Abstract
ElectroEncephaloGram (EEG) signals play an important role to identify epileptic disorders. Epilepsy is a neurological disorder that is an unexpected electrical disruption of the brain, because the activity of nerve cells in the brain becomes disrupted, causing people to experience “seizures.” Nowa-day, researcher works and focuses on automatic analysis of EEG signals to classify epilepsy. The EEG signal recording system produces very long data. Thus, the classification of epileptic seizures requires a time-consuming process. This paper proposes a Support Vector Machine (SVM)-based automated seizure classification system using Approximation Entropy (ApEn). ApEn reduces patient data size without loss of information. ApEn is a statistical parameter that measures the amplitude value of an EEG signal current based on its previous amplitude value. In this paper, we measure sensitivity, specificity, and accuracy using SVM classifiers. The overall score as high as 98.62% can be achieved by using the proposed system to distinguish the epilepsy state (seizure class) from the normal state (non-seizure class) using the time domain method.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- ElectroEncephaloGram (EEG) signal
- Classification of epilepsy seizure
- Approximate entropy (ApEn)
- Support Vector Machine (SVM)
1 Introduction
Spikes of electrical activity in different brain regions are determined by the EEG signal, and we can determine the position and relative strength. Epilepsy is a disease that was taken using electricity unusual EEG activity. Fifty million people suffer from epilepsy seizures [1] around the world. Factors that may cause epilepsy, brain injury, metabolic disorders, alcohol or drug abuse, brain tumors, and genetic disorders.
A moment in time, epilepsy is not possible to predict in most cases. Classification purposes, required for continuous recording of the EEG. In some cases, EEG recording requires a very large duration of time, perhaps until a week or two. Since the traditional method was boring and slow, an automatic epileptic seizure classification system was developed [2]. The proposed work is an automatic epileptic EEG classification system using SVM and feature extraction and reduction by using approximate entropy (ApEn).
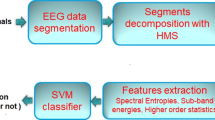
This is shown in the image below, and we will mark the EEG inputs. ApEn technique [3] is used to mark features. Extracted features are then applied to the classifier to classify seizures or non-seizures data (Fig. 1).
Epilepsy since recording began in the mid-1970s, the programmer investigation and discover EEG Signal. Epilepsy and seizures EEG analysis of the placement, the current PC-based test and discuss two issues: Epilepsy seizure classification and EEG analysis. Many feature extraction techniques have been used for the classification of epilepsy seizure. SVM (Support Vector Machine) - based classification system for epilepsy seizure has been proposed by many researchers.
The Lyapunov exponent [4, 5] provides significant details about changes in EEG activity in turn facilitating early detection of epilepsy. The correlation dimension [6] is useful to measure correlation which quantifies complex neural activity of human brain. During epileptic seizure, the value of ApEn has been found to exhibit strong relationship with synchronous discharge of large groups of neurons. The features obtained from complexity analysis and spectral analysis of EEG signals has been effectively used for diagnosis of epilepsy [7]. Recently, the approximate entropy (ApEn) [3] - based methods have been developed for analyzing linear signals for classification of epileptic seizures in epilepsy seizure [8, 9]. The mean frequency parameter of IMFs has been proposed to discriminate well between seizure and seizure-free EEG signals. For classification between healthy and epileptic EEG signals, weighted frequency has been found to be some parameter [10]. Analysis of normal and epileptic seizure EEG signals by using area measured from the trace of analytical signal representation of intrinsic mode function (IMF) has been proposed in [11]. The area parameter and mean frequency of IMFs computed using Fourier–Bessel expansion are used for epileptic seizure classification in EEG signals [12]. Also, IMFs of EEG signals have been used for recognition of epileptic seizure [9].
In first experiment, all 100 time series of F and S are taken for training and testing. For frame size 173, entropy values are 690 for each time series, so if we take 100 time series, entropy values would be 69,000 for one class and it is double (138,000) by considering both seizure and non-seizure class. These procedures followed for all four features. Entropy values of both classes S and F for training and testing dataset for all frames are shown in Table 1 (Fig. 2).
For frame size N = 2048 and m = 2 and r = 0.9, gets optimum accuracy for ApEn. From that, we get highest accuracy 95.25% of the feature ApEn with SD for experiment and frame size N = 2048 and m = 2 and r = 0.0, gets optimum accuracy for ApEn. From that, we get highest accuracy 94.25% of the feature ApEn with mean for experiment.
