Abstract
With increasing usage of medical images for the diagnosis in healthcare sector, the size of the image repository grows enormously. Image retrieval becomes a critical task with increasing size of repository. To address this problem, this article deals with the design of an automated system to predict the modality of medical image. This work then can be incorporated into image retrieval system with a large collection of medical images. Six modalities such as CT (computed tomography), XR (X-ray), PET (positron emission tomography), US (ultrasound), MR (magnetic resonance imaging) and PX (photograph) are considered in this experiment. Dense SIFT (scale-invariant feature transform) features, sampled at regular intervals, are extracted from the images, represented with bag-of-words histogram and classified by SVM (support vector machine). This paper explores three directions to improve the classification accuracy—usage of increasing number of training images, preferring spatial histogram rather than simple histogram and extending kernel map from linear to hellinger in SVM classifier. The obtained results are compared with existing complicated approaches and proved that better classification results are obtained with proposed simple approaches.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Over the last three decades, healthcare sector is ruled by medical imaging where the physician mainly depends on various medical imaging modalities to diagnose and treat the diseases. Based on the type of diseases, different modalities are preferred for different organs. For example, X-rays are suitable for diagnosing lung disease and bone fractures, CT for tumour detection in head and abdominal disease, ultrasound during pregnancy, etc. With the day-to-day invention of latest medical equipment, medical images are also acquired at an increasing rate. These bulk volumes of images are stored in a centralized repository and accessed frequently for diagnosis and study purposes. Retrieving image from such a large repository poses a difficult task, and hence, an effective and efficient computerized system is required to retrieve such images. The survey shows that modality is used as one of the filters to reduce the search space [1]. Hence, an automated system to identify and classify the modality of medical images becomes an emerging area of the research.
To promote the research in this sector, ImageMedCLEF—a forum, organized contest for modality classification task from the year 2010 [1]. Modality classification is the important task of ImageMedCLEF till 2013. Evaluation of the contest is based on the percentage of correctly classified images. Many research groups registered for the contest and submitted their promising results. The latest task proposed by ImageMedCLEF in 2016 such as compound figure separation also requires the results of modality classification task. Hence, the research is continued in modality classification task and explored in multidimensions to outperform the classification results obtained so far.
This paper is organized as follows: literature review is given in Sect. 2. The proposed work is discussed in Sect. 3, and experimental results are reported in Sect. 4. At the end, Sect. 5 concludes the paper and explores the way to extend the work in future.
2 Related Work
Several research groups performed experiments on modality classification tasks and submitted their results in ImageCLEFmed2013 competition [2]. IBM, the research group stood first in modality classification task, adopted sophisticated multimodal fusion techniques and obtained 81.68% classification accuracy [3]. FCSE group ranked second in modality classification task extracted densely sampled SIFT features and employed spatial pyramid [4]. The medGIFT group ranked fourth in modality classification performed feature fusion from many features descriptors and to name a few, colour and edge directivity descriptor (CEDD), bag of visual words (BoVW) using SIFT, fuzzy colour and texture histogram (FCTH) [5].
The fifth position bagged by Image and Text Integration (ITI) group adopted flat and hierarchical classification strategies with SVM [6]. The best classification accuracy in the modality classification task obtained by DEMIR research team was 64.60%, and they preferred mixed approach by combining CEDD, FCTH and colour layout descriptor (CLD) features along with textual information [7]. MIILab (Medical Image Information Laboratory) participated in ImageCLEFmed2013 modality classification task and submitted the results [8]. They extracted features using the fast filtering techniques and SURFContext with classical BoF (bags of features) approach. The overall classification accuracy is around 65%. Dimitrovski I et al. [9] evaluated classification results from different combinations of visual and textual descriptors and obtained 87.10% accuracy which is the best classification result reported so far. In [10], authors extracted different visual and textual features and employed a strategy called joint kernel equal contribution (JKEC) to give equal weightage to all the features used. Kalpathy-Cramer et al. developed neural network-based, hierarchical classifier and achieved greater than 95% classification accuracy with greyscale image [11].
