscriptionally (Chapman and Carrington 2007).
Eukaryotic organisms possess 20–40-nucleotide (nt)-long noncoding RNA molecules called small RNAs, and depending on their biogenesis and precursor structure, small RNAs are placed in two discrete groups: microRNAs (miRNAs) and small interfering RNAs (siRNAs) (Yang and Huang 2014).
8.2.1 MicroRNA
Small noncoding RNA generated from an imperfectly base-paired hairpin structure with 21–24 nt is called miRNA (Chen 2009). MicroRNAs (miRNAs) negatively regulate gene expression at the posttranscriptional level through mRNA degradation or translation repression (Iwakawa and Tomari 2013). Plant miRNAs are derived from the distinct noncoding transcripts of miRNA genes which are transcribed by enzyme RNA polymerase II. The primary miRNAs (pri-miRNAs) form a secondary fold-back structure and thereupon get processed by the RNase III-type enzyme Dicer-Like1 (DCL1) to create the precursor miRNAs (pre-miRNAs) (Rogers and Chen 2013). The miRNA duplexes once formed from pre-miRNA are stabilized by 2°-O-methylation and catalyzed by Hua Enhancer 1 (Yang et al. 2006) and transported to the cytoplasm by HASTY (Bollman et al. 2003). The passenger strand of the miRNA duplexes is often removed by unwinding or cleavage (Kawamata and Tomari 2010), and the guide strand is maintained in the RNA-induced silencing complex (RISC) that defines target recognition. Plant miRNAs exert a considerable effect on gene expression and mediate the cleavage of target mRNAs with near-perfect complementarity (Voinnet 2009).
8.2.2 SiRNA
Small interfering RNAs (siRNAs) are formed from near-perfect complementarity long double-stranded RNAs (dsRNAs) and are generated either from antisense transcription or by the action of RNA-dependent RNA polymerases (RDRs) (Katiyar-Agarwal and Jin 2010). There are many subclasses of siRNA present in plants depending on origin and biogenesis: trans-acting siRNAs (ta-siRNAs), heterochromatic siRNAs (hc-siRNAs), natural antisense transcript-derived siRNAs (nat-siRNAs), and long siRNAs (lsiRNAs).
8.3 RNA Silencing
Communication taking place between organisms whether pathogenic, parasitic, or symbiotic mediates the transport of regulatory molecules across the cellular boundaries between the host and its interacting pathogens/pests/parasites or symbionts. This triggers gene silencing in trans in the non-related species, a mechanism called cross-kingdom or cross-organism RNAi (Knip et al. 2014).
RNA interference (RNAi) is a gene silencing event that regulates sequence-specific gene and gets induced by double-stranded RNA (dsRNA). This results in inhibition of translation or transcription. Gene regulation is initiated by sRNAs in hosts or pathogens by posttranscriptional gene silencing (PTGS) or transcriptional gene silencing (TGS). PTGS is induced by miRNAs and siRNAs through messenger RNA (mRNA) cleavage/degradation or translational inhibition with the help of an RNA-induced silencing complex (RISC), while TGS is induced by siRNAs and some specific miRNAs. TGS is responsible for DNA methylation, histone modification, or chromatin modification (Cui and Cao 2014). A number of pathways are involved in producing regulatory small RNAs using various conserved protein families like the RNA-dependent RNA polymerases (RDRs), the double-stranded RNA-binding proteins (DRBPs), the Dicer-like proteins (DCLs), the small RNA methyltransferase (HEN1), and the Argonaute (AGO) proteins. Plant sRNAs and RNA interference (RNAi) pathway components are major regulatory players in providing immunity to plants against viruses, bacteria, fungi, oomycetes, and pests (Seo et al. 2013). Transposable element (TE) regions transcribe sRNAs in filamentous plant pathogens, and silencing this TE can help in fighting infection (Chang et al. 2012).
8.4 RNA Silencing Suppressors of Pathogens
8.4.1 Viral Suppressors of RNA Silencing
Many viruses cipher specific proteins to suppress the host antiviral silencing response and to cause infection in them. These viral suppressors of RNA silencing (VSRs) perform at three different levels, i.e., they can (a) inhibit generation of viRNAs, (b) inhibit loading of viRNAs in RISC by binding to the viRNA, and (c) inhibit components of RISC. Table 8.1 discusses the mode of action of VSRs in plants.
8.4.2 Bacteria-Encoded Suppressors of RNA Silencing
Bacterial pathogens too have developed similar silencing suppressors to combat antibacterial defense responses in plants as in viruses. Navarro et al. (2008) identified several Pst type III secretion effectors that enhance the disease susceptibility by suppressing host RNA silencing machinery. Effectors include AvrPtoB which represses transcription of miRNA genes and lowers the level of pri-miR393, AvrPto which interferes with miRNA precursor processing and downregulates mature miR393 level, and HopT1 which inhibits the action of the AGO1 protein in the RISC complex. Likewise, fungi and oomycetes too have developed RNAi suppressors to counteract host antipathogen RNA silencing mechanisms.