For training purpose, all 70-time-series data for F and S and 100-time-series data are taken for testing. Entropy values of both classes S and F for training and testing dataset for all frames are shown in Table 2.
Figure 3 are for all optimum results of experiment feature dataset as shown in Tables 2. The figure shows the SVM classification for the seizure and normal class using radial basis kernel function, where seizure is denoted by * and normal by +. The line is describing linear classification of the dataset. The o describes wrongly classify data points of opposite class. Here, we measure the performance parameters like, standard deviation (SD) [13], accuracy [14], sensitivity [14], specificity [14], and mean [13].
2 Experimentation Results
In our work, we have extracted the features from the EEG signal and classification is done using SVM classifier [15] in two class, i.e., seizure-free and seizure patient data. ApEn values are measured in form of m, r, and N. The values of m, r, and N are as follows [3]:
-
1.
Number of samples (m) = 1, 2, 3;
-
2.
Normalization ratio (r) = 0–90% of SD of the data sequence in increments of 10%;
-
3.
Frame size (N) = 173, 256, 512, 1024, and 2048.
Approximation entropy is extracted along with SD and mean. The randomness of EEG signal was extracted in the features, based on different size of frame (N), number of samples values (m), and normalized ratio (k). From the set of features, ApEn with SD and mean are used for classification using the SVM classifier.
We have used BONN dataset for EEG signals which is publicly available online and described by Andrzejak et al. [16]. The EEG dataset contains both seizures and non-seizures. The Bonn dataset consist of five subsets (Z, O, N, F, and S) each containing 100 single-channel EEG signals, each signal of 23.6 s in duration with the sampling rate of 173.61 Hz.
EEG recordings of five healthy volunteers with eyes open (Z) and closed (O) have been recorded on the surface, using standard electrode placement scheme. These two are recorded in seizure-free intervals from five patients in the epileptogenic zone (F-seizure free) and from the hippocampal formation of the opposite hemisphere of the brain (N-seizure free). The set S contained seizures signal which gives an ictal activity by using the same 128-channel amplifier system with an average common reference, and all EEG signals are recorded. In the proposed work, classification of the F (Seizure-free class) and S (Seizure class) is done by using approximate entropy (ApEn) feature extraction and reduction and SVM as classifier.
For all 100 EEG datasets, 70 datasets are used for training and the others are used for testing using SVM classifier. SVM classifier is used to classify unknown data properly. The highest accuracy is 98.625% for the feature set ApEn with SD for frame size N = 1024, sample value m = 1, and normalization ratio r = 0.0. In the proposed method, accuracy is achieved up to 98.625% for the feature set ApEn with SD. For training and testing purpose, we get different accuracy, sensitivity, and specificity as shown in Fig. 4.
As shown in Table 3, all the papers are worked on Bonn dataset and they achieved maximum accuracy 98.27%. In the proposed method, accuracy up to 98.625% is achieved for the feature set ApEn with SD.
3 Conclusion
We have extracted the features from the EEG signal, and classification done using SVM classifier in to two class seizure and normal. Approximation entropy is extracted along with SD and mean. The randomness of EEG signal was extracted in the features, based on different size of frame (N), number of samples values (m), and normalized ratio (r). From the set of features, ApEn with SD and ApEn with mean were used for classification using the SVM classifier. The highest classification accuracy is 98.625% for F and S class. The main classes of the F and S that will be an classes of the seizure data and non-seizure data.
References
Klaus Lehnertz, Florian Mormann, Thomas Kreuz, Ralph G. Andrzejak, Christoph Rieke, Peter David, And Christian E. Elger, “Seizure prediction by nonlinear EEG analysis. IEE Eng Med BiolMag”, article in IEEE engineering in medicine and biology magazine, Research gate January 2003.
N. Mc Grogan (1999). Neural network detection of epileptic seizures in the electroencephalogram, [Online]. Available: http://www.new.ox.ac.uk/˜nmcgroga/work/transfer
Vairavan Srinivasan, “Approximate Entropy-Based Epileptic EEG Detection Using Artificial Neural Networks” IEEE Transactions On Information Technology In Biomedicine, Vol. 11, No. 3, May 2007.
N.F. Güler, E.D. Übeyli, I. Güler, Recurrent neural networks employing Lyapunov exponents for EEG signal classification, Expert Systems with Applications 29 (3) (2005) 506–514, October 5. Foster, I., Kesselman, C., Nick, J., Tuecke, S.: The Physiology of the Grid: an Open Grid Services Architecture for Distributed Systems Integration. Technical report, Global Grid Forum (2002).
