Abstract
Nucleosome, the fundamental unit of chromatin, is histone octamer composed of dimers of each histone H2A, H2B, H3, and H4. Histones are the key epigenetic players and regulate chromatin architecture. During later stages of spermatogenesis, extensive remodeling of chromatin takes place in which somatic histones get replaced by testis-specific histones, which in turn get replaced by transition proteins and finally by protamines. Disturbances that impair this highly orchestrated process may result in loose DNA packing, endangering its integrity. This reflects on sperm morphology and motility, resulting in teratozoospermia and asthenozoospermia and consequently infertility. These sperm are unable to reach the oocyte and, if they do, fail to fertilize. Assisted fertilization in the form of IVF or ICSI may help overcome this hindrance; however, the risk of failure at early embryonic developmental stages or preimplantation loss increases dramatically. This review provides an update on our current understanding of the role of sperm chromatin compaction in sperm function and the impact of its failure on male fertility.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
Key Points
-
Chromatin packaging is an integral part of spermatogenesis, and sperm DNA is packed into almost crystalline status that is at least six times more condensed in comparison to mitotic chromosomes.
-
During spermiogenesis, somatic histones in the haploid spermatid are replaced by testis-specific histones, which in turn are replaced by transition proteins and finally by protamines, leading to dense chromatin compaction in sperm.
-
Sperm histones undergo several posttranslational modifications, predominantly methylation and acetylation, to repress the transcriptional activity in sperm.
-
Compaction or proper packaging of chromatin is essential for shutting down the transcription activity in sperm and also for protecting its DNA from damage during its transit from testis through epididymis into the female reproductive tract.
-
Defects in chromatin packaging affect the morphology of sperm and its transcriptional activity and are associated with infertility or the outcome of ARTs.
-
Significantly higher histone-protamine ratios are observed in sperm from infertile men; a direct correlation exists between sperm protamine levels, DNA integrity, and sperm quality.
1 Introduction
WHO estimates as reported in 2012 indicate that about 50 million couples worldwide suffer from infertility (Mascarenhas et al. 2012). Male infertility accounts for almost 50% of the infertility. Asthenozoospermia, oligoasthenozoospermia, oligozoospermia, teratozoospermia, globozoospermia, azoospermia, and aspermia are the observed manifestations in male infertility. There is a considerable population of infertile individuals where none of these manifestations are observed and thus are referred to as idiopathic. Although research world over has been overwhelming with respect to female infertility, with respect to male infertility, it is limited probably because of the general perception that all problems of male infertility can be bypassed using assisted reproductive technologies (ARTs) such as IVF and ICSI. Increasing evidence is now available on the problems associated with ICSI (Wennerholm et al. 2000; Belva et al. 2007; Bonduelle et al. 2004; Morris et al. 2002). One of the biggest drawbacks of ICSI is that the genetic quality of sperm is overlooked leading to embryonic loss despite successful fertilization following ICSI. Genetic quality of sperm is determined by the integrity of its DNA and its compaction during spermiogenesis. A positive correlation has been observed between chromatin condensation and successful pregnancy in IUI and ICSI couples (Ioannou et al. 2016; Irez et al. 2015; Morris et al. 2002). In order to understand the impact of chromatin compaction on male fertility, it is imperative to understand the process of chromatin condensation.
2 DNA Packaging and Chromatin Compaction During Spermatogenesis
Spermatogenesis is a well-synchronized and tightly regulated process by which haploid male germ cells are formed. In the third and final stage of spermatogenesis, i.e., spermiogenesis, the haploid round spermatids undergo extensive morphological changes and nuclear remodeling to give rise to structurally distinct cell, the spermatozoa. Mature spermatozoon contains nucleus carrying haploid male genome, which is sixfold more compact as compared to any somatic cell of the body. Two major nucleoproteins involved in DNA compaction are nucleosomes and protamines. Nucleosome is the histone octamer composed of dimer of each histone H2A, H2B, H3, and H4. Histone H1 binds to the DNA in between two nucleosomes and is thought to be involved in higher-order chromatin structure formation. Each histone of nucleosome core particle (NCP) is divided into two parts: structured region made up of core and C-terminal region of histone and N-terminal unstructured tail which protrudes out of the nucleosome and interacts with DNA. Approximately 146 bp DNA is wrapped around each nucleosome. Posttranslational modifications (PTMs) like acetylation, phosphorylation, and ubiquitination occurring especially on N-terminus can influence chromatin structure either directly by adding negative or positive charge and altering histone-DNA interaction or indirectly by recruiting modification-specific chromatin remodeling factor (Pivot-Pajot et al. 2003).
