Abstract
Large graphs such as social network, web graph, biological network, are complex and facing the challenges of processing and visualization. Motivated by such issues, Taivonen et al. [1] proposed models and sequential algorithms for weighted graph with the intentions to generate a candidate compress graph. The proposed compression algorithm is expensive in terms of computation time because of the sequential process. The weighted graph compression algorithms can be made faster while adopting parallel processing technique. In this paper, we adopt parallel processing technique for weighted graph compression problem while using multi-selection nodes to perform merge-able technique with various graph clustering algorithms to avoid overlapping between nodes from different threads. For the performance evaluation purposes of the proposed method, we carry out series of tests on the real networks. We perform extensive experiments on parallel graph summarization while using different graph clustering algorithms. Our results demonstrate their effectiveness for parallel graph compression on real networks.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Real-world data are transformed to graph structure with the intention to extract hidden knowledge while exploiting the linked structure. This linked structure is playing a vital role in various domains ranging from Web structure to the data generated by scientists. In the graph structure, the node represents the entities e.g. person, Web page, module etc., and the link is the relationship between entities such as a friend link, hyperlink etc. Below, we look at some of these application domains:
World Wide Web.
The web-graph describes the directed links between webpages of the World Wide Web. The link structure of Web has been playing a significant role in the search engine such as a Google, where quality of search has been improved. Web-graph is a prime example of large-scale graph. Google estimates the total number of Webpages exceeds one trillion; experimental graphs of the World Wide Web contain more than twenty billion nodes (pages) and one hundred and sixty billion edges (hyperlinks) [2].
Social Networking Graph.
In the context of the internet, social graph signifies relations of internet users where node and edge represent users and relations among them respectively. Best known examples of social graphs are the Twitter graph and the Facebook graph, with millions of users. The social networks give birth to various groups and communities of interests, for example, music, car, school, work and much more. Communications between users of such associations are quite strong that allows identifying them easily [3].
Biological Network.
In the biological domain, the nodes represent entities such as genes, proteins, and enzymes, whereas the edges encode the relationships, such as control of the reaction and the correlation, between these objects [4]. The graph generated in such domains consists of million and billions of nodes and edges. Performing various data mining operations on such huge graph are quite challenging and expensive in terms of memory, processing and time.
Contributing in the area of graph summarization Toivonen et al. [1] proposed the randomized semi-greedy method [1] that computes possibility of all pairwise merges and then recursively performs the best merge until the required compression ratio is reached. To understand this approach, we remind that graph is contained many pairs of nodes that gives reduction in cost by merging operation. However, this algorithm is expansive in terms of execution time, because of sequential selection of nodes during graph compression. Therefore, the algorithm [1] can be made more efficient in terms of execution of time adopting the concept of parallelism. To achieve better performance, we use different clustering algorithms to cluster a large graph in to small independent graphs so that to be executed independently.
In this paper, we exploit different clustering algorithms to create independent small graph and then we apply the algorithm of [1] for compression. Then, we compare the performance of each clustering algorithms in the context of graph compression.
2 Related Work
Graph Compression.
The problem of graph compression is relevant in the study of graphs. Most of the works have been done based on common ideas and theories of graph analysis. In fact, those researchers are proposing to utilize similar connections to merge graph nodes. Navlakva et al. [5] and Tian et al. [6] made a fundamental contribution to the graph compression research. They have proposed a compact graph as a major summary model, where nodes represents a set of nodes and edges are the relationships among of the nodes. Moreover, the original graph can be recreated from summary graph by employing the set of edge corrections. The sum of size of the edge correction set and size of the summary graph is minimized by the principle of the Minimum Description Length (MDL) [7]. The most related methods to our work have been proposed by Toivonen et al. [1]. They apply several methods to determine the efficiency of compressing large graphs such as co-authorship graphs. According to the experiments, the randomized semi-greedy method, that is a generalized version of the randomized algorithm of [5] can efficiently compress a weighted graph with a little effect on the node clustering. The authors present notions of supernodes and superedges by 2-hops optimization. The selected node merges a suitable nodes with minimized mean weight in its 2-hop neighborhood to produce a smaller graph [1].
The above-mentioned works have been focused on sequential data processing for graph representation, therefore we have explored parallel data processing and graph partitioning for better performance.
Parallelization.