8.5 Host Endogenous Small RNAs in Plant-Microbe Interactions
When pathogen interacts with its host at first, it triggers the immunity response in plants known as pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI). Now, bacteria too rectify PTI by secreting and injecting effector proteins into plant cells leading to PTI suppression. Finally, host plant releases resistance components such as resistance (R) proteins that can recognize effectors and elicit effector-triggered immunity (ETI) (Chisholm et al. 2006). Unlike viruses that replicate inside the host cell, bacteria, fungi, and other microbes interact with plants without undergoing DNA or RNA replication and transcription inside the plant cell. In such interactions, host endogenous small RNAs play a pivotal role in counteracting these pathogens.
8.5.1 Noncoding Small RNAs
Small noncoding RNAs (sncRNAs), discovered in eukaryotes, are 18–30-nt-long molecules which perform numerous functions such as gene expression control, defense against other parasitic nucleic acids, epigenetic modification, and heterochromatin regulation (van der Krol et al. 1990). There are ample functions and beneficial applications reported so far. Few of them are encompassing cell-to-cell signaling and communication in multicellular organisms (Mittelbrunn and Sanchez-Madrid 2012), trans-generational RNAi (Bond and Baulcombe 2014) and memorization (Rasmann et al. 2012), cell fate differentiation and vascular formation (Benkovics and Timmermans 2014), systemic antiviral immunity (Saleh et al. 2009), environmental RNAi (Zhuang and Hunter 2012), cancer prevention and diagnosis (Salido-Guadarrama et al. 2014), and intercellular immune activation (Robbins and Morelli 2014). MicroRNAs (miRNAs) and small interference RNAs (siRNAs) are the best-studied sncRNAs.
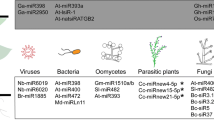
In response to different pathogen stressors, various targets and functions of sRNAs are summarized in Table 8.2.
8.6 Components of the Small RNA Biogenesis Pathway Play an Important Role in Plant Defense
Many plant genomes possess multiple components such as DCLs, RDRs, and AGOs in the RNAi silencing machinery. Arabidopsis has four DCLs, six RDRs, and ten AGOs, many of which are involved in plant defense signaling pathway.
8.6.1 Dicer-Like Proteins and Their Associated Proteins
Four DCLs present in Arabidopsis process dsRNA or fold-back RNA precursors to generate siRNAs and miRNAs, respectively. The role of DCLs and their compensatory functions in the production of virus-derived small RNAs (viRNAs) is well understood using single, double, or triple mutants of DCLs in genetic experiments. A loss of function mutation in both DCL4 and DCL2 is enough to cause viral susceptibility (+ssRNA) in plants (Diaz-Pendon et al. 2007).
Qu et al. (2008) observed that all four DCL proteins, key components of RNA silencing pathway, are involved in providing an antiviral defense in plants with functional hierarchy as (DCL4>DCL2>DCL3>DCL1) in processing viral RNAs into viRNAs (Deleris et al. 2006). Other important cofactors like small dsRNA-binding proteins (DRBs) of DCL proteins are known, but these do not show hierarchical redundancy as do DCLs (Curtin et al. 2008). DRB4 when interacting with DCL4 confers resistance against viruses (Qu et al. 2008). On the contrary, DCL2 and DCL3 do not need interaction with DRB for production of viRNAs (Curtin et al. 2008). Another protein HEN1 containing dsRNA binding domain plays an important role in viral resistance (Park et al. 2002). When mutation was done in hen1 of Arabidopsis, hyper-susceptibility to cauliflower mosaic virus (CMV) was observed in the plant as compared to wild type suggesting that HEN1 contributes to resistance against the virus (Boutet et al. 2003). Along with the abovementioned, DCL proteins are also involved in the production of small RNAs thereby giving antibacterial immunity in plants. The dcl1 mutant showed heightened susceptibility to Pst DC3000 hrcC−, a nonpathogenic strain that can evoke PTI (Navarro et al. 2008). HYL1, the dsRNA-binding protein associated with DCL1, is also involved in bacterial infection resistance as the hyl1 mutant was susceptible to Pst (avrRpt2).
8.6.2 RNA-Dependent RNA Polymerases
Elaborated studies have stated RDRs to be induced by antiviral defense as well as in the presence of defense signaling compounds such as salicylic acid (SA) (Xie et al. 2001). It was observed that when the expression levels of RDR1 are lowered in transgenic antisense Arabidopsis plants, viral RNAs get piled up and susceptibility to TMV and potato virus X (PVX) infection is increased. NtRDR1 is also involved in fighting against potato virus Y (PVY) infection and its ortholog AtRDR1 transmits defense against tobamovirus and tobravirus because Arabidopsis rdr1 mutant plants had enhanced levels of viral RNAs (Yu et al. 2003). A functional homolog of AtRDR6, NbRDR6, provides resistance against viruses (Qu et al. 2005) as downregulation of NbRDR6 increased the susceptibility to many different viruses at high temperatures.
