Abstract
In modern context, integrated approach of science and technology has given new subjects such as bioinformatics. This discipline of informatics gave a pathway to understand the larger data of various biological systems in much simplified manner. The various attributes studied in the form of computational patterns result in phylogenetic tree construction. These phylogenetic trees establish both similarities and dissimilarities among organisms. Different algorithms of clustering were studied and compared on various parameters to establish the best among them and utilities of others methods as well. The current text makes us informative about clustering methods used to generate phylogenetic trees by both distance- and character-based analyses.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The branch of bioinformatics is mainly based on analysis of various biological data such as physiological, biochemical, and genetic information with the help of modern software and data already existing as result of numerous observation made by contributors. The large number of data present helps to generate information about living world [1, 2]. Phylogenetic tree is a pictorial representation of progressive relationship among organism. Their branching shows that how much species is evolved with common ancestor. The distance of one group from the other groups indicates the degree of relationship; i.e., closely related groups are located on branches close to one another and vice versa [3]. To identify these close groups, clusters emerge as more robust method.
Cluster analysis commonly represented as clustering is a method in which we can group a set of objects or characters of similar factors more close to each other. Each cluster formed is viewed as class of objects. The objects thus clustered are based on the phenomenon of intra-class maximization in similarity and minimizing the inter-class similarity. Their grouping based on similarities and dissimilarities in their physical and genetic characteristic. The construction of graphical phylogenetic tree reveals similarity as well as dissimilarity among organisms [4]. To make a cluster or group of similar organism in the tree, various clustering methods are applied. Such observations also make the appearance of organism in relation to time. Phylogenetic tree represents branches and nodes. Basically, phylogenetic tree is categorized as follows [5].
-
(1)
Rooted Tree: Rooted trees are single node consist of a common sector and a uncommon path emerging from it across evolutionary time to any other node.
-
(2)
Un-rooted Tree: Un-rooted trees help in specifying the relationship among nodes and depict nothing about the direction in which evolution happens.
Phylogenetic trees based on sequence data give the more accurate encryption of patterns of relatedness. It also gives the Linnaean classification of new species. Phylogenetical technique is now commonly used to assess DNA evidence presented under law to inform situations. Molecular sequencing techniques with assistance of phylogenetic approaches are now in use to learn more about a new pathogen outbreak. This includes finding out about which species the pathogen is related to and subsequently the likely source of transmission. This gave the direction to new approaches recommended for public health policy. Besides this, phylogenetical recommendations help us by informing conservation policy for various uniquely identified extinct species [6].
2 Methodology
2.1 Distance-Based Method
2.1.1 UPGMA
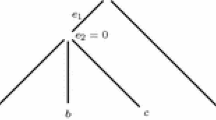
The phonograms are result of agglomerative or hierarchical clustering applied in bioinformatics. Pair-wise distance matrix (similarity index) is major tool which is to study applying algorithm, which results in the structure and construct of a rooted tree, i.e., genogram [7] (Fig. 1).
2.1.2 Neighbor Joining Method
The DNA or protein sequence data obtained after wet laboratory analysis is used for production of trees based on algorithm which generates a pair-wise distances of taxa resulting into construct. Neighbor joining is an application of distance matrix explaining the distance between each pair of taxa. Presently, neighbor joining also known as bottom clustering is frequently used technology to generate phenograms [8, 9] (Fig. 2).
2.2 Character-Based Method
2.2.1 Maximum Parsimony
This technique is the widely accepted and is based on the assumption that the most preferable tree generated depends upon the requirement of minimum number of alteration to depict the data used in alignment [10, 11]. The minimum length of the branches indicates most parsimonious tree and is considered as best representative of the evolutionary pattern. The minimum alterations within the tree construct have always been the basic approach of MP. The minimum number of changes within the tree is generated using post-order traversal activity initiating from the leaf of the tree leading toward the root [12, 13] (Fig. 3).