Csurka et al. [12] used Fisher vector representation of the images from visual aspect and the image captions from textual aspect for classification. The authors in [13] used BoVW, bag of colours (BoC), CEDD, FCTH and fuzzy colour histogram (FCH) descriptors to represent the image. Thus, the detailed survey proves that SIFT features are used in almost many modality classification tasks giving the best classification results. Hence, experiments are conducted to optimize some parameters in the existing SIFT feature extraction and also in the classification methodologies to improve the overall accuracy still better.
3 Proposed Methodology
On seeing the frequent application of SIFT-based BoVW (bag of visual words) representation of images particularly for the classification tasks, we intended to extract dense SIFT features from the image and converted to BoVW histogram but along with some modifications in the parameters normally employed. The changes are introduced based on the contributions from the three works as follows:
-
Akata et al. [14] suggested different ways to improve the classification accuracy with large-scale images. Among them, one suggestion is to have good number of training images.
-
Vedaldi and Zisserman [15], in their assignment on Image Classification Practical, 2011, suggested to include spatial histogram to improve the classification accuracy.
-
Swathi Rao [16] proved that hellinger kernel outperformed linear kernel.
The proposed method combined the advantages of the above-stated three approaches and tested for experiments. The proposed system consists of extraction of densely sampled SIFT descriptors of reasonable size of training images, inclusion of spatial histogram from bag-of-words representation of images and comparison of classification results with SVM classifier using linear and Hellinger kernel mapping. The architecture of the proposed system is illustrated in Fig. 1. The classifier performance is evaluated at three stages, stage I with the number of images, stage II with inclusion of spatial histogram and stage III with the comparison of linear and Hellinger kernels.
The various stages of the proposed system are discussed briefly in subsequent subsections.
3.1 Dense SIFT Feature Extraction
Bag of visual words formed with SIFT features is used traditionally in many classification problems. SIFT keypoints can be extracted in three modes, key point detection, dense sampling and random sampling. SIFT keypoint represents a circle with its centre depicting x and y coordinates, the radius of the circle depicting scale and the angle depicting its orientation.
To obtain keypoints at multiple scales, Gaussian scale space is constructed. The scale space is a collection of images obtained by smoothing the input images progressively. Such a scale space is shown in Fig. 2. Smoothing the image results in reducing the resolution of images.
The keypoints are then extracted at four different scales (sigma = 0.6, 1, 1.3 and 1.6 for the Gaussian filter) and sampled densely with an interval distance of 4 pixels in an image grid. For each keypoint, 128-dimensional descriptor is obtained. To reduce the large dimension of descriptors, the obtained descriptors are then mapped to a codebook containing say 1000 codewords. Then, histogram containing the proportion of the descriptors to that specific codeword is constructed.
3.2 Bag of Visual Words
The origin of BoVW is based on the regular text analysis. Normally any text document is interpreted as the collection of words and to analyse the document; we identify the frequency of occurrences of those words. Similarly, the image can also be interpreted as the collection of visual words and to analyse the image, we identify the frequency of occurrences of those visual words.
Among three modes of SIFT feature extraction, dense sampling approach provides more keypoints as the features are extracted from the whole grid image with an interval of normally 2–4 pixels. Hence, much feature will be obtained with this approach when compared with the other two modes, keypoint detection and random sampling. Thus, to reduce the feature descriptor size appropriately, feature quantization is done by simply running k-means on the obtained descriptors. The centroids of the k-means represent the visual words of the image.
The various steps in forming the visual words of an image are as follows:
-
1.
Dense SIFT features are extracted from the training images.
-
2.
Each feature has its descriptors in 128 dimensions. k-means with say 1000 centroids is run on the obtained SIFT descriptors to end up with 1000 words.
-
3.
