Abstract
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been used for two decades to profile the glycan constituents of biological samples. An adaptation of the method to tissues, MALDI mass spectrometry imaging (MALDI-MSI), allows high-throughput spatial profiling of hundreds to thousands of molecules within a single thin tissue section. The ability to profile N-glycans within tissues using MALDI-MSI is a recently developed method that allows identification and localization of 40 or more N-glycans. The key component is to apply a molecular coating of peptide-N-glycosidase to tissues, an enzyme that releases N-glycans from their protein carrier. In this chapter, the methods and approaches to robustly and reproducibly generate two-dimensional N-glycan tissue maps by MALDI-MSI workflows are summarized. Current strengths and limitations of the approach are discussed, as well as potential future applications of the method.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Formalin-fixed paraffin-embedded tissue
- Fucosylation
- Glycomics
- Glycoprotein
- MALDI imaging mass spectrometry
- N-linked glycosylation
- Sialylation
4.1 Introduction
MALDI-MSI methods have been routinely utilized for spatial profiling and identification of proteins, lipids, and small metabolites directly in tissue (Angel and Caprioli 2013; Baker et al. 2017; Berin and Boughton 2017; Cornett et al. 2007). In 2013, our group was the first to report the use of MALDI-MSI to profile N-linked glycans in frozen tissues (Powers et al. 2013), followed by application of the method to formalin-fixed paraffin-embedded (FFPE) tissue blocks and tissue microarrays (TMA) (Powers et al. 2014; Drake et al. 2017). The key to the approach is the spraying of a molecular layer of recombinant peptide N-glycosidase F enzyme (PNGase F) on the tissue of interest. After digestion and spraying of chemical matrix, the released N-glycans are detected by MALDI-FT-ICR MS or MALDI-TOF MS (Drake et al. 2017). Different imaging software options are used to visualize the two-dimensional distributions of individual N-glycans to specific histopathology regions and features within normal and diseased tissues. In addition to studies from our laboratories in cancer tissues (Powers et al. 2014, 2015; Drake et al. 2017) and cardiac tissues (Angel et al. 2017a), other groups have successfully used the approach to profile N-glycans in cancer (Everest-Dass et al. 2016) and noncancer tissues (Briggs et al. 2016; Gustafsson et al. 2015).
As the method has matured, several advantages of the approach are emerging, as well as continued challenges. The ability to use FFPE tissues and TMA obtained directly from pathology services and/or archives in biorepositories allows for analysis of unlimited sample options, especially for most types of cancers. The requirement for PNGaseF to release the N-glycans results in signal detection that is dependent on enzyme activity, making selection of N-glycans in the spectra straightforward. No purification and enrichment of the glycans are required prior to analysis. For most tissue samples analyzed thus far, 30 or more glycans are detected using standard MALDI-TOF instruments (Heijs et al. 2016; Holst et al. 2016) and 45 or more per sample when using a MALDI-FT-ICR instrument (Drake et al. 2017). This in turn facilitates the generation of different glycan panels associated with specific histopathology features and tissue subregions useful for differentiation of disease from normal regions. There are still limitations associated with MALDI analysis of N-glycans, such as loss of sialic acid residues and differentiation of isomeric structures. Some progress has been made in addressing these limitations as applied to MALDI-MSI of tissues, and these will be discussed in this chapter along with example data. A summary of the optimized techniques used for MALDI-MSI of N-glycans will be provided with example data, along with protocol and troubleshooting notes for new users. Lastly, a summary of new instrumentation options and potential for developing new clinical diagnostics using the method will be described.
4.2 Characterization of Tissue N-Glycans by MALDI-FT-ICR MS and MALDI-TOF MS
The MALDI-MSI workflows build on decades of research using MALDI-TOF MS as a characterization tool for N-glycans (Harvey 1999, 2015; Nishikaze 2017). An experimental flowchart of the basic steps in the MALDI-MSI analysis of N-glycans is shown in Fig. 4.1, and specific experimental details can be found in published studies (Angel et al. 2017b; Briggs et al. 2017; Holst et al. 2016). Relative to electrospray ionization (ESI) MS methods, the advantages of using MALDI-TOF for N-glycan analysis include ease of use and data interpretation, rapidity and throughput, low cost per assay, and accessibility to instrumentation (Nishikaze 2017). For all MALDI analyses, one of the key considerations is chemical matrix type, which influences ionization properties of the N-glycans. For N-glycans, 2,5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA) are the most commonly used in positive ion mode, with CHCA being used primarily by our group (Powers et al. 2014).
