Abstract
Genes need to be investigated either in Gene Interaction Network or in a DNA microarray gene expression data to understand the role they play in complex diseases like cancer. The prioritized genes can help us to know the molecular mechanism, as well as to discover the promising candidates of cancer. Several gene ranking algorithms already have been proposed that produces the top ranked genes according to their importance with respect to a particular disease. In this work, we have developed one Genetic Algorithm (GA) based algorithm, MicroarrayGA, to rank the genes responsible for a particular cancer to occur. The whole research works on six datasets like Colorectal Cancer, Diffuse Large B-Cell Lymphoma, Pediatric Immune Thrombocytopenia (ITP), Small Cell Lung Cancer (SCLC), Breast Cancer and Prostate Cancer, publicly available from NCBI (National Center for Biotechnology Information) online repository. We have validated the outcome of the proposed algorithm by classification step using Support Vector Machine (SVM) classifier and we have also compared the results of MicroarrayGA with three existing methods on the basis of percentage of accuracy, precision, recall, F1-Score and G-Mean metrics.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Uncontrollable growth of cells in an unusual fashion is the primary basis of cancer, which is often classified as a group of diseases [1]. Thousands of several genes are responsible for a particular cancer to occur. But all those genes may not be equally significant in this case. Due to this reason, selection of genes based on their ranks for a specific cancer is highly important [2]. In this work, we try to find the significant mediating genes of six various human cancer datasets and to prioritize them as well.
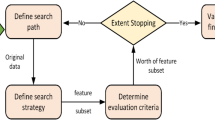
The algorithm used in this paper to rank the genes is primarily constructed by analyzing the expression change of the genes between a diseased and a normal sample. The Genetic Algorithm (GA) based implementations, namely MicroarryGA is applied on microarray gene expression datasets of different cancers to rank genes. Genetic Algorithm can be considered as one of the popular evolutionary algorithms [3, 4]. Later, these ranks are validated by classification step using SVM classifiers [5, 6]. The workflow of the proposed MicroarrayGA method is given in Fig. 1.
2 Related Earlier Works
Ranking genes based on the information from several DNA microarray gene expression data is of fundamental importance [7]. Since data sources contain thousands of several genes, so to determine the set of prioritized genes according to their significance order is an important task. Some of methods from current literature are mentioned briefly as follows:
A soft clustering based microarray expression analysis algorithm is proposed in [8] to analyze genes responsible for acute leukemia. Another method mentioned in [9] uses non-parametric statistical tests - Mann-Whitney U test and k-sample Kruskal-Wallis H test to rank genes. A statistical gene subset selection algorithm using Wilcoxon rank sum test is presented in [10].
Huerta et al. [13] analyzed Leukemia [12] and Colon cancer [14] datasets using their proposed genetic algorithm based gene selection method along with SVM classifier. Mondal et al. presented a multi-objective GA based gene ranking approach in [15]. Another method proposed in [16], selects significant features using genetic algorithms (GAs) and then uses Constructive Neural Networks (CNN) [17], C-Mantec [18] based classification methods to validate the results. A neuro-fuzzy approach is proposed in [19] to select those genes responsible for a particular type of cancers. Another method presented in [20] uses graph theory based approach to identify most significant as well as most non-redundant genes from a DNA gene expression dataset. Dragon Wrapper Feature Selection (DWFS), a web based tool, proposed in [21], is used to select significant features for a various types of problems efficiently.
Another new approach was proposed in [22] to rank the genes of microarray gene expression data using squared Pearson correlation coefficient. Another work presented in [23] developed a fuzzy set theory based index, named as Gaussian Fuzzy Index (GFI) to select a set of cancer mediating genes. GeneRank developed by Morrison et al. [24], is a straightforward generalization of PageRank [25] algorithm used by Google.
3 Methodology
We have developed following GA based approach named as MicroarrayGA. This method initially begins with pre-selection based on filtering, and then generates gene subset \( G_{P} \) of \( P \) genes, where \( P \) is problem dependent. A small set of the most important genes are selected from this reduced subset of genes, which give the highest accuracy when used with classification task. Detailed steps of this approach are given below:
-
1.
In the first step, genes/features form the microarray are sorted using Fisher’s Discriminant Criteria [26] in decreasing order. Then from the sorted genes, top \( P \) genes are selected for our calculation.
-
2.
Then \( N \) different gene subsets are generated randomly from selected \( P \) genes and they are used as initial GA population. To encode each of the gene subsets of the population, binary chromosome representation is used, given in Fig. 2. If a particular gene is present in the gene subset, then the corresponding bit is set to 1, otherwise set to 0 in that chromosome representation.