Protamines are arginine-rich, small, basic, major nucleoproteins in sperm. They are synthesized in late-stage spermatid. Around 80–85% sperm DNA is compact due to protamination. In case of mammals, protamines are of two types protamine 1 (P1) and protamine 2 (P2). The presence of P1 in association with sperm DNA can be observed in nearly all vertebrates, whereas P2 is present only in primates, many rodents, and a subset of other placental mammals (Balhorn 2007). The number of protamine genes and copies present per haploid genome varies from species to species. Mammals have single-copy genes of P1 and P2, located on chromosome 16 (Reeves et al. 1989). P1and P2 are products of gene Prm1 and Prm2, respectively. The precursor protein of Prm2 undergoes proteolytic processing at its N-terminus to give rise to p2, p3, and p4. P2 family proteins, p2, p3, and p4, differ in 3–4 residues at N-terminus. The arginine-rich DNA-anchoring domains by which protamines bind with the negatively charged DNA and the multiple serine and threonine residues that can be used as phosphorylation sites form the structural elements of protamines (Balhorn 2007). The cysteine residues allow disulfide bond formation and thus link two adjacent protamines, which leads to further compaction of DNA.
During the process of spermiogenesis, nucleohistone to nucleoprotamine transition occurs. This transition is not direct but a gradual process, comprising of well-defined events, which involves first replacement of somatic histone by testis-specific histone variants and subsequently by transition proteins and then protamines.
At the round spermatid stage, the DNA compaction is the same as that of any somatic cell of the body. After the completion of second meiotic division, there is a surge of transcription observed characterized by two features not observed in somatic cells, namely, (a) the use of specialized transcriptional machinery and (b) the expression of large numbers of spermatogenic-specific genes which includes transcription of proteins like transition proteins, protamine, etc. required for spermiogenesis. At the same time, hyperacetylation of somatic histone H4 is observed. In vitro studies have indicated the role of hyperacetylated histones in nucleosome disassembly and replacement of histone by protamines (Oliva et al. 1987; Awe and Renkawitz-Pohl 2010). It has also been shown that bromodomain-containing protein (BRDT) binds with the hyperacetylated H4 and initiates nuclear remodeling (Pivot-Pajot et al. 2003; Moriniere et al. 2009).
Hyperacetylated histones then get replaced by testis-specific histone variants. Excepting histone H4, testis variants have been reported till date for core histones H2A, H2B, H3, and linker histone H1. During spermiogenesis, testis-specific histones get replaced by transition proteins (TP). Mammals, including mouse, rat, human, ram, and boar, predominantly have two types of transition proteins, viz., transition protein 1 (TP1) and transition protein 2 (TP2) (Akama et al. 1996; Chevaillier et al. 1998; Steger et al. 1998). Both TP1 and TP2 are encoded by single-copy genes, Tnp1 and Tnp2, respectively (Rathke et al. 2014). TP1 is a 6200 Da protein with about 20% arginine and 20% lysine, distributed uniformly, and no cysteine (Kistler et al. 1975). TP2 is a 13,000 Da protein with about 10% arginine, 10% lysine, and 5% cysteine (Grimes et al. 1975). It has a highly basic C-terminal domain and an N-terminal domain that forms zinc fingers (Meetei et al. 2000). TP1 is abundantly expressed (Heidaran et al. 1988), and its sequence is highly conserved in various mammals as compared to TP2 (Kremling et al. 1989). The role of TPs is not extensively studied. TP1−/− and TP2−/− knockout mice have been shown to be less fertile than normal mice and show abnormal chromatin condensation (Zhao et al. 2001). TP1 and TP2 double-knockout mice are sterile, and spermatogenesis is severely impaired suggesting their important role in spermiogenesis (Zhao et al. 2004).