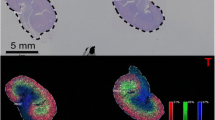
Instrumentation choice is also another key factor, as there are two main groups of instruments for MALDI-IMS: time of flight (TOF) and Fourier transform ion cyclotron resonance (FT-ICR). MALDI-TOF instruments provide high throughput with lower mass resolution, while MALDI-FT-ICR instruments provide a higher mass resolution with less throughput capabilities. The mass resolution capabilities of MALDI-FT-ICR instruments are critical to distinguish the many isotopic peaks of the N-glycans, with on-tissue resolving power of 85,000–160,000, with high sensitivity detection of below 200 femtomoles on-tissue. Recently, advances in instrumentation have resulted in commercially available high-speed MALDI-TOF instrumentation for high-throughput image acquisition from manufacturers like Bruker Daltonics, Shimadzu, and SimulTOF. These high-speed MALDI-TOF can perform MS tissue imaging with a 50x increase in speed over the MALDI-FT-ICR MS without oversampling (Ogrinc Potočnik et al. 2015). As an example, a comparison of one of these new high-speed instruments, the rapifleX MALDI-TOF and MALDI-FT-ICR, is shown in Fig. 4.2 for N-glycan imaging analysis of the same esophageal cancer tissue for a complex bi-antennary N-glycan. High-speed MS imaging should allow for faster 3D spatial rendering of a tissue, as well as potential clinical laboratory applications.
High-speed MALDI-TOF (rapifleX, Bruker) versus MALDI-FT-ICR (solariX Bruker) IMS for N-linked glycans for serial esophageal cancer tissue. RapifleX IMS data (left panel) was acquired at a 50 μm spatial resolution in 2.5 h and SolariX IMS data (right panel) at a 200 μm spatial resolution in 4.1 h. For glycans, red triangle, fucose ; blue square, N-acetylglucosamine ; green circles, mannose ; yellow circles, galactose ; and purple diamonds, sialic acid
4.2.1 Protocol Notes
-
Antigen retrieval of FFPE samples prior to PNGaseF was adapted directly from routine tissue slide protocols used in immunohistochemistry analyses. We have found that a low-cost vegetable steamer available at many retail stores works well for the heating steps during antigen retrieval (Angel et al. 2017b).
-
For PNGaseF enzyme, be sure that there are minimal additives in the buffer (e.g., stabilizers, detergent) and low salt to minimize potential ion suppression effects.
-
A key to MALDI-MSI is the spraying of a thin layer of PNGaseF for digestion and a thin layer of matrix. The automated solvent sprayers typified by those available from HTX Imaging and SunChrom provide reproducibility from sample to sample, as well as optimization of signal detection parameters.
4.3 Structural Classes of Tissue N-Glycans Detected by MALDI-MSI
There are many diverse types and structural classes of N-glycans that are readily detected from tissues by MALDI-MSI. Evaluation of structures is done based on motif and context of the N-linked glycan biosynthesis and processing pathways. A summary of the major components of these pathways is shown in Fig. 4.3. The diversity of the many possible N-glycan structures that are attached to glycoproteins is the result of a series of sequential glycosidase and glycosyltransferase reactions (reviewed in Rini et al. 2009; Stanley et al. 2009). After transfer of a lipid-linked precursor oligosaccharide co-translationally to a glycoprotein carrier, the glycan is trimmed sequentially in the ER and cis-Golgi by glucosidases and mannosidases to Man5GlcNAc2, which serves as the precursor to complex, branched, and hybrid glycan structures (see Fig. 4.3). Glycans that are not processed or incompletely processed by the mannosidases are termed high-mannose glycans, containing 5–9 mannose residues (Man5–9GlcNAc2). To generate complex glycans, Man5GlcNAc2 is processed by a series of N-acetylglucosaminyltransferases to generate an initial bi-antennary complex glycan. Fucosyltransferase 8 (FUT8) catalyzes the addition of α1,6-linked fucose residues to the first GlcNAc residue bonded with asparagine on the glycoprotein (termed core fucosylation ). Triantennary complex glycans are generated from the activity of N-acetylglucosaminyltransferase IV (GnT-IV), which adds a β1,4 GlcNAc branch, and tetra-antennary complex glycans are generated in turn from the activity of N-acetylglucosaminyltransferase V (GnT-V), which adds a β1,6 GlcNAc branch. N-acetylglucosaminyltransferase III (GnT-III) is an additional enzyme that transfers a bisecting GlcNAc residue onto the complex glycan (Fig. 4.3). In the trans-Golgi , further glycans are added to bi-, tri-, or tetra-antennary structures by the activity of different galactosyltransferases, fucosyltransferases (Schneider et al. 2017), and sialyltransferases (Pearce and Läubli 2016). Another common type of complex structure that is detected in many tissues is polylactosamine (LacNAc) glycans (Bern et al. 2013; Kinoshita et al. 2014) (Fig. 4.3). In unpublished studies, our group has detected by MALDI-FT-ICR structures containing as many as 5 LacNAc disaccharides on a tetra-antennary structure.