2.2.2 Maximum Likelihood Felsenstein
It is one of the most computationally intensive approaches. The model of nucleotide evolution and tree topology is optimal requirement of this methodology. ML is used to specify model of evolution using data available. The process also defines the greatest probability of observed data. Since ML depends upon purely probability or likelihood, it makes no similarity with other existing methods. The probability or likelihood is obtained where L = P (data/tree), indicating probability of observing data. The ML provides advantage of statistical comparisons between topologies. It is one of the robust methods used by the scientists. On the other hand the disadvantage of the process that one can observe multiple maximum likelihood for a given phylogenetic tree, which demands more detailed computational exercise [14, 15].
3 Conclusion
In all exercise, generation of construct using given sequence or data requires high degree of accuracy, thus achieving an optimal alignment along with reduction in complexity of sequences. Such results are obtained with the use of distance-based and character-based techniques. The large number of models can be constructed using distance-based method; on the other hand, phenotypic and genotypic attributes are generated using character-based method (Table 1).
References
Egan, A.N., Crandall, K.A.: Theory of Phylogenetic Estimation. Evolutionary Genetics Concepts and Case Studies, 1st edn., pp. 426–436. Oxford University Press, USA (2006)
Fitch, W.: Toward defining the course of evolution: minimum change for a specified tree topology. Syst. Zool. 20, 406–416 (1971)
Rizzo, J., Rouchka, E.C.: Review of phylogenetic tree construction. Bioinform. Rev. (2007)
Kaur, S., Singh, H., Singh, R.: Implementing UPGMA and NJ method for phylogenetic tree construction using hierarchical clustering. IJCST 4 (2013)
Munjal, G., Hanmandlu, M., Gaur, D.: A new alignment free method for phylogenetic tree construction. Int. J. Database Theory Appl. 8, 111–124 (2015)
Singha, R.S., Dasgupta, R., Bagchi, A.: A review on phylogenetic analysis: a journey through modern era. Comput. Mol. Biosci. 4, 39–45 (2014)
Kaur, M., Singh, R.: Phylogenetic tree construction revision. IJARCSSE 6 (2016)
Yadav, D.R.K., Ercal, G.: A comparative analysis of progressive multiple sequence alignment approaches using UPGMA and neighbor join based guide trees. Int. J. Comput. Sci. Eng. Inf. Technol. (IJCSEIT) 5(3/4) (2015)
Needleman, S.B., Wunsch, C.D.: A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443–453 (1970)
Chen, L.: The research of data mining algorithm based on association rules. In: The 2nd International Conference on Computer Application and System Modeling (2012)
Pujari, A.: Data Mining Techniques. Universities Press, Nancy (2001)
Sturn, A., Quackenbush, J., Trajanoski, Z.: Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208 (2002)
Verma, G., Verma, V.: Role and applications of genetic algorithm in data mining. Int. J. Comput. Appl. 4 (2012)
Wodajo, B., Mustefa, F.B., Tesfaye, K.: Clustering analysis of ethiopian safflower (Carthamus tinctorius) using ISSR Markers. Int. J. Sci. Res. Publ. 5 (2015)
Jayapriya, J., Arock, M.: Enhanced bio-inspired algorithm for constructing phylogenetic tree. ICTACT J. Soft Comput. 6 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Sharma, A., Jaloree, S., Thakur, R.S. (2018). Review of Clustering Methods: Toward Phylogenetic Tree Constructions. In: Tiwari, B., Tiwari, V., Das, K., Mishra, D., Bansal, J. (eds) Proceedings of International Conference on Recent Advancement on Computer and Communication . Lecture Notes in Networks and Systems, vol 34. Springer, Singapore. https://doi.org/10.1007/978-981-10-8198-9_50
Download citation
DOI: https://doi.org/10.1007/978-981-10-8198-9_50
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-8197-2
Online ISBN: 978-981-10-8198-9
eBook Packages: EngineeringEngineering (R0)