-
3.
After obtaining the initial population, a minimizing fitness function is applied on each of the chromosomes to find the individual fitness value. For each gene present in chromosome, p-value is calculated from Student’s two-tailed t test between the two types of sample corresponding to that gene (normal and cancerous). After calculating the p-values of each of the genes present in the chromosome, the average p-value is calculated, which is the fitness value of the chromosome.
-
4.
In this step, a temporary population \( N^{\prime } \) is created which will be used as a population \( N \) in the next generation. The new population \( N^{\prime } \), is filled up in three steps. In the first step, the top 40% individuals (based on fitness value; in this method from lower to higher) of \( N \) are copied to \( N^{\prime } \) (elitism). In the next two steps, genetic operators: uniform crossover and binary mutation operators are applied respectively to obtain new chromosomes (child solutions).
-
5.
A random pair of solutions are selected from \( N \) and uniform crossover operator is applied on these two individuals. The resulting new child solution is inserted in \( N^{\prime } \). This step is repeated for \( 0.4* \left| N \right| \) times, where 0.4 is crossover probability (\( P_{c} \)).
-
6.
In this step, mutation operator is applied on individual solution selected randomly from \( N \). This step is repeated for \( 0.2* \left| N \right| \) times, where 0.2 is mutation probability (\( P_{m} \)).
-
7.
Replace \( N \) with \( N^{\prime } \) and repeat the steps from 3 to 7 until fixed number of generations are completed.
-
8.
After the completion of all the generations, each gene is assigned an importance value depending on the frequency of selection among all the \( N \) gene subsets. A gene having higher frequency of appearance gets high importance value and vice versa. Their significance is tested by the classification performance using SVM classifier.
4 Experimental Results
MicroarrayGA based approach is applied on microarray gene expression dataset of six various types of cancers, like Breast Cancer, Prostate Cancer, Pediatric Immune Thrombocytopenia (ITP), Small Cell Lung Cancer, Diffuse Large B-Cell Lymphoma (DLBCL) and Colorectal Cancer available at NCBI online repository.
4.1 Dataset Used
Colorectal Cancer tumor (GDS4382) [27, 32] dataset contains 17 samples are of type normal and 17 are of type diseased. Small Cell Lung Cancer (GDS4794) [28, 32] dataset is divided into diseased and normal sample subsets of size 23 and 42 respectively. For DLBCL-FL (GDS4236) [29, 32] dataset, 8 samples are rapamycin sensitive and 6 are rapamycin resistant. In Prostate Cancer (GDS4824) [30, 32] Dataset, 13 diseased samples and 8 normal samples are present. Breast Cancer (GDS4056) [11, 32] contains 32 diseased samples and 29 normal samples. Pediatric Immune Thrombocytopenia (ITP) [31, 32] dataset has total 13 samples out of which 7 are of type newly diagnosed immune thrombocytopenia samples and 6 are of type chronic immune thrombocytopenia samples.
4.2 Parameter Settings
The parameters of the GA applied on microarray gene expression dataset are represented in Table 1 below:
4.3 Results and Comparison with Other Methods
Classification algorithm is used to validate the proposed GA based algorithm’s effectiveness in gene ranking. The proposed method’s efficiency is compared with some existing ranking methods on the basis of percentage of classification accuracy along with some other metrics.
We take top 20 genes from the ranked set of 1000 genes. These ranking is validated again by the classification process using SVM method with three kernels. Before the classification process starts, we divide the whole sample space into 60:40 ratio. Following Fig. 3 shows how the MicroarrayGA’s performance varies on the basis of percentage of accuracy using only top most 20 genes for six cancer datasets. From that figure it is clear that for which dataset what is the percentage of classification accuracy proposed algorithm achieves by using how many genes. Although we have done our experiment on six publicly available cancer datasets, but to compare our GA based method’s performance with three state-of-the art methods, like Particle Swarm Optimization (PSO) based Graph Theoretic Approach [20], DWFS using K-Nearest Neighbor (KNN) classifier and DWFS using Naïve Bayes (NBC) classifier [21] only two datasets, Prostate Cancer and DLBCL-FL are used. Table 2 shows this comparative study on the basis of percentage of classification accuracy, F1 Score, G-Mean, Recall, Precision [33].
Following Table 3 shows top 15 genes returned by MicroarrayGA algorithm.