Parallel algorithms on graphs [8] are used in various scientific and practical applications. Multi-core systems, provide good localization of data and provide good acceleration with an increase in the number of cores. Acceleration of application processing strongly correlates with the used memory bandwidth. Therefore, parallel processing plays a significant role in many areas of scientific computing. The graph represents the vertex-centric view in the case of parallel processing. Parallel processing of graphs occurs by partitioning the graph data through processing resources and resolving dependencies along edges through iterative computation and communication.
Graph Clustering Algorithms.
We mainly reviewed the clustering algorithms such as MCODE, Graph entropy, IPCA, and COACH that have performed well in previous surveys [9].
Molecular complex detection (MCODE) is well studied method for finding dense regions of the network. Dense regions are determined based on the connectivity data in the network. This algorithm works in three ways: (1) node scoring; (2) cluster finding; and (3) post processing is repeated and clusters that do not contain the maximum connected subgraph are filtered out in the resulting subgraphs.
Proposed by Li et al., Identification of protein complexes (IPCA) algorithm is focused on vertex distance and density subgraphs. The algorithm can be managed by two equations the shortest path between a pair of vertices, and the vertex interaction probability, both in subgraph level. There are four dominant divisions in the algorithm: (1) weighting vertex and computing the weight for every edge and vertex; (2) selecting seed, where the highest weighted vertex is selected as the seed; (3) extending cluster, working on extension of some cluster K; and (4) extending judgment. The IPCA algorithm requires more time to process the data, since it is related to the number, as well as the size of the created clusters.
Core-attachment based method (COACH) operates in two phases; detection a strongly connected area, which calls the “preliminary cores” of the network, and the expansion of regions by highly connected neighbors. The COACH algorithm begins from discovering cores and end with formation complexes from the graph. If the core graph is not dense enough, core nodes will be removed from the core graph. The extended graph can contain a number of connected elements and recursively perform this process to obtain strongly connected sub-graphs in each connected elements. The deleted cores are recursively added back into resulting sub-graph. Then, applying the post-processing of the discovered cores, the maximal dense form can be obtained.
Kenley and Cho presented a novel theoretical term, “Graph entropy” that measures the structural complexity of the given graph. The algorithm includes the method of seed increment. Moreover, this method not requires threshold, due to it searches through examination, representing advantageous solution by reducing graph entropy. This technique based on searching locally the most favorable clusters, which have a minimal value of a graph entropy. Parallelization and clustering algorithms is related to each other because they have the same properties of data partitioning for solving the time complexities. However, the task of clustering is expensive, since many of algorithms require iterative or recursive procedure, and most of real-world networks are huge. Consequently, the parallelization of clustering algorithms should not be avoid.
3 Preliminary
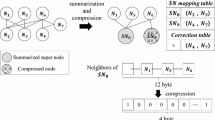
We denote some basic graph notations, which can be useful for understanding this paper. Let G = (V, E, w) be a directed weighted graph, where V is the set of vertices, E ⊂ V × V is the set of edges, and w: E → R + assigns positive weight to each edge e∈ E. \( S = (V',E',w') \) is defined as a compressed weighted graph of G if \( V' \) is a partition of V, where nodes \( v' \in V' \) are grouped to supernodes, and edges \( e' \in E' \) are grouped to superedges. A supernode is a node v ∈ VS, correspond to a set of Av nodes in G, and each edge (u, v) ∈ ES is called a superedge, which represents the edges between all pairs of nodes in Au and Av [5].
4 Parallel Compression of Graphs Using Clustering Algorithms
In this section, we introduce a parallel graph compression approach to improve the existing work [1].
4.1 Using Graph Clustering Towards Summarization
Graph clustering is the clustering of data in the form of graphs. The graph clustering algorithms are focused on summarization graph nodes by their similarities. The main purpose of clustering is the division of the graph into two or more partitions based on the similarity of objects. Clustering affects the simplification of extracting the meaningful information from the graph. The clustering algorithm generates clustering for any input data taking into account the fact that not all graphs have natural clusters. The discovery of clusters makes it possible to identify certain hidden structures in the graph.
4.2 Proposed System Overview
The task of our parallel algorithm is to divide the problem into sub-problems and executed in parallel to obtain outputs independently. For our implementation, we use more than one processor cores to run a program for processing computational intensive jobs. The program is split and executed by several CPUs at the same time, where processed results are then recombined.