To represent a particular image using the visual vocabulary, again dense features are extracted from it and assigned to the visual vocabulary. The assignment is based on calculating the Euclidean distance (L2 distance) between a word and a given descriptor.
-
4.
Finally, a histogram of visual words is built to represent that particular image.
The procedure for representing BoVW histogram for one image is visually summarized in Fig. 3.
3.3 Bag of Visual Words with Spatial Information
Another approach to improve classification accuracy is incorporating spatial information on the existing plain BoVW histogram containing 1000 words. To achieve this, the given image is divided into 2 × 2 subregions and the histogram is computed for each subregion. Thus, 4 histograms with 1000 words are obtained and they are then stacked to form an array of single dimension of size 4000 (1000 × 4). Figure 4 shows the partition of an image into 2 × 2 subregions.
3.4 SVM Classifier with Linear and Hellinger Kernel
The support vector machine (SVM) introduced by Boser, Guyon and Vapnik in 1992 is used as the classifier along with kernel trick to maximize the margin of hyperplanes [17]. This algorithm just plots the feature in feature space, and using hyperplane, it identifies the boundary of each class. Kernel trick is employed to identify the best hyperplane segregating the different classes. Two SVM classifiers one with linear and other with Hellinger kernel are used for classification. Square root of the histogram is considered for implementing Hellinger kernel.
To classify the images of multiple classes, two flavours of SVM, one-versus-one and one-versus-all approaches, can be used. We preferred one-versus-all approach in which a classifier is built for each modality/class. The examples pertaining to that class are assigned positive labels and the remaining examples are assigned as negative labels. SVM with linear and Hellinger kernel mapping is used.
The one-vs-all SVM classifier classifies the feature vector as positive or negative using the Eq. 3.1.
where x, w and b are the feature vector to be classified, weight vector and bias, respectively. The values of w and b are determined during training process and the equations are then used to obtain decision hyperplane which classify the images as positive or negative. The crucial aspect is to find a set of weight and bias such that the margin is maximized. Kernel tricks are employed to obtain the best margin. The kernel makes the data linearly separable.
4 Results and Discussion
Data set
The experiments are carried out on 780 images of six different modalities. The training set consists of 50% of images while the testing set forms 50% of images in the data set. Table 1 contains the detailed split up of the images into training and testing set.
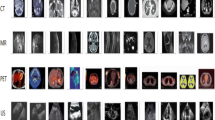
The images are collected from open-i biomedical image search engine filtered by image modality and PubMed collections [18]. Examples of images from the training data set are shown in Fig. 5.
The images obtained are of different size, and it is resized not to exceed 480 pixels in the row, and the column is adjusted automatically such that image aspect ratio is preserved. In all experiments, densely sampled SIFT features on the whole image grid with an interval of 4 pixels are extracted at 4 scales with sigma of 0.6, 1, 1.3 and 1.6. k-means with 1000 centroids is then applied on the extracted features.
As a next level, the image is partitioned into 2 × 2 subregions and again the histogram is computed separately for each subregion.
The visual words of training image from each modality are formed and their histograms are constructed as shown in Fig. 6.
This histogram is the signature of the image, and because of its uniqueness for each modality, the classifiers are trained with different histograms. One-vs-all SVM classifiers for all modalities are tested for all the test images with two variants of SVM classifiers—linear and hellinger kernels.
The proposed system is evaluated by identifying the overall classification accuracy. The overall accuracy of the system is the ratio of the number of correctly classified images to the number of all images. This is the commonly used evaluation strategy for any classification problem.
The results are tabulated as the confusion matrix for the test set and the main diagonal depicts the number of images correctly classified.
Evaluation with similar training and testing sets was performed for the following choices:
-
1.
Varying number of training images.
-
2.
BoVW histogram and BoVW spatial histogram.
-
3.
SVM with linear and Hellinger kernel.
In this section, the results of the proposed system for automatic classification of medical imaging modalities are reported. Six runs are performed for modality classification task. The classification result of all runs for each modality classifier is shown as confusion matrix.