8.6.3 Argonautes
Silencing of target genes is activated by AGOs as these are associated with small RNAs and form RISC complexes (Hannon 2002). In Arabidopsis, 10 AGOs are found to take part in plant immunity. hc-siRNAs promote transcriptional gene silencing (TGS) by guiding RNA-directed DNA methylation (RdDM) and histone modification in plants (Vaistij et al. 2002). AGO4 is a leading nuclear RNAi effector associated with hc-siRNAs or ra-siRNAs that allows DNA methylation (Li et al. 2008) which links DNA methylation and plant defense together. Using both cytosine and histone methyltransferases, Arabidopsis plants silence viral chromatin of cabbage leaf curl virus (CaLCuV) and beet curly top virus (BCTV) (Raja et al. 2008). Viral suppressors AL2 and L2 stop adenosine kinase (ADK) activity which otherwise generates S-adenosylmethionine (a methyltransferase cofactor). Therefore, plants infected with virus in the absence of L2 had hypermethylation of viral DNA, and to recover from viral infection, AGO4 is needed (Raja et al. 2008). AGO4 also helps in antibacterial defenses. In addition to AGO4, AGO1 and AGO7 play a pivotal role in slicing viral RNAs (Qu et al. 2008). AGO1 is the primary slicer because it targets viral RNAs with more compact structures, but AGO7 is an alternate slicer which targets RNAs with less complexity. The biogenesis of AtlsiRNA-1 involved AGO7, as ago7 mutant that does not accumulate AtlsiRNA-1 (Katiyar-Agarwal et al. 2006). However, other ago mutant plants, including ago3, ago4, and ago9, showed no significant change in the level of AtlsiRNA-1 as compared with wild type. AGO7 is also associated with TAS3 ta-siRNA (Fahlgren et al. 2006). AGO7 accumulates bacteria-induced AtlsiRNA-1 hence suggesting its role in antibacterial defense.
8.7 Cross-Kingdom RNAi and sRNA Trafficking
When two unrelated interacting organisms communicate with each other, it is called cross-kingdom RNAi. This process is observed in both animal and plant systems. Plants transfer RNAi signals into interacting organisms, such as filamentous fungi, oomycetes, nematodes, parasitic plants, and pests, to restrain their growth. This process is known as HIGS, the most noticeable example of cross-kingdom RNAi in plants (Koch et al. 2013). In order to develop pest- and pathogen-resistant crops, scientists have engineered diverse plant species, from model plants to commercial crops, so as to express exogenous artificial RNAi signals that suppress the gene of parasitic nematodes, herbivores, and fungal and oomycete pathogens by targeting their mRNAs (Koch and Kogel 2014). HIGS is functional and successfully used against parasitic plants such as Orobanche and Cuscuta spp. and in model plants such as Arabidopsis thaliana and tobacco Nicotiana benthamiana as well as in important crops, including wheat, barley, Medicago, and banana, to efficiently work against a variety of fungal and oomycete pathogens, such as Blumeria graminis, Puccinia tritici, Fusarium spp., and Phytophthora capsici (Koch and Kogel 2014). Basic mechanism of HIGS is that it alters the fungal morphology and growth inhibition in plants, thereby reducing virulence. Additionally, HIGS is also used to study gene function in non-transformable species (Yin et al. 2014). A HIGS approach was carried out on Glomus spp. to study gene function of the monosaccharide transporter 2 (Helber et al. 2011), showing that HIGS is functional on arbuscular mycorrhiza, which forms symbiotic relationship with hosts. Successfully applying HIGS helps plants to deliver mobile gene silencing signals for communication and manipulating diverse interacting organisms.
There are evidences of RNAi signaling taking place in the opposite direction. Advanced pathogens and parasites use cross-kingdom RNAi to suppress host immunity for infection (Weiberg et al. 2015). Three Bc-sRNAs in Botrytis-host interaction suppress Arabidopsis and tomato immunity genes in vivo (Mayoral et al. 2014). It is also estimated that sRNAs are also likely to be exchanged between the parasitic plant and its host, but the study still awaits the research output. Secretion and uptake of protein and other macromolecules participate in providing barrier against pathogens and parasites (Huckelhoven 2007) and in pathogenesis and effector-triggered suppression of host plant immunity (Kale and Tyler 2011).
8.8 Small RNA Biogenesis Pathways in Plants
Arabidopsis is taken as a model plant to study small RNA pathways in plants. Generative work involves both forward and reverse genetic screens to study the cellular proteins participating in biogenesis and function of miRNAs and siRNAs. A brief review of different kinds of small RNA pathways known in Arabidopsis is discussed below.
8.8.1 Biogenesis and Mechanism of miRNAs in Plants
The very first observation of microRNAs (miRNAs) took place in a nematode Caenorhabditis elegans (Lee et al. 1993). These are also known as short temporal RNAs (stRNAs) because they were expressed temporally in a mutant nematode. These endogenous noncoding small RNAs accelerate the growth, development, and survivability of plants. Transcription of miRNA gene is carried out by RNA polymerase II forming primary transcripts (pri-miRNAs) as a stem-loop structure of 1000-bp-long nucleotides (Chen 2005). Two processing steps are involved in the formation of mature miRNAs. The first step is carried out inside the nucleus where the microprocessor complex acts on pri-miRNAs to pre-miRNAs (precursor miRNAs) of 60–70 nt long. Two proteins, Drosha (169 kDa, RNAse III protein) and Pasha (dsRNA binding protein/DGCR8), constitute the microprocessor complex (Creelman and Mullet 1997). Two orthologs of Drosha and Pasha, namely, Dicer-like 1 (DCL-1) and Hyponastic Leaves 1 (HYL-1), are engaged in preliminary processing step of miRNA biogenesis pathway in plants (Schauer et al. 2002). To allow second processing step occuring in the cytoplasm, HASTY transport protein (ortholog of exportin-5) is required to transport pre-miRNAs from nucleus to cytoplasm. In subsequent step, ATP-dependent RNAse III protein (Dicer) converts hairpin dsRNA (pre-miRNA) into 21–24-nt-long mature miRNA-miRNA∗ duplex with 2-nt 3′ overhangs. This enzyme recognizes 2-nt 3′ overhangs and eliminates about ~21-nt sequence from its ends (Du et al. 2011). Out of two strands in miRNA duplex, one is called as antisense miRNA (miRNA) which has G:U base pairs, mismatches, and unpaired base pairs at its 5′ end, while the other strand is known as sense strand (miRNA∗). A complex is formed between Argonaute 1 (AGO1) protein and one strand of miRNA to guide miRNA to target its complementary mRNA sequence. The destiny of target mRNA depends on the degree of its complementarity with associated miRNA sequence. Complete degradation occurs from near-perfect complementarity, while repression of protein translation occurs from partial complementarity. This miRNA biogenesis pathway is under the feedback regulation by two principal miRNAs, miR162 and miR168, causing cleavage of DCL1 mRNA and AGO1 mRNA (Zhang et al. 2011), respectively.