5 Conclusion and Future Scope
In the proposed approach, a genetic algorithm based implementation, MicroarrayGA is used to rank the genes causing various cancers. Ranking is validated by SVM classifier method with three kernels. From Fig. 3 it is clear that for which dataset what is the percentage of classification accuracy proposed algorithm achieves by using how many genes. Three different existing methods from current literature are used in this paper along with MicroarrayGA method for the performance comparison purpose. From the performance scores given in Table 2, it can be concluded that our GA based approach surpasses the other methods. Since MicroarrayGA uses gene expression dataset, the results shows that this approach uses top most 3 genes to top most 9 genes for various cancer datasets to achieve percentage of classification accuracy close to 100%.
We have used only microarray dataset to rank genes. But it is quite possible that gene dataset is obtained in terms of gene interaction network. In that case we need to think about new ways to rank the genes involved in the network. Another modification that can be done is the generation of multiple gene subsets (non-dominated solutions) rather than a single gene subset, which can be used for further biological analysis. For that we can think about multi-objective genetic algorithm. That may improve our results further.
References
Defining Cancer: National Cancer Institute, June 2014
Zhang, C., Lu, X., Zhang, X.: Significance of gene ranking for classification of microarray samples. IEEE/ACM Trans. Comput. Biol. Bioinform. (TCBB) 3(3), 312–320 (2006)
Goldberg, D.E.: Genetic Algorithms in Search, Optimization and Machine Learning. Addison-Wesley, Boston (1989)
Holland, J.H.: Adaptation in Natural and Artificial Systems, 2nd edn. MIT Press, Cambridge (1975)
Boser, B.E., Guyon, I., Vapnik, V.: A training algorithm for optimal margin classifiers. In: Proceedings of 5th Annual Workshop on Computational Learning Theory, pp. 144–152. ACM Press (1992)
Zisserman, A.: The SVM Classifier. Lecture Notes (2015)
Wang, Y., et al.: Gene selection from microarray data for cancer classification—a machine learning approach. Comput. Biol. Chem. 29(1), 37–46 (2005)
Yoo, C.K., Leeb, I.B., Vanrolleghema, P.A.: Interpreting patterns and analysis of acute leukemia gene expression data by multivariate fuzzy statistical analysis. In: Proceedings of 14th European Symposium on Computer Aided Process Engineering. ESCAPE-14, vol. 29, no. 6, pp. 1345–1356 (2005)
Peterson, L.E., Coleman, M.A.: Comparison of gene identification based on artificial neural network pre-processing with k-means cluster and principal component analysis. In: Bloch, I., Petrosino, A., Tettamanzi, A.G.B. (eds.) WILF 2005. LNCS (LNAI), vol. 3849, pp. 267–276. Springer, Heidelberg (2006). https://doi.org/10.1007/11676935_33
Liao, C., Li, S., Luo, Z.: Gene selection using Wilcoxon rank sum test and support vector machine for cancer classification. In: Wang, Y., Cheung, Y.-M., Liu, H. (eds.) CIS 2006. LNCS (LNAI), vol. 4456, pp. 57–66. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-74377-4_7
West, M., Blanchette, C., Dressman, H., et al.: Predicting the clinical status of human breast cancer using gene expression profiles. Proc. Natl. Acad. Sci. 98, 11462–11467 (2001)
Golub, T.R., Slonim, D.K., Tamayo, P., Huard, C., Gaasenbeek, M., et al.: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(1999), 531–537 (1999)
Huerta, E.B., Duval, B., Hao, J.-K.: A Hybrid GA/SVM approach for gene selection and classification of microarray data. In: Rothlauf, F., et al. (eds.) EvoWorkshops 2006. LNCS, vol. 3907, pp. 34–44. Springer, Heidelberg (2006). https://doi.org/10.1007/11732242_4
Alon, U., Barkai, N., Notterman, D.A., Gish, K., Ybarra, S., et al.: Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 96, 6745–6750 (1999)
Mondal, K.C., Mukhopadhyay, A., Maulik, U., Bandhyapadhyay, S., Pasquier, N.: MOSCFRA: a multi-objective genetic approach for simultaneous clustering and gene ranking. In: Rizzo, R., Lisboa, P.J.G. (eds.) CIBB 2010. LNCS, vol. 6685, pp. 174–187. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-21946-7_14