E.D. Übeyli, Lyapunov exponents/probabilistic neural networks for analysis of EEG signals, Expert Systems with Applications 37 (2) (2010), pp. 985–992, March.
A. Accardo, M. Affinito, M. Carrozzi, F. Bouquet, Use of the fractal dimension for the analysis of electroencephalographic time series, Biological Cybernetics 77 (5) (1997),pp. 339–350, November 2.
S.F. Liang, H.C. Wang, W.L. Chang, Combination of EEG complexity and spectral analysis for epilepsy diagnosis and seizure detection, EURASIP Journal on Advances in Signal Processing (2010), vol. 2010, Article ID 853434.
R.B. Pachori, Discrimination between ictal and seizure-free EEG signals using empirical mode decomposition, Research Letters in Signal Processing 2008 (2008), Article ID 293056.
S. Li, et al., Feature extraction and recognition of ictal EEG using EMD and SVM, Computers in Biology and Medicine 43(7) (2013), pp. 807–816.
R.J. Oweis, E.W. Abdulhay, Seizure classification in EEG signals utilizing Hilbert–Huang transform, Bio Medical Engineering On Line 10 (2011) 38, December.
R.B. Pachori, V. Bajaj, Analysis of normal and epileptic seizure EEG signals using empirical mode decomposition, Computer Methods and Programs in Biomedicine 104 (3) (2011), pp. 373–381, December 7.
V. Bajaj, R.B. Pachori, EEG signal classification using empirical mode decomposition and support vector machine, in: Proceedings International Conference on Soft Computing for Problem Solving, AISC 131, 20–22 December, 2011, Roorkee, India, 2011, pp. 623–635.
Ram Bilas Pachori, Shivnarayan Patidar, “Epileptic seizure classification in EEG signals using second-order difference plot of intrinsic mode function,” in Elsevier Transactions on Computer methods and programming in biometric(2014), vol. 9, pp. 494–502.
Varun Bajaj and Ram Bilas Pachori, “Classification of Seizure and Non seizure EEG Signals Using Empirical Mode Decomposition”, IEEE transactions on information technology in biomedicine (2012), vol. 16, no. 6, November.
Nicoletta Nicolaou, Julius Georgiou, “Detection of epileptic electroencephalogram based on Permutation Entropy and Support Vector Machines,” in Elsevier Transactions on Expert Systems with Applications (2012), vol. 8, pp. 202–209.
R.G. Andrzejak, et al., Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: dependence on recording region and brain state, Physical Review E (2001), vol. 64, Article ID 061907.
Guohun Zhu, Yan Li, Peng (Paul) Wen, “Epileptic seizure detection in EEGs signals using a fast weighted horizontal visibility algorithm,” in Elsevier Transactions on Computer method in programming biometric(2014), vol. 12, pp. 64–75.
Siulya, Yan Li a, Peng (Paul) Wenb, “Clustering technique-based least square support vector machine for EEG signal classification,” in Elsevier Transactions on Computer method in programming biometric(2011), vol. 15, pp. 358–372.
Giorgos Giannakakis, Vangelis Sakkalis, Matthew Pediaditis, And Manolis Tsiknakis “Methods For Seizure Detection And Prediction: An Overview” Springer Science+business Media New York 2014.
A.S. Muthanantha Murugavel And S. Ramakrishnan “Multi-class SVM For EEG Signal Classification Using Wavelet Based Approximate Entropy” Institute For Computer Sciences, Social Informatics And Telecommunications Engineering 2012.
U. Rajendra Acharya “Automatic Detection Of Epileptic Eeg Signals Using Higher Order Cumulant Features” International Journal Of Neural Systems, Vol. 21, No. 5 (2011).
Ling Guo, Daniel Rivero, Alejandro Pazos “Epileptic Seizure Detection Using Multiwavelet Transform Based Approximate Entropy And Artificial Neural Networks” Journal Of Neuroscience Methods 193 (2010) 156–163.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Gabani, H.B., Paunwala, C.N. (2018). EEG Signal Classification for Epileptogenic Zone and Seizure Zone. In: Kher, R., Gondaliya, D., Bhesaniya, M., Ladid, L., Atiquzzaman, M. (eds) Proceedings of the International Conference on Intelligent Systems and Signal Processing . Advances in Intelligent Systems and Computing, vol 671. Springer, Singapore. https://doi.org/10.1007/978-981-10-6977-2_5
Download citation
DOI: https://doi.org/10.1007/978-981-10-6977-2_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-6976-5
Online ISBN: 978-981-10-6977-2
eBook Packages: EngineeringEngineering (R0)