Abstract
EEGLAB, a widely used toolbox in MATLAB (The Mathworks, Inc.), uses Independent Component Analysis (ICA) to decompose the EEG signal into sub-signals, and localizes brain sources of those sub-signals prior to independent component (IC) clustering for group study. In 2013, the Measure Projection Toolbox (MPT) was introduced as a new data-driven IC clustering toolbox for EEGLAB. Despite the numerous features and advantages offered by EEGLAB and the MPT, they both have limitations for statistical analyses with more than two independent variables. In order to work around those limitations, this paper introduces StaR, an EEGLAB framework for the MPT statistical analyses to be performed in R. StaR initially exports the data from different clusters generated by the MPT for different measures of interest (e.g., Event-Related Potentials (ERPs) and Event-Related Spectral Perturbations (ERSPs)) and formats the data such that further statistical analyses can be performed in R. Once in R, StaR uses linear mixed models as its default method to better handle missing values and intra-subject variability. Finally, StaR brings the results back into MATLAB to plot the results with the well-known and easy to interpret EEGLAB graphics. To make the whole process easy, StaR also offers an intuitive user interface that integrates into EEGLAB’s menu.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
EEGLAB [1] is an open-source toolbox running in MATLAB (The Mathworks, Inc.) made for EEG signal analysis and conceived to go beyond traditional mean peak analysis. One of its major preprocessing steps is the Independent Component Analysis (ICA) [2]. A dipole fitting plugin, DIPFIT [3], is then used to locate the corresponding potential brain sources (modeled as dipoles) in a brain template.
EEGLAB uses the Independent Component (IC) clustering approach for multi-subject analysis in the brain source domain. The number of adjustable parameters available in its GUI to cluster the ICs might, however, introduce subjectivity into the process. To address this issue, Bigdely-Shamlo, Mullen, Kreutz-Delgado, and Makeig (2013) developed a plugin for EEGLAB, called the Measure Projection Toolbox (MPT), which proposes a simpler and objective clustering procedure.
Even though EEGLAB and MPT offer performing algorithms intended for processing individual datasets and for IC clustering, they both have some important limitations when it comes to creating and running multi-subject studies with complex statistical designs such as longitudinal designs.
These limitations are (1) EEGLAB does not allow one to create a “studyset” (dataset gathering all the individual datasets) including different ICA decompositions for a given subject; (2) the STUDY feature in EEGLAB, which handles the statistical modules for multi-subject comparisons (i.e., “study design”), which does not support more than two independent variables; (3) the MPT statistical feature only supports two independent variables (Group and Dynamic); (4) in the MPT, once the domains (i.e., clusters) are computed, only two types of univariate analysis are available (t-test and permutation); and (5) in the MPT, in the case of comparison of two different groups for one condition, missing values are replaced with zeros (0) in order to keep the number of datasets equal in both groups, and in the case of comparison of two different conditions for both groups gathered together, the datasets with even a single missing value are completely removed from the statistical analysis, which can have a significant impact on the results.
Since its release in 2013, the MPT has been used in only a few published studies [4,5,6,7,8]. We believe that the limitations mentioned above are one of the reasons why few researchers have used it.
As a solution to these limitations, we developed a framework for EEGLAB called StaR (Statistics in R). StaR provides a complete pipeline extracting the data from EEGLAB, the MPT, and MATLAB altogether, in order to perform statistical analyses in R framework, and bringing the results back in MATLAB to easily plot them in various ways using the StaR UI that leverages the EEGLAB graphics [9,10,11,12].
2 Methodology
This section describes the StaR framework that we have developed. The code can be downloaded from https://github.com/FaubertLab/StaR. Questions can also be sent to yannick.roy@umontreal.ca. If we were to receive positive feedback and requests to provide a complete toolbox, we would happily seek funding to do so.
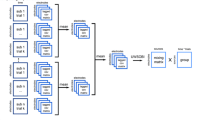
2.1 StaR Framework Steps
Steps #1 and #2 consist of preprocessing the data and creating a studyset in EEGLAB, then performing the clustering in the MPT (see EEGLAB (sccn.ucsd.edu/wiki/EEGLAB) and MPT wiki pages (sccn.ucsd.edu/wiki/MPT).