The ability of miRNAs in crop improvement can be well documented as transgenic plants harbor miRNAs under constitutive and inducible promoters that can specifically downregulate target genes of interest with limited non-autonomous effect.
8.8.2 siRNA
Antisense transcription or cellular RNA-dependent RNA polymerase (RDR) is used to derive siRNAs. In plants, there are four discrete siRNAs present: trans-acting siRNAs (ta-siRNAs), natural antisense transcripts (NATs)-derived siRNAs (nat-siRNAs), heterochromatic siRNAs (hc-siRNAs) or repeat-associated siRNAs (ra-siRNAs), and long siRNAs (lsiRNAs). For the initiation of ta-siRNA formation, RNA Pol II transcribes noncoding TAS genes where long primary transcript products upon cleavage by miRNAs and RNA-induced silencing complexes (RISCs) produce a 5′ fragment or a 3′ fragment which acts as a template for complementary strand synthesis, also coordinated by RDR6 and SGS3 (Vazquez 2006). DCL4 and DRB4 act consecutively on dsRNA molecule to form ta-siRNAs (Gasciolli et al. 2005). Intersecting regions of sense and antisense transcripts of cis-NATs give rise to nat-siRNAs. RNA interference is exploited in order to accomplish desirable traits in crops by operating the gene expression (Table 8.3). After the identification of the target genes, RNAi construct with hairpin cassette was created. Plant transformation and later screening and traits evaluation take place.
8.8.3 miRNA vs. siRNA
The most important regulators of gene expression are microRNAs (miRNAs) and short-interfering RNAs (Vazquez 2006) having size of 20–24 nt long. The difference between the two lies in precursor structures, pathway of biogenesis, and modes of action (Axtell 2013) (Table 8.4). Both are processed from long RNA precursors by Dicer-like ribonucleases (Bernstein et al. 2001) and regulate the target gene repression (Hammond et al. 2000).
8.8.4 Transposon-Associated sRNAs in Eukaryotic Plant
Eukaryotic pathogens are capable of silencing TEs by producing transposable element (TE)-associated sRNAs. The transcription of sRNA effectors in Botrytis cinerea takes place via TEs to suppress host immunity-related genes. In return, host plant resistance (R) genes get clustered in genomic loci embellished with TEs. TEs show epigenetic control of R-gene expression by R-gene sRNAs. Likewise, pathogen protein effector genes occur as clusters and scatter with TEs. In another example, protein effector gene-derived sRNAs in Phytophthora spp. control the expression levels of effector. For both pathogen protein effector genes and host plant R genes, sRNAs play the crucial regulators assisted with TE transposition. TEs are a core source of sRNA production where pathogens allow regulation of TEs and TE-associated protein effector gene expression by sRNAs, delivering sRNA effectors into host cells to change host defense gene expression. In plants, the advent of sRNAs upon infection epigenetically controls R-gene expression thereby activating defense genes. There are chances that plants may deliver their own RNA or protein molecules into pathogen cells. These events affect plant-pathogen interaction to provide host resistance, pathogen virulence, and host adaptation.
8.9 sncRNAs and Viruses: New Frontiers of Defense
For universal gene expression changes, current studies affirm the use of sncRNAs in plant-virus interactions. It has been proposed that plant miRNA expression that targets plant transcripts changes its response virus recognition affecting both viral replication and spreading. Numerous plant miRNAs after viral infection get either up- or downregulated (Pacheco et al. 2012). For example, when turnip mosaic virus infects Brassica rapa, miR1885 is induced in its response and targets a TIR-NBS-LRR (TNL) disease resistance gene (He et al. 2008).
8.10 Biotic Stress Resistance
Ample economic loss is posed by plant pathogens due to depletion in crop production. Therefore, several RNAi strategies are on the board to provide improvement in crop defense mechanisms against various biotic stresses (viruses, bacteria, fungi, nematodes, and insects).