Improving and verifying detection of these many types of glycan structures has required much optimization. Thus far, structures containing mannose, galactose, glucose , N-acetylglucosamine, fucose, and sialic acids have been readily detected in mammalian tissues. In general, the base structures in regard to mannose, GlcNAc, and galactose content are easiest to detect in tissues. These same structures with a core fucose are also readily detectable, which was confirmed by on-tissue CID using the MALDI-FT-ICR MS (Powers et al. 2014). Other possible modifications that include sulfation, phosphorylation, and N-acetylgalactosamine can be detected but still require further optimization strategies to facilitate their routine detection. Xylose and other sugar modifications associated with N-glycans present in plants and other nonmammalian organisms remain an under-evaluated area of study in regard to MALDI-MSI approaches. Detection of branched N-glycans with multiple fucose and sialic acid constituents presents their own challenges in regard to stability, identification, and isomeric structures. In the next sections, observations and optimization strategies for different classes of N-glycans are presented.
4.3.1 Protocol Notes
-
Using the preparation protocols noted for both frozen and FFPE tissues followed by MALDI-TOF MS, the glycans are detected as sodiated species, so an additional 22 m.u. (i.e., M + H/M + Na) for MALDI is included and reported for each total glycan mass. Additional sodium ions were detected with each sialic acid present. This shift in mass associated with sodium atoms can be used to distinguish single sialylated glycans (291 m.u. plus Na) from those with two fucose residues (292 m.u., no extra Na).
-
The MALDI-FT-ICR configuration is most advantageous for identifying as many N-glycans present in a given tissue sample. The high mass accuracy, sensitivity, and isotope patterns that can be detected for each N-glycan are critical features provided for each N-glycan imaging analysis. This resolution and sensitivity comes at the cost of analysis time and data file sizes.
-
Another advantage of using the MALDI-FT-ICR instrument is the ability to perform collision-induced dissociation (CID) on glycans directly from tissue. Ions of interest can be specifically isolated in a quadrupole followed by subsequent CID. The CID energies used primarily release constituent glycans intact, and not with cross-ring fragmentation (Powers et al. 2014). Released glycan masses represent hexoses (162.053 m.u.), HexNAc (203.079 m.u.), sialic acid (291.095 m.u.), fucose (146.058 m.u.), fucose-HexNAc (349.137 m.u.), and Hex-HexNAc (365.132 m.u.).
-
It is important to confirm the N-glycan structures determined by MALDI-MS imaging using other carbohydrate analysis tools like lectins or glycosidase digestions with HPLC (Powers et al. 2015). Correlative LC-MS identification methods for N-glycans from adjacent tissue slices are helpful in further defining structural features (Briggs et al. 2017; Holst et al. 2016).
4.4 N-Linked Glycans Associated with Cancer and Cell Proliferation
MALDI-MSI applied to FFPE tumor tissue slides is very effective for identifying N-glycans that are localized to regions of tumor (Drake et al. 2017). Example MS images of the most abundant type of N-glycans detected in prostate cancer tissues are shown in Fig. 4.4. The first two structures in this figure represent high-mannose glycans. While N-glycans with high-mannose structures (GlcNAc2-Man5–9) represent an early stage in normal biosynthetic processing, as in Fig. 4.2, these high-mannose glycans are frequently detected in high abundance in most all solid tumor tissues analyzed by MALDI-MS imaging (Drake et al. 2017; Powers et al. 2014; Everest-Dass et al. 2016; Holst et al. 2016; Heijs et al. 2016). For normal biosynthesis, the high-mannose structures are degraded by mannosidases to only three mannose residues and then further processed to complex N-glycans. In tumor cells, it is apparent that high-mannose structures are bypassing normal ER and Golgi glycan processing. In sera from prostate cancer patients, autoantibodies to high-mannose N-glycans have been reported (Wang 2012; Wang et al. 2013), and specific C-type lectin receptors, especially in liver and on macrophages, are known to bind to them. Also in tumor tissues, glycan structures with three or less mannose residues, with and without core fucose, are termed pauci-mannose glycans, which are also readily detected on secreted glycoproteins and on the cell surface (Loke et al. 2016; Nyalwidhe et al. 2013). Defining the functional roles of high-mannose and pauci-mannose N-glycans and their glycoprotein carriers is at the nexus of understanding basic tumor biology, glycoprotein processing, and tumor immunity.
The other main structural classes of tumor N-glycans detected are the representative tetra-antennary structures shown in Fig. 4.4. These tetrasaccharides are the result of GnT-V activity, and the increase in β1–6 GlcNAc branching of N-linked glycans is associated with the metastatic phenotype of multiple cancer types (Miwa et al. 2012; Pinho and Reis 2015; Schultz et al. 2012; Taniguchi and Kizuka 2015). Structurally, increased branching of glycans in cancers is typified by increased detection of sialyl Lewis X and sialyl Lewis A antigens, as well increases in polylactosamine modifications. These structures in turn are recognized by selectins and other carbohydrate lectins expressed on different tissues involved with immune cell binding and innate immunity and, in the case of extravasation, binding to cells in distant organ/tissue sites.
4.4.1 Protocol Notes
-
It is critical to have a pathologist involved in studies to confirm the location and presence of tumor in a given sample. It is the ability to directly overlay the N-glycan distributions directly by MALDI that provides the location of glycans associated with tumor and non-tumor regions.