Step #3 consists of exporting the data from the MPT to R. StaR tags missing values with a NaN (Not a Number) value when exporting them from the MPT.
Step #4 consists of creating the complex data frame in R.
Step #5 consists of carefully placing the MPT values in the long data structure previously created. The dimensionality of the data will automatically be switched around by StaR to facilitate its usability by the user and developer while also optimizing for parallel computing. In order to study the EEG signal in greater detail, the different measures of interest (e.g., ERP, ERSP, etc.) are analyzed in a point-by-point fashion (i.e., considering each data point constituting a given graphical representation of the measure for each subject). Thus, for each subject, the data frame may hold tens of thousands of values, and hence there is the necessity for parallel computing.
Step #6 and #7 consist of the actual statistical analysis and post hoc tests. We opted for the linear mixed model procedure (lme4 package) to capture the within-subjects variability and to use as much of the data as possible, instead of removing subjects having missing values [13, 14]. Nevertheless, other tests can be used in R with small modifications in the code. Options of correction methods for multiple comparisons are also offered such as False Discovery Rate (FDR) [15] or Bonferroni.
Step #8 consists of bringing the results back into MATLAB to use EEGLAB graphics that are easy to recognize and interpret [10, 16]. One of the challenges here was the poor protocol of exchange between R and MATLAB. The data had to be linearized by carefully reshaping the matrices.
Step #9 consists of plotting the results. Handling more than two independent variables is not possible in EEGLAB and the MPT, therefore, even if the graphics are generated with EEGLAB functions, having the ability to plot this kind of graphics from a combination of three or four independent variables is new. The illustrations presented here come from a subset of datasets used in a longitudinal study initially comprising of four factors (Group, Session, and two condition-related crossed factors such as Dynamic and Modulation). In order to better handle the missing values, we have included a cluster-related factor (Domain).
Figure 1 shows an example of the data and statistical results for a given cluster (domain 2) from a combination of two factors (Group × Dynamic), both with two levels, while all other factors were kept combined. Any other pair of two variables could have been selected and would have produced a similar output. One could also fix the other variables to a specific value (e.g., Session = 1) instead of leaving them combined. It is also possible to select the option to only plot the significance mask showing only two colors (i.e., significant or not) instead of a color scale representing the difference between the two graphs.
ERSP 2 × 2. In this figure, the two factors are Group (2 levels: 3 and 4) and Dynamic (2 levels: M and F, where M stands for motion and F for flicker) for Domain 2. The effect of the factor Dynamic within a specific group is figured in the first two rows and the effect of the factor Group within a specific condition is figured in the first two columns. In the outer graphs, the green regions mean that it is not significant and the values are set to 0
Figure 2 shows an example of the data and statistical results for a given cluster (domain 1) from a combination of two factors (Group × Modulation), both with two levels, while all other factors were kept combined. Any other pair of two variables could have been selected and would have produced a similar output. One could also fix the other variables to a specific value (e.g., Session = 1) instead of leaving them combined.
ERP 2 × 2. In this figure, the two factors are Group (2 levels: 3 and 4) and Modulation (2 levels: FO, SO). Colored dots or lines below the curves indicate where the differences are significant. The legend indicates the independent variables being compared. For example, “gr = 3–gr = 4 (do = 1; mo = SO)” means that datasets from group 3 for domain 1 and second-order modulation were compared to datasets from group 4 for the same domain and modulation levels, all other factors combined
2.2 StaR UI
Creating an intuitive user interface (UI) that makes it easy to plot different layouts of graphs based on the interaction of multiple independent variables, all with a different number of values (e.g. 2 x 2, 2 x 3), is a challenge. Actually, when supporting multiple independent variables the combinations and possibilities quickly become manually unmanageable. The simple and intuitive StaR UI simplifies all these possible combinations and options.