-
Run 1: SVM with linear kernel considering 10% of training images and 2 × 2 spatial histogram.
-
Run 2: SVM with linear kernel considering 50% of training images and 2 × 2 spatial histogram.
-
Run 3: SVM with linear kernel considering 100% of training images and 2 × 2 spatial histogram.
-
Run 4: SVM with linear kernel considering 100% of training images and histogram without spatial information.
-
Run 5: SVM with hellinger kernel considering 100% of training images and histogram without spatial information.
-
Run 6: SVM with hellinger kernel considering 100% of training images and spatial histogram.
In all the above runs, in addition to the overall classification accuracy, the following metrics are calculated:
The kappa is another metric that is also used to evaluate the classifiers. It compares observed accuracy with expected accuracy from a random classifier. It is calculated using the formula
The classifier for each modality is trained with 65 images of each modality. The entire test image set consisting of 65 images of each modality is given to all the classifiers to classify the corresponding modality images. The confusion matrix for the six runs is tabulated as shown in Tables 2, 3, 4, 5, 6, 7. The various metrics are calculated to assess the performance of the classifier as given in Eqs. 4.1–4.4.
The overall classification accuracy and kappa for the six runs are tabulated in Table 8. According to Fliess, kappa > 0.75 is the best classifier, 0.40–0.75 is as fair as good and <0.40 is the worst classifier [19].
Table 8 shows that both classification accuracy and kappa keeps on increasing in the consecutive 6 runs. The 6th run, the combination of SVM with Hellinger kernel, spatial histogram and 100% training images gives the better classification accuracy of 73.077% and kappa of 0.677. Even though the classifier cannot be rated as the best, it is as fair as good, according to Fliess. The overall classification accuracy and kappa for the different runs are plotted and shown in Figs. 7 and 8, respectively.
The comparison of the proposed work with the results submitted by the research groups in the conference organized by ImageCLEF 2013 for modality classification task is tabulated in Table 9.
The output of the best run for each class is shown in Fig. 9. It can be seen from the output that some images are misclassified in each class. XR and US classifiers perform much better compared with other modality classifier. The reason behind that is PET mostly comes in combination with CT which is misclassified as CT. As visual similarities among CT, MR and PET are confusing even for human, the system predicts many images from these groups in a wrong manner. Hence, further tuning of the parameters is still required to improve the classification accuracy still better. Perhaps if the training set is built strongly including similar type of images which are wrongly misclassified, the classification accuracy can be improved still better. But that approach also should not end up in overfitting. Hence, deep analysis of wrongly misclassified images should be taken into consideration and the changes in the parameters from multiple views can be performed to achieve the goal.
5 Conclusion
The experimental results are reported for the proposed system to classify the modalities of medical images. This work is mainly to integrate into medical image retrieval system where the medical images are retrieved based on its modality. Using a data set of 780 images, six approaches are evaluated and the approach combining densely sampled SIFT descriptors and bag-of-words spatial histogram along with Hellinger kernel mapping of SVM gives the best overall classification accuracy. The maximum overall classification accuracy obtained is 73.077%. In the experiments, we have shown that increasing training images, incorporating spatial histogram and extending linear to Hellinger kernel mapping of SVM produce good results. As an extension to existing work, we plan to tune other parameters in future to improve classification results.
References
Alba Garcia Seco de Herrera, Jayashree Kalpathy-Cramer, Dina Demner-Fushman, Sameer Antani and Henning Muller, Overview of the ImageCLEF 2013 medical tasks, in: CLEF working notes 2013, Valencia, Spain, (2013).
Mani Abedini, Liangliang Cao, Noel Codella, Jonathan H. Connell, Rahil Garnavi, Amir Geva, Michele Merler, Quoc-Bao Nguyen, Sharathchandra U. Pankanti, John R. Smith, Xingzhi Sun, and Asaf Tzadok, IBM Research at ImageCLEF 2013 Medical Tasks, IBM Multimedia Analytics ImageCLEF (2013).