4.5 N-Linked Glycans Most Commonly Associated with Tissue Stroma Regions
The most abundant and easily detectable glycans by MALDI-MSI are those represented in the central portion of the biosynthetic pathway shown in Fig. 4.3, representing the spectrum from high-mannose to sialylated bi-antennary structures, with and without the core fucose. This statement is based on peak intensity comparisons across hundreds of tissues analyzed by MALDI-MSI. Core-fucosylated bi-antennary glycans are frequently the most abundant N-glycans detected in areas of tissue containing smooth muscle and collagen, while bi-antennary glycans lacking core fucose are found in the adjacent stroma areas (an example is shown in Fig. 4.5). In a given tissue, these glycans can be detected in tumor regions, but not with the frequency observed for the high-mannose and branched chain glycans. Because their molecular masses range from 1100 to 2100 daltons, these stroma glycans are easily detectable by MALDI-TOF, with Hex5HexNAc4, Hex4HexNAc4Fuc1, Hex5HexNAc4Fuc1, Hex5HexNAc4NeuAc1, and Hex5HexNAc4Fuc1NeuAc1 glycans being the most abundant detected from tissues.
Stabilization of sialic acids by in-tissue ethyl esterification. Ethyl esterification (EE) was adapted from a published protocol (Holst et al. 2016) for a prostate cancer FFPE tissue and analyzed by MALDI-FT-ICR IMS. Shown are the examples for a mono-sialylated core fucose bi-antennary glycan, without EE (left panel) or with EE for α2,6 (+28 amu) (middle panel) or α2,3 (+18 amu) (right panel) linkages
4.6 Stabilizing Terminal Sialic Acids for MALDI Analysis
N-glycans containing a terminal sialic residue in either α2,3 or α2,6 linkages are common tissue components and readily detectable by MALDI-MSI. A well-described complication in the detection of sialylated glycans by MALDI is their well-known lability due to in-source decay during the ionization process, especially in positive ion mode. The negative charge also leads to different Na salt adducts and lowered sensitivity (Reiding et al. 2014). When native glycans are analyzed by MALDI-TOF MS, modifications like permethylation or amidation that stabilize and help ionize sialic acid containing glycans are routinely used (Bern et al. 2013; Sekiya et al. 2005). For free glycans derived from fluids or cells, these common chemical modification methods require organic solvents and the need to separate reaction products, thus they are not feasible for on-tissue derivatization or maintenance of spatial distributions. An ethyl esterification approach that differentially modifies α2,3- or α2,6-linked sialic acids (Reiding et al. 2014) was adapted to MALDI-MSI for N-glycans in FFPE tissues, allowing distinct tissue distributions of sialylated glycans with both isomers to be determined (Holst et al. 2016). Relative to the unmodified parent sialylated glycans, an ethylation reaction results in an increase in 28 amu when an α-2,6 linkage is involved, and when an α-2,3 linkage is involved, a lactonization reaction will occur with a decrease of 18 amu. These mass shifts are easily detectable by MALDI-TOF MS and MALDI-FT-ICR MS.
Example MALDI-FT-ICR MSI N-glycan data for an ethyl esterified prostate cancer FFPE tissue using an adapted protocol from Holst et al. is shown in Figs. 4.5 and 4.6. In Fig. 4.5, images for a core-fucosylated bi-antennary glycan with one sialic acid (Hex5HexNAc4Fuc1NeuAc1) are shown without modification and with both α2,6 and α2,3 linkages. In particular for MALDI-MSI of tissues, detection of glycans with more than one sialic acid has proven difficult. This is effectively illustrated in the non-ethyl esterified image examples shown in Fig. 4.6 for three glycans: a Hex5HexNAc4NeuAc1, a Hex5HexNAc4NeuAc2, and a Hex6HexNAc5NeuAc3. The mono-sialylated glycan is readily detected by MALDI-FT-ICR MSI (top panel), as also evident in Fig. 4.5. However, without stabilization, it is clear that detection of the di-sialylated is decreased relative to the mono-sialylated glycan and the tri-sialylated glycan is undetectable. Ethyl esterification stabilization greatly facilitates detection of these multi-sialylated glycans (Fig. 4.6).
Stabilization of multi-sialylated tissue glycans by ethyl esterification. MALDI-FT-ICR IMS data from the same ethyl esterified prostate tissue prepped as in Fig. 4.5 is shown for mono-, di-, and tri-sialylated bi-antennary glycans. Images in each panel on the left side represent non-ethyl esterified glycans, and the right side images are after EE. Also shown for the EE examples are the α-2,6 sialic acid linkage structures
4.6.1 Protocol Notes
-
The MALDI-FT-ICR instruments incorporate a cooling nitrogen gas stream within the source that can decrease sialic acid cleavage (O’Connor and Costello 2001), but it does not fully eliminate the problem (powers et al. 2013). Sialic acid derivatization and stabilization are still the most effective way to maximize detection of sialylated glycans by MALDI-IMS.