Transition proteins remain associated with DNA for a short period of time and rapidly get replaced by protamines. Immediately after their synthesis, protamines get phosphorylated. Phosphorylation is thought to be essential for their nuclear transport as protamines can bind to their nuclear receptor and get transported only when phosphorylated (Mylonis et al. 2004). After binding of protamine to DNA, dephosphorylation takes place, and the disulfide bond formed between protamine further compacts the DNA. Chromodomain helicase DNA-binding protein 5 (Chd5) has a key role in the DNA compaction. It is involved in H4 hyperacetylation, histone variant expression, and removal and replacement of the histones with nucleoprotamines, and Chd5 deficiency in mice leads to defective sperm chromatin compaction and infertility (Li et al. 2014). Low expression of Chd5 has also been observed in the testis of infertile men by the same group.
3 Testis-Specific Histones
Replacement of histone by protamine is not 100%, and about 5–15% histones are still retained in mature human spermatozoa (Tanphaichitr et al. 1978; Gatewood et al. 1987; Zalensky et al. 2002). Retained histones have been found to be specifically enriched in the regulatory region of genes that are important for the earliest development stages postfertilization (Hammoud et al. 2009). Later it was shown by MNase sequencing that infertile males have random distribution of retained histones in spermatozoa (Hammoud et al. 2011). Testis-specific histone variants are thought to have specific biological function during spermiogenesis, as demonstrated by knockout studies with different variants. Table 17.1 summarizes the testis-specific histone variants known to date and their localization and influences on fertility.
Among the retained histones, TH2B is the major testis-specific histone variant present in mature sperm. TH2B differs from H2B mainly at its N-terminus. N-terminus of H2B has been shown to be associated with chromosome condensation in meiotic cell (de la Barre et al. 2001). N-terminus of TH2B has additional three potential phosphorylation sites (Ser12, Thr23, and Thr34) and repositioning of two others (ser5 and ser60), which are not seen in H2B, resulting in a different phosphorylation map of the N-terminal tail for TH2B (Pradeepa and Rao 2007).
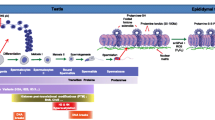
Presence of TH2A/TH2B in nucleosome has been shown to induce a more open chromatin structure (Padavattan et al. 2015). This open chromatin structure facilitates the removal of histones and their replacement by protamines, thus enabling further compaction of DNA (Montellier et al. 2013). We have earlier reported reduced TH2B in asthenozoospermic individuals (Parte et al. 2012). However, TH2B knockout male mice have been shown to be fertile as the absence of TH2B is compensated by overexpression of somatic H2B variants and modifications on other histones. But Th2b+/tag mice show arrest at condensing spermatid stage leading to lack of sperm in epididymis and consequently infertility (Montellier et al. 2013). However, double knockout for TH2A/TH2B causes defect in spermatogenesis in males. Histone replacement during spermiogenesis is also affected. The mice showed reduced testis and epididymis weight and are sterile. Secondary spermatocytes at interkinesis (the interphase between meiosis I and II) are more abundant in the mutant testis than in the wild type, suggesting extended interkinesis in mutant mice (Shinagawa et al. 2015). Interestingly, it is the TH2A and TH2B from oocyte that is involved in activation of paternal genome postfertilization (Shinagawa et al. 2014). The dynamic changes in chromatin structure during spermiogenesis, epididymal maturation, and up to early embryonic development are summarized in Fig. 17.1.
Chromatin dynamics in the sperm during its journey from a round spermatid to the formation of an embryo. Circles with no pattern represent somatic histones, circles with pattern indicate respective testis-specific histones, while circles with diagonal lines indicate the respective maternally expressed testis-specific histones. TSHV testis-specific histone variant, TP transition protein
4 Histone Modifications in Sperm and Their Influence on Sperm Fertilizing Ability/Embryonic Development
Several posttranslational modifications (PTMs) have been observed in mouse and human sperm (Fig. 17.2). In mouse sperm, 26 PTMs have been reported in specific residues of core histones and linker histone and 11 PTMs on PRM1 and PRM2 (Brunner et al. 2014). Comprehensive assessment of the histone modifications in normal human sperm revealed 102 modifications (Schon et al. 2015). Modifications are observed on the linker histone H1, the canonical histones, as well as their variants. While modifications on H4 are conserved, those on H3 vary between individuals. The modifications are not altered on cryopreservation of the sperm. Some PTMs of histones are uniquely distributed in human sperm, and this distribution varies among individuals and also between the sperm of a single individual (Krejci et al. 2015). Variations among individuals have been observed in the levels of H3K9me1, H3K9me2, H3K27me3, H3K36me3, and H3K79me1 in the sperm-head fractions. Levels of acetylated (ac) histones H4 are relatively stable. Lower levels of H3K9ac, H3K9me1, H3K27me3, H3K36me3, and H3K79me1 are seen in sperm with P2 deficiency. H3K9me2 and levels of P2 show a strong correlation. While the localization of H3 lysine 4 methylation (H3K4me) or H3 lysine 27 methylation (H3K27me) is highly similar in the gametes of infertile men compared with fertile men, a reduction in the amount of H3K4me or H3K27me retained at developmental transcription factors and certain imprinted genes has been noted. Also, the methylation status of certain developmental promoters and imprinted loci are altered in a subset of infertile men (Hammoud et al. 2011). Recently, histone PTMs and their relative abundance in distinct stages of mouse spermatogenesis and in human spermatozoa have been identified (Luense et al. 2016). They observed a strong conservation of histone PTMs for histone H3 and H4 between mouse and human sperm; however, H1, H2A, and H2B showed very little conservation (Luense et al. 2016).