2.3 Exploratory
We added a feature called specific Dipoles (Fig. 3) allowing the visualization of the domains generated by the MPT with the contributing dipoles from different groups and sessions represented with different colors. This feature does not compute anything but simply plots the dipoles already identified and located (i.e., source localization) with EEGLAB plug-in such as DIPFIT using a brain template such as the Montreal Neurological Institute, one prior to using the MPT and therefore before using StaR.
Specific Dipoles UI and output. The UI (at the top) allows for the selection of a specific domain to show its contributing dipoles. The user can either select all groups for all sessions to see all the contributing dipoles of a specific domain, or can break down the analysis to a specific group and/or session
Such a tool gives the opportunity to quickly check if the data seem well-balanced in order to validate results. By selecting a specific domain with the groups and/or sessions of interest using different colors, one can see at a glance if (1) the groups seem to be equally represented and/or if (2) the sessions seem to be equally represented.
3 Discussion and Conclusion
StaR, the statistical framework introduced in this article is mainly targeting EEGLAB users, especially those interested in the use of the MPT as IC clustering tool. StaR creates a link between EEGLAB/MPT in MATLAB and R. It uses the “studyset” created in EEGLAB to perform statistical analyses of EEG signal characteristics of interest (e.g., ERPs, ERSPs, power spectra, etc.) in R, and finally brings the results back into MATLAB in order to plot them using EEGLAB graphics.
The necessity for creating such a tool came from the limitations we encountered using EEGLAB and the MPT to create a longitudinal statistical study design with four factors (including two crossed condition-related factors).
The recent increase in computing power has made possible the use of more complex statistical designs like mixed-effect models, which are better at modeling within-subject variability and at dealing with missing values and unbalanced datasets than the classical ANOVA [17, 18]. Furthermore, contrary to the classical ANOVA, mixed-effect models do not violate underlying assumptions (e.g., linearity, sphericity, etc.) [19]. Else, mixed-effect models are well-suited for the analysis of longitudinal data containing missing values [20,21,22], and were thus particularly relevant in our case.
StaR was developed to be flexible and to easily support different statistical tests in R with minimum changes in the code while keeping all other steps untouched (e.g., exporting the data, plotting results with EEGLAB, etc.).
Finally, by making StaR freely accessible, we hope to encourage the research community to explore their EEG data beyond traditional ERP curves (peak amplitude and latency) obtained from the electrodes, especially by using the MPT as IC clustering tool.
References
Delorme, A., Makeig, S.: EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 134, 9–21 (2004). https://doi.org/10.1016/j.jneumeth.2003.10.009
Hyvärinen, A., Oja, E.: Independent component analysis: algorithms and applications. Neural Netw. 13, 411–430 (2000). https://doi.org/10.1016/s0893-6080(00)00026-5
Oostenveld, R., Delorme, A., Makeig, S.: DIPFIT: equivalent dipole source localization of independent components (2003)
Chung, J.W., Ofori, E., Misra, G., Hess, C.W., Vaillancourt, D.E.: Beta-band activity and connectivity in sensorimotor and parietal cortex are important for accurate motor performance. Neuroimage 144, 164–173 (2017). https://doi.org/10.1016/j.neuroimage.2016.10.008
Bogost, M.D., Burgos, P.I., Little, C.E., Woollacott, M.H., Dalton, B.H.: Electrocortical sources related to whole-body surface translations during a single- and dual-task paradigm. Front. Hum. Neurosci. 10, 524 (2016). https://doi.org/10.3389/fnhum.2016.00524