-
These derivatization protocols represent an active area of research that is still evolving. The initial approach has proven to be robust and adaptable to different FFPE tissues. We have found that doing the chemical reactions in tissues prior to antigen retrieval and PNGaseF release allows extensive rinsing after reactions and more flexibility in the type of reactions that can be done while maintaining spatial localization as the N-glycans are still attached to the protein structure. Doing these modifications in fresh tissue is likely to result in other detection and side-reaction challenges and remains to be optimized.
4.7 Glycan Isomers and Fucosylation
The glycan structures shown in Fig. 4.3 for the different tri- and tetra-antennary fucosylated and sialylated glycans are compositional representations of what each glycan could be, and these do not account for the many different anomeric glycan linkages that are possible. Distinguishing these structural isomers has always been a challenge for mass spectrometry analysis of N-glycans and especially for MALDI-MS approaches. While accurate masses and glycan compositions can be easily determined, the location and anomeric linkages associated with fucose and sialic acid modifications are difficult to establish. The ethyl esterification examples highlight that multiple α2,6- or α2,3-linked sialic acids can be detected, but this still does not identify which branch of the glycan is occupied. For fucose modifications, this is particularly challenging as there are 13 known fucosyltransferase (FUT1–13) genes (Schneider et al. 2017), and fucose residues can be attached via α1,2 (FUT1,2), α1,3 (FUT3,4,6,7,9,10,11), or α1,4 (FUT3) linkages for outer arm modifications or α1,6 for core fucosylation linkages (FUT8). There are no differences in masses for these fucose linkages, and using MALDI-MS approaches, the presence of two or more fucose residues precludes identification of the specific linkages or which arm of the glycan the attachment occurs. There are two approaches that we expect can begin to address these fucosylation challenges in the context of MALDI-MSI workflows. One is the use of ion mobility MS instruments, which measures the mobility of gas-phase ions through an electric field in the presence of a buffer gas. The mobility of ions is based on their charge, shape, and size, and the method is increasingly being applied to the separation of glycan isomers (Fenn and McLean 2011; Gray et al. 2016), as well as imaging MS (Škrášková et al. 2016). The second approach is based on the ability to effectively spray enzymes on tissue (analogous to PNGaseF). Linkage-specific fucosidase s could be applied to tissues, before PNGaseF digestion for comparative analysis with non-fucosidase treated tissues. This is feasible if the linkage specificity of the fucosidase is established, an area that still needs improvement and demonstration for the glycomic field in general.
4.7.1 Protocol Notes
-
Using the MALDI-FT-ICR instrument, collision-induced dissociation (CID) of N-glycans under 2300 m.u. can be used to effectively distinguish core-fucosylated glycans (Powers et al. 2014) based on disaccharide fragmentation patterns. Demonstrating this for multi-fucosylated glycans has not been reported.
4.8 Current and Emerging Applications of N-Glycan Tissue Imaging
Our laboratories have cumulatively evaluated over 1000 different human tissue samples from multiple organ types using the basic PNGaseF digestion and MALDI-MS imaging workflows described herein. These methods have also been applied to animal models used in biomedical research, allowing studies on mechanisms and therapeutics that influence N-glycosylation changes over a wide range of applications including cancer treatments, transplant optimization, and cardiovascular diseases. Based on this experience, there are several research applications that we have identified, summarized in Fig. 4.7, grouped as different color codes. The major application of the technique is linked with the core premise of using an MS imaging approach for analysis of tissues, i.e., determine the location and number of N-glycans in a given tissue (Fig. 4.7, green panel). This data is linked with histopathology correlates of the tissue, which can be standard pathology stains like hematoxylin and eosin or immunohistochemical stains of specific protein targets. Also inherent in this determination is a list of N-glycans associated with different tissue regions that can be used as potential biomarker panels when multiple tissues are analyzed. This is particularly amenable to screening of many samples using tissue microarrays (Drake et al. 2017). The rapid analysis capabilities of the high-speed MALDI-TOF instrumentation in turn can be adapted to clinical chemistry laboratory assays as possible diagnostics. Emerging capabilities in this grouping include the ability to link molecular glycan data for specific tissue slices co-registered with clinical images obtained by magnetic resonance imaging. Determining 3D MS imaging of N-glycans in a given tissue remains to be reported, but it is certainly feasible and will be done in the near future.