8.10.1 Virus Resistance
Virus-induced gene silencing (VIGS) is an RNA-mediated PTGS mechanism that allows plants to protect themselves from foreign gene invasion (Ding 2010). Pathogen-derived resistance (PDR) provides plants resistance against virus through genetic engineering (Simon-Mateo and García 2011). This PDR is either protein mediated where transgene encodes the protein or RNA mediated where transgene forms the transcript. To attain PDR, hairpin dsRNAs including small hairpin RNA (shRNA), self-complementary hpRNA, and intron-spliced hpRNA are produced in vivo using inverse repeat sequences from viral genomes. This approach was used successfully to anchor resistance in cassava plants against African cassava mosaic virus (ACMV) (Vanderschuren et al. 2009). Another means of providing resistance against viruses is targeting the coat protein (CP) gene through RNAi. This strategy was shown by Powell-Abel et al. (2006) in transgenic tobacco expressing the CP gene of tobacco mosaic virus (TMV) thus giving resistance to TMV. This method was further utilized to generate resistance against many different viruses such as potato resistant to potato virus Y (PVY) (Missiou et al. 2004), tobacco resistant to beet necrotic yellow vein virus (BNYVV) (Andika et al. 2005), Cucumis melo resistant to papaya ring spot virus type W (PRSV-W) (Krubphachaya et al. 2007), N. benthamiana resistant to cucumber green mottle mosaic virus (CGMMV) (Kamachi et al. 2007), and N. benthamiana and Prunus domestica resistant to plum pox virus (PPV) (Hily et al. 2007). RNA silencing approach is not restricted to RNA viruses alone but also seen in DNA viruses. For example, following infection with geminivirus Vigna mungo yellow mosaic virus (VMYMV), blackgram plant recovers back when inoculated with hpRNA construct containing the promoter sequence of VMYMV under the control of the 35S promoter (Pooggin et al. 2003). On the advent of infection by turnip mosaic virus (TuMV) in Brassica rapa, two miRNAs, bra-miR158 and bra-miR1885, were greatly upregulated (He et al. 2008), the condition only seen in this particular interaction.
8.10.2 Bacterial Resistance
Bacteria spread at a speedy rate and therefore it is tough to control diseases caused by them. Supression of two genes of Agrobacterium tumefaciens carried out by RNAi involved in crown gall tumor formation (iaaM and ipt) also helps in reducing the production of tumors in Arabidopsis (Dunoyer et al. 2006). This approach could be further spread out to other plants. Resistance to plants from bacterial disease is negatively regulated by fatty acids and their derivatives (Jiang et al. 2009). Multiple pathogens can be resisted in Arabidopsis and soybean plants by RNAi-mediated suppression of SACPD gene that encodes for fatty acid desaturase (Jiang et al. 2009). In Arabidopsis, miR393 is said to repress auxin signaling by negatively regulating the F-box auxin receptors like TIR1, hence restricting the infection by bacteria Pseudomonas syringae (Navarro et al. 2006). Thus, transgenic Arabidopsis plants where miR393 is overexpressed have enhanced bacterial resistance with some developmental alterations (Navarro et al. 2006). But two different miRNAs, miR398 (Jagadeeswaran et al. 2009) and miR825 (Fahlgren et al. 2007), are said to be downregulated by bacterial infections. miR398 expression targets coding for two Cu/Zn superoxide dismutases that are CSD1 and CSD2 were analyzed, and it was observed that CSD1 was upregulated on the outburst of bacterial infection in accordance with the downregulation of miR398 under biotic stress (Jagadeeswaran et al. 2009).
MiR482/2118 family of miRNAs were shown to target a number of NBS-LRR mRNAs encoding disease resistance proteins in tomato (Solanum lycopersicum) and other members of Solanaceae (Shivaprasad et al. 2012). MiR482-mediated silencing of R genes gets affected by viral and bacterial invasion. These miRNAs are either upregulated or downregulated and affect gene expression by either suppressing negative regulators or inducing positive regulators of immune responses.
8.10.3 Fungal Resistance
Fungal resistance is regulated by posttranscriptional gene silencing (PTGS). In Arabidopsis RNA silencing mutants sgs2, sgs3, ago7, dcl4, nrpd1a, and rdr2 displayed exhibited heightened susceptibility to Verticillium strains (Ellendorff et al. 2009). In another example, RNAi-mediated suppression of a rice gene OsSSI2 embellished resistance to blast fungus Magnaporthe grisea and leaf blight bacterium Xanthomonas oryzae (Jiang et al. 2009) by suppressing two genes, namely, OsFAD7 and OsFAD8 (omega-3 fatty acid desaturases) (Yara et al. 2007). Similarly, RNAi-mediated targeting of genes for lignin production led to enhanced resistance in soybean against phytopathogen Sclerotinia sclerotiorum due to reduced lignin content (Peltier et al. 2009). However, in case of wheat, 24 miRNAs are known to get affected by the fungus Blumeria graminis f. sp. tritici (Bgt) which is causing the deadly disease of wheat powdery mildew (Xin et al. 2010). On the other hand, rice miRNA osa-miR7695 negatively regulates a natural resistance-associated macrophage protein 6 (OsNramp6) against the blast fungus Magnaporthe oryzae. To overcome this disease, overexpression of Osa-miR7696 was carried out (Campo et al. 2013).