The parallel compression on a single system can be achieved by using two different approaches. In the first approach, parallel processing can be performed on a single large graph. The idea is to select multiple nodes according to the available CPU cores and then merge it with a suitable node in its 2-hop neighborhood. In the second approach, the large graph can be partitioned into independent subgraphs and then can be executed in parallel by compressing each sub-graph independently. In this paper, we use the graph partitioning technique for parallel graph compression while leaving the former one for future work.
In order, to avoid the node’s overlap among the nodes in the procedure of 2-hop optimization in parallel processing and generating independent sub-graphs, we use available graph clustering algorithms which we mentioned in above section. Once we generate independent sub-graphs, then we randomly choose subgraphs accordingly to the number of CPU cores and process them in parallel.
The architecture of the proposed parallel graph compression is shown in Fig. 1. In the first stage, we partation a large graph into small independent subgraphs. Then we apply above algorithms. There are well-known methods attempt to detect highly interconnected subgraphs within the datasets. It can be applied to various datasets such as social networks, literature and other interaction network.
After this step, we select randomly different number of subgraph according to the number of available CPU cores and execute for compression independently.
4.3 The Proposed Algorithm
Algorithm 1 is the proposed algorithm for parallel compression of the weighted graph. The input to the algorithm is G where the output is S. The proposed algorithm consists of four main parts. In the part first part, we set the main required variables i.e. graphPartitioner (select the algorithm to partition the graph), cpuCount (get the numbers of processes), and executeService (create a thread pool according to the available CPUs). In the second part of the algorithm, we call a function called graphPartition which is responsible to partition the graph according to the graphPartitioner parameter (being set in step 1) while returning the numbers of partitions numOfPartitions.
One of the challenges in parallel processing is load balancing. In our algorithm, step 5 solves the problem of load balancing with the aim to utilize the resources efficiently. Step 1 gives us the number of processors (cpuCount) while step 4 returns the number of graph partitions (numOfPartations). We divide the numOfPartations by cpuCount and get the partationSlab which means that how many numbers of partitions should be executed per CPU. Finally, the submit function is used to assign the number of partitioned graphs to each processor and execute in parallel.

5 Experiments
In this section, we describe the experimental setup being used for the evaluation of the proposed algorithm. Furthermore, we present results showing favorable performance for weighted graph compression in parallel processing.
5.1 Experimental Setup
The purpose of our experimental study is to show the effectiveness and compare the scalability of various methods. For the evaluation purpose we have implemented our approach as an extension to the compression graph. We used four different clustering algorithms that are intended for partitioning the huge amount of data that is needed for parallel processing.
All the algorithms have been implemented framework program by using Java (JDK 1.7) by using NetBeans IDE 8.2 and Python 27, and all experiments were run on a standard PC with 8 GB of main memory and Intel Core i3-4130 3.40 GHz CPU processor.
5.2 Datasets
The following Table 1 introduces the datasets for our experiments. We used four real-world networks i.e., Web graph, Social Networking graph and Biological graphs [10]. The size of graph increases from 1000 to 50000 nodes. The dataset of 1 K consist of 1000 nodes and 7692 edges. The dataset of 10 K has 10000 nodes and 153308 edges, whereas 20 K dataset has 20000 nodes and 164586 edges. The biggest dataset consist of 50000 nodes with 764032 edges. Each edge of graphs has an associated numerical value as a weight. The weighted graphs are used to represent structures, where pairwise connections have some numerical value.
5.3 Results
In this section, we present experimental results of our proposed data cauterization techniques for parallel processing of compression of weighted graphs with real data sets.
Depending on algorithms properties number of generated clusters might be different. In our proposed algorithm, each thread store almost equal number of clusters. The Fig. 2 illustrates the number of clusters from datasets with 50000 nodes and 764032 edges generated by various algorithms. It can be seen that MCODE algorithm produced only 7687 number of clusters, whereas IPCA has comparable number of clusters with 19784. By contrast, the algorithms of CoAch and Graph entropy produce approximately the same amount of clusters 12084 and 10690 respectively.