Application options for the N-glycan MALDI-MSI method. Different current and future applications of the method are summarized in four color-coded groups: (1) Tissue localization and disease biomarkers (green panels), (2) glycoproteomic and glycomic analyses (blue panel), (3) Genomic and metabolomic correlates (orange panel), and (4) potential applications to plant, microbial, and other nonmammalian systems (pale yellow panel)
Highlighted in the second panel of Fig. 4.7 (blue) are applications related to the analysis of N-linked glycoproteins and other types of complex carbohydrates. The tissue N-glycans described herein were all released by PNGaseF from a protein carrier. Therefore, the presence of N-glycans detected by MALDI-MSI in a particular tissue location, e.g., in a tumor region, also informs on tumor-specific glycoproteins. Identifying these glycoprotein carriers are also of great interest, and thus the glycan distribution tissue profiles can be used as localization maps. The most direct approach would be to target regions of interest for digestion with trypsin, followed by extraction and enrichment of glycopeptides from these regions. The biggest limitation of this approach is the minimal amount of glycopeptides that are extractable from small focal areas of a thin tissue slice. A current alternative approach is to use MS imaging of peptides in situ, combined with extracted peptide sequencing identification, to map protein expression to tissue localization. This distribution map of the peptides can be combined with the N-glycan map to correlate areas of overlap (Angel et al. 2017b). Recent studies (Angel et al. 2017b; Heijs et al. 2016) describe a sequential approach, one in which a FFPE tissue is first treated with PNGaseF to release N-glycans, which are then imaged, followed by trypsin digestion of the same tissue and peptide imaging. An adjacent tissue slice is treated similarly, but glycans and peptides are extracted for tandem MS analysis and peptide sequencing. A new MALDI imaging approach that uses collagenase digestion to map the distribution of collagens and extracellular matrix proteins (Angel et al. 2017c) could also be applicable with PNGaseF analyses. Alternatively, other carbohydrate digestion enzymes could be applied to tissues to map fine structures of heparan sulfates or chondroitin sulfate.
The third panel in Fig. 4.7 (orange) highlights the connection of N-glycans to their underlying genetic control and metabolic pathways. The different N-glycans shown in the biosynthetic schematic in Fig. 4.3 are the result of the sequential activity of glycosyltransferases and glycosidases . It is feasible that the activity of these enzymes is controlled at the transcriptome level or other genomic components, which in turn regulates the composition of the N-glycome in each region (Neelamegham and Mahal 2016). A previous study comparing the transcriptome of glycan biosynthetic genes in different mouse organs with the most abundant N-glycans detected in each tissue determined that the levels of many transcripts could be correlated with the levels of specific glycan structures (Nairn et al. 2008). Several studies have been published that have evaluated the transcriptome profiles of glycan biosynthesis genes to predict what glycan structures would be present (Kawano et al. 2005), including glycosylation reaction network analysis strategies (Liu and Neelamegham 2014; Neelamegham and Mahal 2016). The N-glycome is also linked to basic glucose and N-acetylglucosamine metabolism via the hexosamine biosynthetic pathway (Ryczko et al. 2016). There are now comprehensive metabolomic networks available for tissue metabolites following in vivo 13C-glucose labeling (Bruntz et al. 2017), which can readily be linked to metabolic precursors of N-linked biosynthesis and the related hexosamine pathway. Additionally, there is an increasing number of genetic mouse models emerging from CRISPR/Cas9 technology, as well as related models with organ-specific inducible regulation. The ability to examine individual organs that have gene knockout or knock-ins, or multiple organs from full knockout mice, represents many new opportunities to evaluate the role of glycosylation in these models. In addition to glycosyltransferases and glycosidase genes, any gene target that is linked to glycan metabolism, glycoproteins, and extracellular matrix proteins can potentially affect the N-glycome in these models. It is feasible to now link multiple genetic models with comprehensive transcriptome and metabolome information to evaluate the effect of the N-glycome in target tissues.
The last panel in Fig. 4.7 represents the vast opportunity to apply this approach to nonmammalian samples. Descriptions of N-glycan imaging by MSI have yet to be reported for any other species beyond human and rodent tissues. Beyond evaluation of different plants, amphibians, or reptiles, there is an opportunity to evaluate the effect that microbial infections or microbiome constituents have on the glycomes of affected tissues. It is expected that application and adaptation of the glycan imaging workflow to these systems will provide important new information about the diversity, distribution, and localization of novel N-glycomes.