Misra, G., Ofori, E., Chung, J.W., Coombes, S.A.: Pain-related suppression of beta oscillations facilitates voluntary movement. Cereb Cortex:bhw061. https://doi.org/10.1093/cercor/bhw061 (2016)
Balkan, O., Virji-Babul, N., Miyakoshi, M., Makeig, S., Garudadri, H.: Source-domain spectral EEG analysis of sports-related concussion via measure projection analysis. 2015 37th Annual International Conference IEEE Engineering Medicine Biological Society, vol. 2015, pp. 4053–4056. IEEE (2015). https://doi.org/10.1109/embc.2015.7319284
Misra, G., Wang, W., Archer, D.B., Roy, A., Coombes, S.A.: Automated classification of pain perception using high-density electroencephalography data. J. Neurophysiol. 117, 786–795 (2017). https://doi.org/10.1152/jn.00650.2016
Delorme, A., Mullen, T., Kothe, C., Akalin Acar, Z., Bigdely-Shamlo, N., Vankov, A., et al.: EEGLAB, SIFT, NFT, BCILAB, and ERICA: new tools for advanced EEG processing. Comput. Intell. Neurosci. 130714 (2011). https://doi.org/10.1155/2011/130714
Onton, J., Makeig, S.: Information-based modeling of event-related brain dynamics. Prog. Brain Res. 159, 99–120 (2006). https://doi.org/10.1016/s0079-6123(06)59007-7
Onton, J., Delorme, A., Makeig, S.: Frontal midline EEG dynamics during working memory. Neuroimage 27, 341–356 (2005). https://doi.org/10.1016/j.neuroimage.2005.04.014
Makeig, S., Debener, S., Onton, J., Delorme, A.: Mining event-related brain dynamics. Trends Cogn. Sci. 8, 204–210 (2004). https://doi.org/10.1016/j.tics.2004.03.008
Bates, D.: Computational Methods for Mixed Models, pp. 1–29 (2010)
Bates, D.M., Maechler, M., Bolker, B., Walker, S.: Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). https://doi.org/10.1177/009286150103500418
Benjamini, Y., Hochberg, Y.: Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). https://doi.org/10.2307/2346101
Makeig, S.: Auditory event-related dynamics of the EEG spectrum and effects of exposure to tones. Electroencephalogr. Clin. Neurophysiol. 86, 283–293 (1993). https://doi.org/10.1016/0013-4694(93)90110-h
Rissling, A.J., Miyakoshi, M., Sugar, C.A., Braff, D.L., Makeig, S., Light, G.A.: Cortical substrates and functional correlates of auditory deviance processing deficits in schizophrenia. Neuro Imag. Clin. 6, 424–437 (2014). https://doi.org/10.1016/j.nicl.2014.09.006
Hasenstab, K., Sugar, C.A., Telesca, D., Mcevoy, K., Jeste, S., Şentürk, D.: Identifying longitudinal trends within EEG experiments. Biometrics 71, 1090–1100 (2015). https://doi.org/10.1111/biom.12347
Picton, T.W., Bentin, S., Berg, P., Donchin, E., Hillyard, S.A., Johnson, R., et al.: Guidelines for using human event-related potentials to study cognition: recording standards and publication criteria. Psychophysiology 37, 127–152 (2000). https://doi.org/10.1111/1469-8986.3720127
Gibbons, R.D., Hedeker, D., DuToit, S.: Advances in analysis of longitudinal data. Ann. Rev. Clin. 6, 79–107 (2010). https://doi.org/10.1146/annurev.clinpsy.032408.153550.advances
Krueger, C., Tian, L.: A comparison of the general linear mixed model and repeated measures ANOVA using a dataset with multiple missing data points. Biol. Res. Nurs. 6, 151–157 (2004). https://doi.org/10.1177/1099800404267682
Ibrahim, J.G., Molenberghs, G.: Missing data methods in longitudinal studies: a review. TEST 18, 1–43 (2009). https://doi.org/10.1007/s11749-009-0138-x
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Roy, Y., Piponnier, JC., Faubert, J. (2019). StaR: An EEGLAB Framework for the Measure Projection Toolbox (MPT) Statistical Analyses to be Performed in R. In: Ray, K., Sharan, S., Rawat, S., Jain, S., Srivastava, S., Bandyopadhyay, A. (eds) Engineering Vibration, Communication and Information Processing. Lecture Notes in Electrical Engineering, vol 478. Springer, Singapore. https://doi.org/10.1007/978-981-13-1642-5_66
Download citation
DOI: https://doi.org/10.1007/978-981-13-1642-5_66
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1641-8
Online ISBN: 978-981-13-1642-5
eBook Packages: EngineeringEngineering (R0)