Abstract
Electroencephalography (EEG) can be used to study various brain activities related to human responses and disorders. EEG signal is prone to noises which are caused due to eye movements, power-line interference, muscle movements, etc. Therefore, to obtain refined EEG signals for further processing, it should be denoised. There are several methods by which EEG signals can be denoised, among which we have used Independent Component Analysis (ICA), Principal Component Analysis (PCA)-based Equivariant Adaptive Separation by Independence (EASI), and Wavelet-based unsupervised denoising methods. The performance of these methods is compared using Signal-to-Noise Ratio (SNR) and Percentage Root-mean-square Difference (PRD).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
EEG is a noninvasive technique of recording brain’s spontaneous electrical activities generated by neurons. The quality of recorded EEG signals depends on various factors such as external noises like power-line interference and artifacts (muscle movements, eye movements, etc.). For analysis and processing of EEG signals, it is necessary to eliminate such noises to obtain more accurate and appropriate results. The corrupted EEG signal is, hence, preprocessed using denoising techniques which increases the reliability of EEG data. For removal of artifacts various denoising techniques can be used, among which ICA, PCA-based EASI, and wavelet denoising are prominently selected for this study.
Cardoso and Laheld [1] have given the algorithm for blind source separation known as EASI. The components obtained after PCA are used for extraction of sources using EASI algorithm. An unsupervised algorithm for artifactual component identification is proposed by Mahajan and Morshed [2], which is used here for components obtained after ICA and EASI. Hazra and Guhathakurta [3] measured the performance of different denoising algorithms and is measured using SNR and Mean Square Error (MSE). EEG signal artifacts (ocular) are normally in low-frequency region, hence, denoising is applied on low-frequency Senthil Kumar et al. [4] implemented a method to remove ocular artifacts from EEG data using wavelet transform without an EOG reference channel. Most studies have done analysis on different techniques for denoising EEG signal but not many have compared ICA, PCA-based EASI method, and Wavelet denoising.
2 Materials and Methods
In this work, the EEG data taken into consideration has been acquired by Jadhav et al. [5] using wireless EMOTIV EPOC+ with 14 electrodes (placed according to standard 10–20 system) and sampling frequency of 128 Hz. The protocol shown to subjects has a part of meditation initially followed by slides of pictures depicting four types of emotion. The experimental protocol is given in Fig. 1. Images (I1, I2, I3, and I4) depicting emotions happy, sad, angry, and relax are shown with blank spaces in between.
2.1 Independent Component Analysis (ICA)
EEG signals follow all the assumptions of ICA model and use of ICA model on EEG has been validated [6]. If we have n different observed signals, namely x1, x2, x3, … xn and some random variables as s1, s2, s3, … sn. These observed signals can be expressed in linear combination as [7]
For i = 1, 2, 3 … n where, aij are real constants, where j = 1, 2, 3… n.
ICA technique is used to estimate the random components s1, s2, s3, s4, …, sn which are also known as the independent components (ICs). In vector-matrix notation, this mixing model can be expressed as
To estimate sources, a separation matrix W should be found. Based on experimental results on statistical and computational load analysis of various ICA algorithms, fixed-point Fast-ICA algorithm using tanh nonlinearity with symmetrical orthogonalization is chosen for finding ICs [7].
Fast-ICA implementation for finding Independent Components.
Preprocessing. For better conditioning and making IC estimation simpler, preprocessing is done by centering and whitening [7] on the acquired EEG data x.
Centering—Zero-mean data \( {\bar{\text{x}}} \) is computed as follows:
Whitening—Eigen Value Decomposition (EVD) of covariance matrix of \( {\bar{\text{x}}} \) is used to compute whitened data z
where E is orthogonal matrix of eigenvectors, D is diagonal matrix of its eigenvalues.
Fast-ICA algorithm. Algorithm of fixed-point fast-ICA for several units using symmetrical orthogonalization is given by Hyvärinen and Oja [8] using which separation matrix W such that \( {\tilde{\text{s}}} \) = Wz is computed. Here, \( {\tilde{\text{s}}} \) is the matrix of estimated sources or independent components. W is updated using the following equation:
where, g(y) = tanh(y), W = (w1, w2, w3 …, wm)T and m is the number of independent components to be estimated.
Artifactual component identification and elimination. After finding ICs using fast-ICA, for identification and elimination of artifactual components, kurtosis property of ICs and DWT is used as given in Sect. 2.3.