Luque-Baena, R.M., Urda, D., Subirats, J.L., Franco, L., Jerez, J.M.: Analysis of cancer microarray data using constructive neural networks and genetic algorithms. In: 1st International Work-Conference on Bioinformatics and Biomedical Engineering-IWBBIO, Granada, Spain (2013)
Parekh, R., Yang, J., Honavar, V.: Constructive neural-network learning algorithms for pattern classification. IEEE Trans. Neural Netw. 11(2), 436–451 (2000)
Subirats, J.L., Franco, L., Jerez, J.M.: C-Mantec: a novel constructive neural network algorithm incorporating competition between neurons. Neural Netw. 26, 130–140 (2012)
Ghosh, A., Dhara, B.C., De, R.K.: Selection of genes mediating certain cancers, using neuro-fuzzy approach. Neurocomputing 133, 122–140 (2014)
Mandal, M., Mukhopadhyay, A.: A novel PSO-based graph-theoretic approach for identifying most relevant and non-redundant gene markers from gene expression data. Int. J. Parallel Emerg. Distrib. Syst. 30(3), 175–192 (2015)
Soufan, O., Kleftogiannis, D., Kalnis, P., Bajic, V.B.: DWFS: a wrapper feature selection tool based on a parallel genetic algorithm. PLoS One (2015). https://doi.org/10.1371/journal.pone.0117988
Demidenko, E.: Microarray enriched gene rank. BioData Min. 8, 2 (2015). https://doi.org/10.1186/s13040-014-0033-1
Ghosh, A., De, R.K.: Identification of certain cancer mediating genes using Gaussian Fuzzy cluster validity index (GFI). J. Biosci. 40, 741–754 (2015)
Morrison, J.L., Breitling, R., Higham, D.J., Gilbert, D.R.: GeneRank: using search-engine technology for the analysis of microarray experiments. BMC Bioinform. 6(2015), 233–247 (2015)
Page, L., Brin, S., Motwani, R., Winograd, T.: The PageRank Citation Ranking: Bringing Order to the Web. Stanford InfoLab, Stanford (1999)
Iatan, I.F.: The fisher’s linear discriminant. In: Borgelt, C., et al. (eds.) Combining Soft Computing and Statistical Methods in Data Analysis, vol. 77, pp. 345–352. Springer, Heidelberg (2010). https://doi.org/10.1007/978-3-642-14746-3_43
Khamas, A., Ishikawa, T., Shimokawa, K., Mogushi, K., et al.: Screening for epigenetically masked genes in colorectal cancer using 5-Aza-2′-deoxycytidine, microarray and gene expression profile. Cancer Genom. Proteom. 9(2), 67–75 (2012)
Sato, T., Kaneda, A., Tsuji, S., Isagawa, T., et al.: PRC2 over-expression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci. Rep. 3, 1911 (2013)
Singh, D., et al.: Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2002), 203–209 (2002)
Hans, C.P., Weisenburger, D.D., Greiner, T.C., Gascoyne, R.D., Delabie, J., et al.: Confirmation of the molecular classification of diffuse large B-cell lymphoma by immune histo-chemistry using a tissue microarray. Blood 103(2004), 275–282 (2004)
Shad, A.T., Gonzalez, C.E., Sandler, S.G.: Treatment of immune thrombocytopenic purpura in children: current concepts. Paediatr. Drugs 7(5), 325–336 (2005)
Seal, D.B., Saha, S., Mukherjee, P., Chatterjee, M., Mukherjee, A., Dey, K.N.: Gene ranking: an entropy & decision tree based approach. In: IEEE 7th Annual Ubiquitous Computing, Electronics & Mobile Communication Conference (UEMCON), New York City, NY, USA, pp. 1–5 (2016). https://doi.org/10.1109/UEMCON.2016.7777837
Powers, D.M.W.: Evaluation: from precision, recall and F-measure to ROC, informedness, markedness & correlation. J. Mach. Learn. Technol. 2(1), 37–63 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Saha, S., Das, P., Ghosh, A., Dey, K.N. (2018). Ranking of Cancer Mediating Genes: A Novel Approach Using Genetic Algorithm in DNA Microarray Gene Expression Dataset. In: Singh, M., Gupta, P., Tyagi, V., Flusser, J., Ören, T. (eds) Advances in Computing and Data Sciences. ICACDS 2018. Communications in Computer and Information Science, vol 906. Springer, Singapore. https://doi.org/10.1007/978-981-13-1813-9_13
Download citation
DOI: https://doi.org/10.1007/978-981-13-1813-9_13
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1812-2
Online ISBN: 978-981-13-1813-9
eBook Packages: Computer ScienceComputer Science (R0)