8.11 Biotechnological Use of Mobile sRNAS in Plants
Plant defenses against pathogens and pests get accelerated by the discovery of sRNAs as mobile gene regulators thereby providing alluring and new strategies for crop improvement (Koch and Kogel 2014). HIGS, too, has played a great role in efficiently providing resistance against distinct plant herbivores, nematodes, and filamentous pathogens, when targeting important virulence genes. HIGS is a well-known tool under controlled lab conditions when applied to specific host and definite pathogen, but in field conditions, their suitability is compromised due to fluctuating environmental stresses and humungous variation in genes of pathogen and pest populations. Thus, more advanced studies and experimentation are needed to carry forward. Transportation of sRNA in different interactions such as plant-pathogen, plant-parasite, or plant-symbiont has made it feasible to construct the beneficial fungi or disarmed pathogens (with essential virulence genes deleted) and alter plant physiology via trans-kingdom gene silencing. Moreover, when the target pathogen mRNAs are emphasized, a broad range of pathogens and pests can be controlled in a transgene-free plant framework via RNAi signals. RNA silencing-based technique can be further strengthened when a decent knowledge on molecular mechanisms of RNA communications and transport between plants and interacting organisms is attained. While genetically engineered crops have always been under domain of public eye, an understanding of cross-kingdom RNAi may help relieve public concerns. Some more applications of mobile sRNAs in plants are in metabolic engineering and systemic-induced resistance (Saurabh et al. 2014). Even food RNAi might become an important part of plant food-based technologies in the future (Hirschi 2012). Feeding studies stated that oral uptake of sRNA-containing nutrients led to accumulation of food-borne sRNAs in body fluids and organs, indicating their partial survival inside the intestinal tract (Liang et al. 2014). Research is ongoing to see if food-borne sRNAs have any negative or positive impacts on the physiology of the individual who consumes foods with plentiful sRNAs (Dickinson et al. 2013).
8.12 Conclusion
Research in sncRNAs is ultimately one of the most effective and encouraging fields in plant defense biology, and many more advances are waiting to be explored in this area of research. A large number of studies discussed here emphasize on the significance of sncRNAs in gene regulation in response of plants to pathogens (viruses, bacteria, and fungi). The induction and repression of sncRNAs in plants toward pathogens depend upon the incompatible and compatible interactions indicating that these RNAs can both act as positive and negative regulators of plant immunity. Biotechnological tools and strategies need to be implemented to speed up the resistance studies in plants against various pathogens. During symbiotic interactions, relevance of repression of R genes provides a bridge between pathogenic and beneficial interactions. When effectors interact with the plant silencing machinery, pathogens can surpass the plant immunity mechanisms. Since the complete annotation of sequence of miRNAs involved in biotic stresses still needs to be carried out in crop plants like rice, maize, soybean, mustard, Jatropha, barrelclover, etc., genes of small RNAs (miRNAs) can be used for analysis of stress tolerance in biotic conditions. Computational methods and high-throughput techniques like miRNA microarray, real-time PCR, or northern blot are utilized to identify expressed miRNAs and their target(s) which provide plant defense against various biotic stresses. Studying the complexity of regulation these proteins had to undergo in order to provide crops resistance against pathogens is required. Comparing the antiviral and the antibacterial roles of the small RNA biogenesis factors may shed light on the complex modes of regulation these proteins have to undergo to confer plants’ disease resistance. The study of VSRs and BSRs along with their targets may help to solve redundancy in the activity of several RNA silencing components during plant-microbe interactions. An insight into plant defense mechanisms will help to improvise crops of economic importance which should be pathogen-free too.
References
Andika IB, Kondo H, Tamada T (2005) Evidence that RNA silencing-mediated resistance to beet necrotic yellow vein virus is less effective in roots than in leaves. Mol Plant-Microbe Interact 18:194–204
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159
Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, Vaughn T, Roberts J (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326
Bonfim K, Faria JC, Nogueira EO, Mendes EA, Aragao FJ (2007) RNAi-mediated resistance to bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol Plant-Microbe Interact 20:717–726. https://doi.org/10.1094/MPMI-20-6-0717
Bebber DP, Gurr SJ (2015) Crop-destroying fungal and oomycete pathogens challenge food security. Fungal Genet Biol 74:62–64
Benkovics AH, Timmermans MC (2014) Developmental patterning by gradients of mobile small RNAs. Curr Opin Genet Dev 27:83–91
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A et al (2006) Four plant dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34:6233–6246
Bollman KM, Aukerman MJ, Park MY et al (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130:1493–1504
Bond DM, Baulcombe DC (2014) Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol 24:100–107
Boutet S, Vazquez F, Liu J, Beclin C, Fagard M et al (2003) Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus re-sistance. Curr Biol 13:843–848
Campo S, Peris-Peris C, Sire C, Moreno AB, Donaire L, Zytnicki M et al (2013) Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol 199:212–227
Chang SS, Zhang Z, Liu Y (2012) RNA interference pathways in fungi: mechanisms and functions. Annu Rev Microbiol 66:305–323
Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8:884–896
Chellappan P, Vanitharani R, Fauquet CM (2005) MicroRNA-binding viral protein interferes with Arabidopsis development. PNAS 102:10381–10386
Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579:5923–5931
Chen XM (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44
Chen J, Li WX, Xie D, Peng JR, Ding SW (2004) Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16:1302–1313
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124(4):803–814
Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48:355–381
Cuellar WJ, Kreuze JF, Rajamaki ML, Cruzado KR, Untiveros M, Valkonen JP (2009) Elimination of antiviral defense by viral RNase III. Proc Natl Acad Sci U S A 106(25):10354–10358