2.2 Principal Component Analysis Based EASI
In PCA algorithm, data dimension is reduced by getting component with maximum weight without losing more information. From principal components, i.e., compressed data, the individual components found using (EASI).
PCA algorithm. The N-channel EEG signal XMxN is centered to \( {\bar{\text{X}}} \). By applying rotated orthogonal coordinate system, the data is uncorrelated. Correlation matrix Rxx is obtained. The eigenvalues and eigenvectors are found. Eigenvalues are arranged such that λ1 > λ2 > … λN and accordingly eigenvectors are arranged c1, c2 …, cN. Where, λ = [ λ1, λ2, …, λN] and C = [c1, c2, …, cN]. Equation 6 is used to obtain the matrix of principal components [7, 9]
where Y = [y1, y2 … yN]T. To select the number of principal components, the contribution rate of each component is to be considered using eigenvalues. The contribution rate of each component is found by \( \frac{{\uplambda{\text{i}}}}{{\mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{N}}\uplambda{\text{i}}}} \).
The cumulative contribution rates are plotted in Fig. 2 and eliminated according to its weight. Components of cumulative contribution rate below 99.604 are selected to avoid unwanted data loss. Hence, 12 components among 14 are selected [9].
EASI algorithm. For Blind Source Separation (BSS), EASI is used. EASI algorithm is based on serial updation where transformation on data as well as parameter is equivalent. Assuming source signal to be s and x is matrix of selected principal components, then x can be given as x = As, where, A is the mixing matrix.
To estimate these sources, we find a separating matrix W such that
Here, y is very close to s. Ideally, y = s for obtaining accurate sources which implies A = W−1. The separating matrix for EASI algorithm is updated as follows [9, 10]:
where µ is learning rate, I is identity matrix, W = [W1, W2, … Wn]T is separation matrix and g(Y) = [g(y1), g(y2) …, g(yn)]T, g(.) is considered as any nonlinear function. The estimation of sources is given by Y = WX. If source is super-Gaussian, then nonlinear function tanγh (γ > 1) is used [9]. Main interference in this experiment is EOG which is super-Gaussian in nature. Hence, nonlinear function used is tan10 h.
Artifactual component identification and elimination. Noise containing components are selected by kurtosis values of each component. Further, the thresholding is done using DWT (Discrete Wavelet Transform) instead of direct source elimination to avoid the data loss as given in Sect. 2.3.
2.3 Artifact Elimination for ICA and PCA-Based EASI
Kurtosis and confidence interval. Data having high kurtosis depict high tailed nature similar to ocular noise [2].
Procedure for finding artifactual components.
-
(a)
Compute kurtosis for each component
$$ {\text{kurtosis}} = \sum\limits_{{{\text{i}} = 1}}^{\text{n}} {\left( {\frac{{\left( {{\text{y}}_{\text{i}} - {\bar{\text{y}}}} \right)/{\text{n}}}}{{{\text{s }}^{4} }}} \right)} $$(9)
where n is number of samples, s is standard deviation of n sample data y, and \( {\bar{\text{y}}} \) is mean of samples.
-
(b)
Find upper bound of 95% Confidence Interval (CI) [2]
$$ {\text{CI}} = {\bar{\text{y}}} + \frac{\text{s}}{{\sqrt {\text{N}} }}{\text{t}} $$(10)
where N is number of components, t is the t-value for specific percentage of confidence interval. By calculating CI, threshold value is set, above which the signals are considered to be artifactual.
Further, the artifactual components detected are thresholded using DWT.
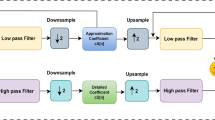
Discrete Wavelet Transform (DWT). For denoising of signal, n level DWT is applied on signal. Levels are number of times that DWT is applied on approximate coefficient of previous level. Here, four-level mother wavelets with db4 kernel are applied on signal. Thresholding of DWT coefficient is done using hard thresholding. For thresholding, threshold value is defined using universal threshold technique [4].
Universal Threshold. It is a good approach for statistical smoothness whose asymptotic behavior is better the MSE [4]. If N is number of coefficient in series and X is series of wavelet coefficients, it is formulated as
After thresholding of coefficients, reconstruction of signal is carried out by applying inverse DWT on modified wavelet coefficients. That reconstructed signal is thresholded signal using DWT.
Reconstruction of ICA and PCA.