Ivan Kitanovski, Ivica Dimitrovski, and Suzana Loskovska, FCSE at Medical Tasks of ImageCLEF 2013, CLEF working notes (2013).
Alba G. Seco de Herrera, Dimitrios Markonis, Roger Schaer, Ivan Eggel, Henning Muller, The medGIFT Group in ImageCLEFmed 2013, CLEF working notes (2013).
Matthew S. Simpson, Daekeun You, Md. Mahmudur Rahman, Dina Demner-Fushman, Sameer Antani, and George Thoma, ITI’s Participation in the 2013 Medical Track of ImageCLEF, CLEF working notes (2013).
Okan Ozturkmenoglu, Nefise Meltem Ceylan, Adil Alpkocak, DEMIR at ImageCLEFMed 2013: The Effects of Modality Classification to Information Retrieval, Procs. of ImageCLEFMed (2013).
Xin Zhou, Miaofei Han, Yanli Song, Qiang Li, Fast filtering techniques in medical image classification and retrieval, CLEF working notes (2013).
Ivica Dimitrovski, Dragi Kocev, Ivan Kitanovski, Suzana Loskovska, Saso Dzeroski, Improved medical image modality classification using a combination of visual and textual features, Computerized Medical Imaging and Graphics 39, pp 14–26, (2015).
Xian-Hua Han and Yen-Wei Chen, Biomedical Imaging Modality Classification Using Combined Visual Features and Textual Terms, International Journal of Biomedical Imaging, vol. 2011, Article ID 241396, 7 pages, doi:10.1155/2011/241396, (2011).
Jayashree Kalpathy-Cramer, William Hersh, Automatic Image Modality Based Classification and Annotation to improve Medical Image Retrieval, MEDINFO (2007).
Gabriela Csurka, Stéphane Clinchant and Guillaume Jacquet, XRCE’s Participation at Medical Image Modality Classification and Ad-hoc Retrieval Tasks of ImageCLEF 2011, CLEF working notes (2011).
Jacinto Arias, Jesus Martinez-Gomez, Jose A. Gamez, Alba G. Seco de Herrara, Henning Muller, Medical images modality classification using discrete Bayesian Networks, Computer Vision and Image Understanding, Volume 151, Pages 61–71, October (2016).
Zeynep Akata, Florent Perronnin, Zaid Harchaoui, Cordelia Schmid, Good Practice in Large-Scale Learning for Image Classification. IEEE Transactions on Pattern Analysis and Machine Intelligence, Institute of Electrical and Electronics Engineers, 36 (3), pp. 507–520, (2014).
Andrea Vedaldi and Andrew Zisserman, Image Classification Practical, (2011).
Swathi Rao G., Effects of Image Retrieval from Image Database using Linear Kernel and Hellinger Kernel Mapping of SVM International Journal of Scientific & Engineering Research, Volume 4, Issue 5, May (2013).
B. E. Boser, I. M. Guyon, and V. N. Vapnik. A training algorithm for optimal margin classifiers. In D. Haussler, editor, 5th Annual ACM Workshop on COLT, pages 144–152, Pittsburgh, PA, ACM Press, (1992).
J. Fleiss, Statistical Methods for Rates and Proportions, 2nd ed. New York: Wiley, (1981).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Sundarambal, B., Bommanna Raja, K. (2018). Parameter Optimization for Medical Image Modality Classification. In: Dash, S., Das, S., Panigrahi, B. (eds) International Conference on Intelligent Computing and Applications. Advances in Intelligent Systems and Computing, vol 632. Springer, Singapore. https://doi.org/10.1007/978-981-10-5520-1_54
Download citation
DOI: https://doi.org/10.1007/978-981-10-5520-1_54
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5519-5
Online ISBN: 978-981-10-5520-1
eBook Packages: EngineeringEngineering (R0)