In Fig. 3, we compared the results of supernodes obtained by the sequential method and our parallel cluster approach for compressing the weighted graph. It can be observed that the outperforming results for graph compression are from MCODE and Graph entropy clustering algorithms. There are less number of supernodes than existing work. The minimum number of supernodes leads to the fact that the compressed graph will be small. A small size of graph allows users to better understand and analyze the graph. Whereas, the MCODE algorithm has a directed node that allows fine-tuning of clusters of interest without considering the rest of the network and allows examination of cluster interconnectivity. The graph entropy produce a high quality clusters that can be evaluated by connectivity. A cluster has a higher quality if it has denser intra-connections within the cluster and sparser interconnections to vertices outside of the cluster. The graph entropy definition is formulated to measure the cluster quality effectively. A graph with lower entropy indicates that the vertices in the cluster have more inner links and less outer links. The IPCA and CoAch methods detect the clusters from each node and it has high probability of overlapping. In comparison with the previous listed algorithms, they produce more number of clusters due to the fact that some predicted clusters are similar and match the same benchmark clusters. However, MCODE and CoAch algorithms lost many nodes by reducing nodes, which do not have any relations. IPCA and CoAch clustering algorithm have higher number of supernodes from existing work in every dataset. Based on the given results, we can conclude, that MCODE and Graph entropy algorithms is more suitable for our approach of parallel graph partitioning and show the improvement for graph compression method of existing work.
In Fig. 4, we have been illustrated the comparison of running time (sec) of existing work with sequential compression and parallel compression. There is significant difference of computational time between sequential and four threads in parallel processing, so it is clearly seen the efficiency of parallel compression method.
Additional scalability experiments were run with different dataset size and different number of threads from one to four. As our system has four threads for each clustering algorithm, it have been a maximum number of threads in our experiments in Fig. 5.
A comparative experiment conducted between a parallel processes with optimization of cluster algorithms shows that the running time decreases by more than two times compared to a sequential process with the same clustering algorithms.
6 Conclusion
In this paper, we compared the methods of sequential data processing and parallel data processing, as well as comparison of four clustering methods in a parallel process, to analyze a large-scale of real graph dataset. We have presented the problem of parallel compression graph and gave the solution by means of graph clustering algorithms and experimental results on multiple graph datasets. In this work, we have expanded the existing work by parallel processing, which shows significant difference between sequential and parallel processing for graph compression. Our experiments illustrates that MCODE clustering method processed 764032 edges graph in 15 s in parallel, and Graph entropy method processed same size of data in 22 s, which are suitable for our approach.
Our proposed method has several advantages such as, notable results in saving the memory space and reducing the computational time. However, during multiple selection nodes in parallel, it is possible, that same nodes can be selected and for some of the query node, all CPU cycles will be wasted, but, the probability of this scenario is very low.
References
Toivonen, H., et al.: Compression of weighted graphs. In: Proceedings of the 17th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM (2011)
Sakr, S.: Processing large-scale graph data: a guide to current technology. IBM Developerworks, p. 15 (2013)
Baruah, T.D.: Effectiveness of Social Media as a tool of communication and its potential for technology enabled connections: a micro-level study. Int. J. Sci. Res. Publ. 2(5), 1–10 (2012)
Cline, M.S., et al.: Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2(10), 2366 (2007)
Navlakha, S., Rastogi, R., Shrivastava, N.: Graph summarization with bounded error. In: Proceedings of the 2008 ACM SIGMOD International Conference on Management of Data. ACM (2008)
Tian, Y., Hankins, R.A., Patel, J.M.: Efficient aggregation for graph summarization. In: Proceedings of the 2008 ACM SIGMOD International Conference on Management of Data. ACM (2008)
Rissanen, J.: Modeling by shortest data description. Automatica 14(5), 465–471 (1978)
Nielsen, F.: Introduction to HPC with MPI for Data Science. Springer, Switzerland (2016)
Price, T., Peña, F.I., Cho, Y.-R.: Survey: Enhancing protein complex prediction in PPI networks with GO similarity weighting. Interdisc. Sci. Comput. Life Sci. 5(3), 196–210 (2013)
Stanford Large Network Dataset Collection.html. http://snap.stanford.edu/data/
Acknowledgment
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. 2015R1A2A2A01008209).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
En, E., Alam, A., Khan, K.U., Lee, YK. (2018). Parallel Compression of Weighted Graphs. In: Lee, W., Choi, W., Jung, S., Song, M. (eds) Proceedings of the 7th International Conference on Emerging Databases. Lecture Notes in Electrical Engineering, vol 461. Springer, Singapore. https://doi.org/10.1007/978-981-10-6520-0_8
Download citation
DOI: https://doi.org/10.1007/978-981-10-6520-0_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-6519-4
Online ISBN: 978-981-10-6520-0
eBook Packages: EngineeringEngineering (R0)