Abbreviations
- FFPE:
-
Formalin-fixed paraffin-embedded
- FT-ICR:
-
Fourier transform ion cyclotron resonance
- Fuc:
-
Fucose
- GlcNAc:
-
N-acetylglucosamine
- GnT-III:
-
N-acetylglucosaminyltransferase 3
- GnT-IV:
-
N-acetylglucosaminyltransferase 4
- GnT-V:
-
N-acetylglucosaminyltransferase 5
- Hex:
-
Hexose (e.g., mannose, glucose, galactose)
- HexNAc:
-
N-acetylhexosamine (e.g., N-acetylglucosamine or N-acetylgalactosamine)
- LacNAc:
-
Lactosamine (galactose and N-acetylglucosamine disaccharide)
- MALDI-TOF MS:
-
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- Man:
-
Mannose
- MS:
-
Mass Spectrometry
- MSI:
-
Mass spectrometry imaging
- NeuAc:
-
N-acetylneuraminic acid (sialic acid)
- NeuGc:
-
N-glycolylneuraminic acid
- PNGaseF:
-
Peptide-N-glycosidase F
- TFA:
-
trifluoroacetic acid
- TMA:
-
Tissue microarray
Literature Citations
Angel PM, Caprioli RM (2013) Matrix-assisted laser desorption ionization imaging mass spectrometry: in situ molecular mapping. Biochemistry 52(22):3818–3828
Angel PM, Baldwin HS, Gottlieb Sen D, Su YR, Mayer JE, Bichell D, Drake RR (2017a) Advances in MALDI imaging mass spectrometry of proteins in cardiac tissue, including the heart valve. Biochim Biophys Acta 1865(7):927–935
Angel PM, Mehta A, Norris-Caneda K, Drake RR (2017b) MALDI imaging mass spectrometry of N-glycans and tryptic peptides from the same formalin-fixed, paraffin-embedded tissue section. Methods Mol Biol In press
Angel PM, Comte-Walters S, Ball LE, Talbot K, Mehta A, Brockbank KGM, Drake RR (2017c) Mapping extracellular matrix proteins in formalin-fixed, paraffin-embedded tissues by MALDI imaging mass spectrometry. J Proteome Res 17(1):635–646
Baker TC, Han J, Borchers CH (2017) Recent advancements in matrix-assisted laser desorption/ionization mass spectrometry imaging. Curr Opin Biotechnol 43:62–69
Berin A, Boughton BA (2017) Spatial metabolite profiling by matrix-assisted laser desorption ionization mass spectrometry imaging. Adv Exp Med Biol 965:291–322
Bern M, Brito AE, Pang PC, Rekhi A, Dell A, Haslam SM (2013) Polylactosaminoglycan glycomics: enhancing the detection of high-molecular-weight N-glycans in matrix-assisted laser desorption ionization time-of-flight profiles by matched filtering. Mol Cell Proteomics 12(4):996–1004
Briggs MT, Kuliwaba JS, Muratovic D, Everest-Dass AV, Packer NH, Findlay DM, Hoffmann P (2016) MALDI mass spectrometry imaging of N-glycans on tibial cartilage and subchondral bone proteins in knee osteoarthritis. Proteomics 16(11–12):1736–1741
Briggs MT, Ho YY, Kaur G, Oehler MK, Everest-Dass AV, Packer NH, Hoffmann P (2017) N-Glycan matrix-assisted laser desorption/ionization mass spectrometry imaging protocol for formalin-fixed paraffin-embedded tissues. Rapid Commun Mass Spectrom 31(10):825–841
Bruntz RC, Lane AN, Higashi RM, Fan TW (2017) Exploring cancer metabolism using stable isotope-resolved metabolomics (SIRM). J Biol Chem 292(28):11,601–11,609
Cornett DS, Reyzer ML, Chaurand P, Caprioli RM (2007) MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods 4(10):828–833
Drake RR, Powers TW, Jones EE, Bruner E, Mehta AS, Angel PM (2017) MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. Adv Cancer Res 134:85–116
Everest-Dass AV, Briggs MT, Kaur G, Oehler MK, Hoffmann P, Packer NH (2016) N-Glycan MALDI imaging mass spectrometry on formalin-fixed paraffin-embedded tissue enables the delineation of ovarian cancer tissues. Mol Cell Proteomics 15(9):3003–3016
Fenn LS, McLean JA (2011) Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Physical Chemistry Chemical Physics 13(6):2196–2205
Gray CJ, Thomas B, Upton R, Migas LG, Eyers CE, Barran PE et al (2016) Applications of ion mobility mass spectrometry for high throughput, high resolution glycan analysis. Biochim Biophys Acta 1860(8):1688–1709
Gustafsson OJ, Briggs MT, Condina MR, Winderbaum LJ, Pelzing M, McColl SR et al (2015) MALDI imaging mass spectrometry of N-linked glycans on formalin-fixed paraffin-embedded murine kidney. Anal Bioanal Chem 407:2127–2139
Harvey DJ (1999) Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom Rev 18:349–450
Harvey DJ (2015) Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: an update for 2009–2010. Mass Spectrom Rev 34(3):268–422
Heijs B, Holst S, Briaire-de Bruijn IH, van Pelt GW, de Ru AH, van Veelen PA et al (2016) Multimodal mass spectrometry imaging of N-Glycans and proteins from the same tissue section. Anal Chem 88:7745–7753
Holst S, Heijs B, de Haan N, van Zeijl RJ, Briaire-de Bruijn IH, van Pelt GW et al (2016) Linkage-specific in Situ Sialic Acid derivatization for N-Glycan mass spectrometry imaging of formalin-fixed paraffin-embedded tissues. Anal Chem 88(11):5904–5913
Kawano S, Hashimoto K, Miyama T, Goto S, Kanehisa M (2005) Prediction of glycan structures from gene expression data based on glycosyltransferase reactions. Bioinformatics 21:3976–3982
Kinoshita M, Mitsui Y, Kakoi N, Yamada K, Hayakawa T, Kakehi K (2014) Common glycoproteins expressing polylactosamine-type glycans on matched patient primary and metastatic melanoma cells show different glycan profiles. J Proteome Res 13(2):1021–1033