For ICA. After artifact selection using kurtosis and thresholding data using DWT, the obtained result is multiplied to inverse of separation matrix to obtain denoised signal. The equation is given by [8]
where \( {\text{x}}_{\text{d}} \) is the denoised signal, \( {\text{s}}_{\text{t}} \) is artifact free source signal, and W is the separation matrix of ICA.
For PCA. As noise-free components are obtained, further reconstructed principal components can be obtained by multiplying inverse of separation matrix inverse with thresholded component matrix.
where \( {\text{Y}}_{\text{re}} \) is reconstructed components, W is the separation matrix of EASI, and Y is the source after artifact removal.
Further reconstruction of noise-free signal can be obtained from reconstructed principal component matrix by multiplying it with weight matrix of principal components [9].
where \( {\text{X}}_{\text{re}} \) is denoised signal, C is the PCA weight matrix.
2.4 Wavelet Denoising
Identification of Noise Region (Method 1). To identify the spike region, Stationary Wavelet Transform is used. SWT provides translational invariance which is important to identify random noise. As EEG data samples at 128 samples per sec (27), sixth (j − 1) level SWT is applied which gives detailed and approximate coefficient. The noise region is identified using the following method which automatically marks spikes in EEG data.
If detailed coefficient of sixth level exceeds 40% of maximum value of detailed coefficient present, then mark that as spike. Similarly, the whole spikes region is marked.
Thresholding Technique for method 1. In order to remove spikes from EEG data, thresholding is applied on it. This only removes ocular noise from data. Threshold is defined as
where N is a positive integer, ranging from 100 to 150, \( {\text{x}}^{{\prime }} \)—Mean of all samples, σ—Standard deviation of all samples.
From that spike, EEG data is separated and ocular noise data is removed using above threshold. The thresholding function that is used is as follows [4, 11]:
Regeneration of ocular noise-free EEG data. After thresholding of noise region in EEG data, it is necessary to regenerate noise-free EEG data. To regenerate this, inverse of stationary wavelet transform is applied on approximate and new series of detailed coefficient. By applying ISWT, ocular noise-free EEG data is obtained.
Method 2. After the applying above method, only spike region is eliminated. Now, the non-spiked noise must be removed for analysis further using DWT and thresholding method. Further, decomposition of four levels is calculated.
Thresholding Techniques for method 2. Threshold selection for the denoising is one of the important tasks [3, 12, 13].
Soft Thresholding. In soft thresholding, if coefficient value exceeds the threshold, then coefficient is modified or otherwise kept as it is.
Stein Unbiased Risk Estimator. In statistics, Stein’s unbiased risk estimate (SURE) is an unbiased estimator of the mean-squared error of a nearly arbitrary, nonlinear biased estimator.
where \( \upsigma \) is standard deviation and \( \hat{\upmu} \) is mean of wavelet coefficient X of each level.
In regaining of signal, Inverse Wavelet Transform of modified wavelet coefficient is used. Applying IDWT on wavelet coefficient, results in denoised EEG signal. The signal obtained at output is not only an ocular free but also other noise free.
2.5 Performance Parameters
The performances of the denoising methods are checked by SNR and PRD [14].
where N is number of samples in signal x, e is the error (difference between original and denoised signal), and x is the original signa
3 Results
The value of 95% CI over mean is 17.9118. Therefore, independent components IC6, IC8, IC9, IC11, and IC12 are selected as artifactual components.
The value of 95% CI over mean is 14.8248. Therefore, sources S-5, S-6, and S-12 are selected as artifactual components (Tables 1 and 2).
The average SNR values and PRD for ICA, PCA-based EASI, and wavelet denoising are 15.8844 ± 4.6606, 55.9983 ± 16.2655, 53.4595 ± 14.1662, and 24.2067 ± 8.5159, 1.6965 ± 0.9650, 8.9043 ± 4.6342, respectively.
Denoised signals by all the methods are given in Fig. 3 which shows elimination of noise. By Fig. 3 it can be observed that ocular noise elimination is more than others. Wavelet method 1 eliminates ocular noise and method 2 gives high-frequency noise-free data.
4 Conclusion
PCA-based EASI gives a better result than ICA and wavelet denoising when SNR and PRD are considered. But PCA-based EASI has a problem with convergence which does not give well-separated sources and can lead to data loss. Experimentally, to obtain well-separated sources, ICA is better method and it gives reliable output. As in wavelet denoising, the noise is removed only from selected segments, and it has better performance in all aspects.