Cui X, Cao X (2014) Epigenetic regulation and functional exaptation of transposable elements in higher plants. Curr Opin Plant Biol 21:83–88
Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM (2008) The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett 582:2753–2760
Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68–71
Diaz-Pendon JA, Li F, Li WX, Ding SW (2007) Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19(6):2053–2063
Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS (2013) Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 31:965–967
Ding SW (2010) RNA-based antiviral immunity. Nat Rev Immuno 10:632–644
Dominissini D et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206
Dominissini D et al (2016) The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530:441–446
Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, Wei L, Li Y (2011) Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog 7:e1002176
Dunoyer P, Himber C, Voinnet O (2006) Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat Genet 38(2):258–263
Ellendorff U, Fradin EF, de Jonge R, Thomma BP (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot 60:591–602
Enrique R, Siciliano F, Favaro MA, Gerhardt N, Roeschlin R, Rigano L, Sendin L, Castagnaro A, Vojnov A, Marano MR (2011) Novel demonstration of RNAi in citrus reveals importance of citrus cal-lose synthase in defence against Xanthomonas citri subsp. citri. Plant Biotechnol J 9:394–407
Eschen-Lippold L, Landgraf R, Smolka U, Schulze S, Heilmann M, Heilmann I, Hause G, Rosahl S (2012) Activation of defense against Phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol 193:985–996
Escobar MA, Civerolo EL, Summerfelt KR, Dandekar AM (2001) RNAi-mediated oncogene silencing confers resistance to crown gall tumorigen-esis. Proc Natl Acad Sci U S A 98:13437–13442
Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK et al (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16:939–944
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2(2):e219
Feng F, Zhou JM (2012) Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 15:469–476
Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15:1494–1500
Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C (2007) Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol 48(7):1050–1060
Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M et al (2008) Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J 27:2102–2112
Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296
Hannon GJ (2002) RNA interference. Nature 418(6894):244–251
He XF, Fang YY, Feng L, Guo HS (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582(16):2445–2452
Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N (2011) A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 23:3812–3823
Hily JM, Ravelonandro M, Damsteegt V, Basset C, Petri C, Liu Z et al (2007) Plum pox virus coat protein gene intron-hair pin-RNA (ihpRNA) con-structs provide resistance to Plum pox virus in Nicotiana bethamiana and Prunus domestica. J Am Soc Hortic Sci 132:850–858
Hirschi KD (2012) New foods for thought. Trends Plant Sci 17:123–125
Huang G, Allen R, Davis EL, Baum TJ, Hussey RS (2006) Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci U S A 103:14302–14306
Huckelhoven R (2007) Transport and secretion in plant-microbe interactions. Curr Opin Plant Biol 10:573–579
Hussain S et al (2013) Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol 14:215
Iwakawa H, Tomari Y (2013) Molecular insights into microRNA-mediated translational repression in plants. Mol Cell 52:591–601
Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229:1009–1014
Jiang CJ, Shimono M, Maeda S, Inoue H, Mori M, Hasegawa M, Sugano S, Takatsuji H (2009) Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant Microbe Int 22:820–829
Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kamachi S, Mochizuki A, Nishiguchi M, Tabei Y (2007) Transgenic Nicotiana benthamiana plants resistant to cucumber green mottlemosaic virus based on RNA silencing. Plant Cell Rep 26(1283):1288
Kale SD, Tyler BM (2011) Entry of oomycete and fungal effectors into plant and animal host cells. Cell Microbiol 13:1839–1848
Katiyar-Agarwal S, Jin H (2010) Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol 48:225–246
Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A Jr, Zhu JK, Staskawicz BJ, Jin H (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A 103:18002–18007
Kawamata T, Tomari Y (2010) Making RISC. Trends Biochem Sci 35:368–376
Knip M, Constantin ME, Thordal-Christensen H (2014) Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet 10:e1004602
Koch A, Kogel KH (2014) New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol J 12:821–831
Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel KH (2013) Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci U S A 110:19324–19329
Krubphachaya P, Jurícek M, Kertbundit S (2007) Induction of RNA mediated resistance to papaya ring spot virus type. W J Biochem Mol Biol 40:401–411
Lee RC, Feinbaum RL, Ambros V (1993) The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Li S, Mason CE (2014) The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet 15:127–150
Li CF, Henderson IR, Song L, Fedoroff N, Lagrange T, Jacobsen SE (2008) Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet 4:e27
Li DH, Liu H, Yang Y, Zhen PP, Liang JS (2009) Down-regulated expression of RACK1 gene by RNA interference enhances drought tolerance in rice. Rice Sci 16:14–20
Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C (2015) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11:592–597
Liang G, Zhu Y, Sun B, Shao Y, Jing A, Wang J, Xiao Z (2014) Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr 2:380–388
Mackey D, Holt BF, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108(6):743–754
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313
Mayoral JG, Hussain M, Joubert DA, Iturbe-Ormaetxe I, O’Neill SL, Asgari S (2014) Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc Natl Acad Sci U S A 111:18721–18726
Missiou A, Kalantidis K, Boutla A, Tzortzakaki S, Tabler M, Tsagris M (2004) Generation of transgenic potato plants highly resistant to potato virus Y (PVY) through RNA silencing. Mol Breed 14:185–197
Mittelbrunn M, Sanchez-Madrid F (2012) Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 13:328–335
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Navarro L, Jay F, Nomura K et al (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321:964–967
Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD et al (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24:1420–1428. https://doi.org/10.1038/nbt1255
Pacheco R, Garcia-Marcos A, Barajas D, Martianez J, Tenllado F (2012) PVX-potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res 165(2):231–235
Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495
Park GG, Park JJ, Yoon J, Yu SN, An G (2010) A ring finger ligase gene, Oryza sativa delayed seed germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol Bio 74:467–478
Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B et al (2006) F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci U S A 103:1994–1999
Peltier AJ, Hatfield RD, Grau CR (2009) Soybean stem lignin concen expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6:3343–3353
Pooggin M, Shivaprasad PV, Veluthambi K, Hohn T (2003) RNAi target-chalcone synthase gene into Petunia results in reversible cosuppression of ing of DNA virus in plants. Nat Biotechnol 21:131–132
Powell-Abel P, Nelson RS, De B, Hoffmann N, Rogers SG, Fraley RT (2006) A plant miRNA contributes to antibacterial resistance by repressing delay of disease development in transgenic plants that express the auxin signaling. Science 312:436–439
Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ (2005) RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol 79:15209–15217
Qu F, Ye X, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci U S A 105:14732–14737
Raja P, Sanville BC, Buchmann RC, Bisaro DM (2008) Viral genome methylation as an epigenetic defense against geminiviruses. J Virol 82:8997–9007
Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G (2012) Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158:854–863
Riechen J (2007) Establishment of broad-spectrum resistance against Blumeria germination and stress responses in rice MLO. J Verbrauch Lebensm 2:120
Robbins PD, Morelli AE (2014) Regulation of immune responses by extracellular vesicles. Nat Rev Immunol 14:195–208
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R (2009) Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458:346–350
Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodriguez-Dorantes M (2014) MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther 7:1327–1338
Saurabh S, Vidyarthi AS, Prasad D (2014) RNA interference: concept to reality in crop improvement. Planta 239:543–564
Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7:487–491
Seo JK, Wu J, Lii Y, Li Y, Jin H (2013) Contribution of small RNA pathway components in plant immunity. Mol Plant-Microbe Interact 26:617–625
Shimizu T, Yoshii M, Wei T, Hirochika H, Omura T (2009) Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of Rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol J 7:24–32
Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24:859–874
Simon-Mateo C, García JA (2011) Antiviral strategies in plants based on RNA silencing. Biochim Biophys Acta 1809:722–731. https://doi.org/10.1016/j.bbagrm.2011.05.011
Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ et al (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J Virol 79:2517–2527
Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 3(4):459–465
Vaistij FE, Jones L, Baulcombe DC (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14:857–867
Van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2(4):291–299
Vanderschuren H, Alder A, Zhang P, Gruissem W (2009) Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol Biol 70:265–272
Vazquez F (2006) Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci 11:460–468
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wang M, Abbott DC, Waterhouse PM (2000) A single copy of a virus-derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol Plant Pathol 1:347–356
Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79:7410–7418
Wang Y, Li P, Cao X, Wang X, Zhang A, Li X (2009) Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem Biophys Res Commun 378(4):799–803
Wang T, Chen L, Zhao M, Tian Q, Zhang WH (2011a) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12:367–378
Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW (2011b) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell 23:1625–1638
Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H (2015) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123
Wu HW, Lin SS, Chen KC, Yeh SD, Chua NH (2010a) Discriminating mutations of HC-pro of zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol Plant Microbe Interact 23(1):17–28
Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y (2010b) DNA methylation mediated by a microRNA pathway. Mol Cell 38:465–475
Xie Z, Fan B, Chen C, Chen Z (2001) An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc Natl Acad Sci U S A 98:6516–6521
Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z (2010) Diverse set of micro RNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10:123–134. https://doi.org/10.1186/1471-2229-10-123
Yadav BC, Veluthambi K, Subramaniam K (2006) Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol Biochem Parasitol 148:219–222
Yang L, Huang H (2014) Roles of small RNAs in plant disease resistance. J Integr Plant Biol 56:962–970
Yang Z, Ebright YW, Yu B et al (2006) HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34:667–675
Yara A, Yaeno T, Hasegawa M, Seto H, Montillet JL, Kusumi K et al (2007) Disease resistance against Magnaporthe grisea is enhanced in transgenic rice with suppression of o-3 fatty acid desaturases. Plant Cell Physiol 48:1263–1274
Yin C, Park JJ, Gang DR, Hulbert SH (2014) Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol Plant Microbe Interact 27:227–235
Yu D, Fan B, MacFarlane SA, Chen Z (2003) Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol Plant-Microbe Interact 16:206–216
Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H (2011) Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(∗)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell 42:356–366
Zhuang JJ, Hunter CP (2012) RNA interference in Caenorhabditis elegans: uptake, mechanism, and regulation. Parasitology 139:560–573
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Singh, M.P., Singh, P., Singh, R.K., Sayyed, R.Z., Sharma, A. (2019). Plant Small RNAs: Big Players in Biotic Stress Responses. In: Sayyed, R. (eds) Plant Growth Promoting Rhizobacteria for Sustainable Stress Management . Microorganisms for Sustainability, vol 13. Springer, Singapore. https://doi.org/10.1007/978-981-13-6986-5_8
Download citation
DOI: https://doi.org/10.1007/978-981-13-6986-5_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-6985-8
Online ISBN: 978-981-13-6986-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)