Liu G, Neelamegham S (2014) A computational framework for the automated construction of glycosylation reaction networks. PLoS One 9(6):e100939
Loke I, Kolarich D, Packer NH, Thaysen-Andersen M (2016) Emerging roles of protein mannosylation in inflammation and infection. Mol Asp Med 51:31–55
Miwa HE, Song Y, Alvarez R, Cummings RD, Stanley P (2012) The bisecting GlcNAc in cell growth control and tumor progression. Glycoconj J 29(8–9):609–618
Nairn AV, York WS, Harris K, Hall EM, Pierce JM, Moremen KW (2008) Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J Biol Chem 283(25):17,298–17,313
Neelamegham S, Mahal LK (2016) Multi-level regulation of cellular glycosylation: from genes to transcript to enzyme to structure. Curr Opin Struct Biol 40:145–152
Nishikaze T (2017) Sensitive and structure-informative N-Glycosylation analysis by MALDI-MS; ionization, fragmentation, and derivatization. Mass Spectrom 6(1):A0060
Nyalwidhe JO, Betesh LR, Powers TW, Jones EE, White KY, Burch TC et al (2013) Increased bisecting N-acetylglucosamine and decreased branched chain glycans of N-linked glycoproteins in expressed prostatic secretions associated with prostate cancer progression. Proteomics Clinical Applications 7:677–689
O’Connor PB, Costello CE (2001) A high pressure matrix-assisted laser desorption/ionization fourier transform mass spectrometry ion source for thermal stabilization of labile biomolecules. Rapid Commun Mass Spectrom 15:1862–1868
Ogrinc Potočnik N, Porta T, Becker M, Heeren RM, Ellis SR (2015) Use of advantageous, volatile matrices enabled by next-generation high-speed matrix-assisted laser desorption/ionization time-of-flight imaging employing a scanning laser beam. Rapid Commun Mass Spectrom 29(23):2195–2203
Pearce OM, Läubli H (2016) Sialic acids in cancer biology and immunity. Glycobiology 26(2):111–128
Pinho SS, Reis CA (2015) Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 15(9):540–555
Powers TW, Jones EE, Betesh LR, Romano PR, Gao P, Copeland JA et al (2013) Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal Chem 85:9799–9806
Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS et al (2014) MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 9(9):e106255
Powers TW, Holst S, Wuhrer M, Mehta AS, Drake RR (2015) Two-Dimensional N-Glycan distribution mapping of hepatocellular Carcinoma tissues by MALDI-Imaging mass spectrometry. Biomolecules 5(4):2554–2272
Reiding KR, Blank D, Kuijper DM, Deelder AM, Wuhrer M (2014) High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Analytical Chemistry 86:5784–5793
Rini J, Esko J, Varki A (2009) Chapter 5: Glycosyltransferases and Glycan-processing Enzymes. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME (eds) Essentials of Glycobiology, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Ryczko MC, Pawling J, Chen R, Abdel Rahman AM, Yau K, Copeland JK, Zhang C, Surendra A, Guttman DS, Figeys D, Dennis JW (2016) Metabolic reprogramming by Hexosamine biosynthetic and Golgi N-Glycan branching pathways. Sci Rep 6:23043
Schneider M, Al-Shareffi E, Haltiwanger RS (2017) Biological functions of fucose in mammals. Glycobiology 27(7):601–618
Schultz MJ, Swindall AF, Bellis SL (2012) Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev 31(3–4):501–518
Sekiya S, Wada Y, Tanaka K (2005) Derivatization for stabilizing sialic acids in MALDI-MS. Anal Chem 77(15):4962–4968
Škrášková K, Claude E, Jones EA, Towers M, Ellis SR, Heeren RM (2016) Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation. Methods 104:69–78
Stanley P, Schachter H, Taniguchi N (2009) Chapter 8: N-Glycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME (eds) Essentials of Glycobiology, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Taniguchi N, Kizuka Y (2015) Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res 126:11–51
Wang D (2012) N-glycan cryptic antigens as active immunological targets in prostate cancer patients. J Proteomics Bioinformatics 5:90–95
Wang D, Dafik L, Nolley R, Huang W, Wolfinger RD, Wang LX et al (2013) Anti-oligomannose antibodies as potential serum biomarkers of aggressive prostate cancer. Drug Dev Res 74:65–80
Acknowledgment
This work was supported by the National Institutes of Health/National Cancer Institute R21 CA185799 to RRD and the National Institutions of Health/National Institute of General Medical Sciences P20GM103542 to PMA.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Drake, R.R., West, C.A., Mehta, A.S., Angel, P.M. (2018). MALDI Mass Spectrometry Imaging of N-Linked Glycans in Tissues. In: Yamaguchi, Y., Kato, K. (eds) Glycobiophysics. Advances in Experimental Medicine and Biology, vol 1104. Springer, Singapore. https://doi.org/10.1007/978-981-13-2158-0_4
Download citation
DOI: https://doi.org/10.1007/978-981-13-2158-0_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-2157-3
Online ISBN: 978-981-13-2158-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)