References
Cardoso, J.-F., Laheld, B.H.: Equivariant adaptive source separation. IEEE Trans. Sig. Process. 44(12) (1996)
Mahajan, R., Morshed, B.I.: Unsupervised eye blink artifact denoising of EEG data with modified multiscale sample entropy, kurtosis, and wavelet-ICA. IEEE J. Biomed. Health Inform. 19(1) (2015)
Hazra, T.K., Guhathakurta, R.: Comparing wavelet and wavelet packet image denoising using thresholding techniques. Int. J. Sci. Res. (IJSR) 5(6) (2016)
Senthil Kumar, P., Arumuganathan, R., Sivakumar, K., Vimal, C.: Removal of ocular artifacts in the EEG through wavelet transform without using an EOG reference channel. Int. J. Open Prob. Comput. Math. 1(3) (2008)
Jadhav, N., Manthalkar, R., Joshi, Y.: Effect of meditation on emotional response: an EEG-based study. Biomed. Sig. Process. Control 34, 101–113 (2017)
Makeig, S., Bell, A.J., Jung, T.-P., Sejnowski, T.J.: Independent component analysis of electroencephalographic data. In: Proceedings of Advances in Neural Information Processing Systems (NIPS 1995), vol. 8 (1995)
Hyv¨arinen, A., Karhunen, J., Oja, E.: Independent component analysis. Wiley (2001)
Hyvärinen, A., Oja, E.: Independent component analysis: algorithms and applications. Neural Netw. 13(4–5) (2000)
Dong Kang, F., Luo Zhizeng, S.: A method of denoising multi-channel EEG signals fast based on PCA and DEBSS Algorithm. 2012 International Conference on Computer Science and Electronics Engineering, (2012)
Simranpreet Kaur, F., Sheenam Malhotra, S.: Various Techniques for Denoising EEG signal: A Review. International Journal Of Engineering and Computer Science ISSN:2319-7242 Volume 3 Issue Page No. 7965-7973, (2014)
Tibshirani, R.: Stein’s unbiased risk estimate. Statistical Machine Learning. Springer (2015)
Khatwani, P., Tiwari, A.: Removal of noise from EEG signals using cascaded filter—wavelet transforms method. Int. J. Adv. Res. Electr. Electron. Instrum. Eng. 3(12) (2014)
Estrada, E., Nazeran, H., Sierra, G., Ebrahimi, F., Mikaeili, M.: Wavelet EEG denoising for automatic sleep stage classification (2011). https://www.researchgate.net/publication/221632560
Walters-Williams, J., Li, Y.: Using invariant translation to denoise electroencephalogram signals. Am. J. Appl. Sci. 8(11), 1122–1130 (2011)
Princy, R., Thamarai, P., Karthik, B.: Denoising EEG signal using wavelet transform. Int. J. Adv. Res. Comput. Eng. Technol. (IJARCET) 4(3) (2015)
Al-Qazzaz, N.K., Hamid Bin Mohd Ali, S., Ahmad, S.A., Islam, M.S., Escudero J.: Selection of mother wavelet functions for multi-channel EEG signal analysis during a working memory task. Sensors 15, 29015–29035 (2015). https://doi.org/10.3390/s151129015
Garg, S., Narvey, R.: Denoising & feature extraction of EEG signal using wavelet transform. Int. J. Eng. Sci. Technol. 5(6) (2013)
Zheng-you, H.E., Xiaoqing, C., Guoming, L.: Wavelet entropy measure definition and its application for transmission line fault detection and identification. In: International Conference on Power System Technology (2006)
Delorme, A., Sejnowski, T., Makeig, S.: Enhanced detection of artifacts in EEG data using higher-order statistics and independent component analysis. NeuroImage 34(4) (2007)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Bhatnagar, A., Gupta, K., Pandharkar, U., Manthalkar, R., Jadhav, N. (2019). Comparative Analysis of ICA, PCA-Based EASI and Wavelet-Based Unsupervised Denoising for EEG Signals. In: Iyer, B., Nalbalwar, S., Pathak, N. (eds) Computing, Communication and Signal Processing . Advances in Intelligent Systems and Computing, vol 810. Springer, Singapore. https://doi.org/10.1007/978-981-13-1513-8_76
Download citation
DOI: https://doi.org/10.1007/978-981-13-1513-8_76
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1512-1
Online ISBN: 978-981-13-1513-8
eBook Packages: EngineeringEngineering (R0)