Posttranslational modifications (PTMs) reported on the core histones in mouse and human spermatozoa. Colors red, blue, green, and yellow represent histones H2A, H2B, H3, and H4, respectively. The extension outside the circle represents the N-terminal region, and that inside it represents the C-terminal region. The segments on the circle represent core region of the respective histone. The PTMs reported on histones in spermatozoa of mouse, human, or both are as shown in the legend to the figure
In sperm, genes relevant to spermatogenesis are marked by H3K4me2, and the genes involved in developmental regulation are marked by H3K27me3 (Brykczynska et al. 2010). While H3K4me2 is an activating mark, H3K27me3 has been shown to be a repressor of genes. This means that prior to fertilization, the genes involved in early embryonic development are repressed by H3K27me3, while those involved in spermatogenesis are maintained in an active state by H3K4me2. Reduction in H3K4me2 induced by human KDM1A histone lysine 4 demethylase transgene overexpression during mouse spermatogenesis has been shown to severely impair development and survival of the offspring, a defect which is also seen in two subsequent generations (Siklenka et al. 2015). H3K4me2 was reduced at the CpG islands of genes involved in development. While this region is majorly marked by H3K27me3, work of Brykczynska has shown that some of the developmentally regulated genes are also marked by H3K4me2. H3K4me3 has also been demonstrated to be important for spermatogenesis; loss of H3K4 methyltransferase MII2 reduces H3K4me3 consequently rendering the male mice sterile (Glaser et al. 2009). H3K9me2/1-specific demethylase JHDM2A also known as JMJD1A is essential for spermatogenesis, and its loss causes infertility in male mice due to incomplete chromatin condensation (Okada et al. 2007).
Stage-specific modifications have been identified for TH2B with acetylated TH2B being most abundant in spermatogonia (28.9%) compared to spermatocytes (8.3) and spermatids (11.2%). At the C-terminus, phosphorylation at K116 and methylation at K117 were observed in combination in TH2B isolated from these stages (Lu et al. 2009). However, its functional relevance is not known. Various PTMs like acetylation, methylation, and phosphorylation have been identified on TH2B from tetraploid spermatocyte and haploid spermatid. LC–MS/MS analysis of TH2B from spermatocytes identified six acetylation, three monomethylation, and one phosphorylation site, while that of TH2B from round spermatids identified four acetylation and two monomethylation sites. In silico analysis showed altered histone-histone as well as histone-DNA interactions in TH2B-bearing nucleosome. Also acetylation on N-terminal tail of TH2B has been shown to weaken its interactions with the DNA (Pentakota et al. 2014). Its physiological relevance remains to be determined.
5 Protamines and Male Infertility
Protamines and histones are the two major nuclear proteins in many vertebrate species including mice, rat, human, etc. These proteins play major roles during chromatin condensation at spermiogenesis. Several reports indicate that P1 and P2 are expressed in nearly the same amount in fertile human sperm and alteration in P1/P2 ratio is associated with male infertility (Aoki et al. 2005a, 2006; Zhang et al. 2006; Hammoud et al. 2009).
The first documented report highlighting the importance of protamines revealed the absence of protamines in the spermatozoa of infertile (oligozoospermic) patients (Silvestroni et al. 1976). This was followed by a study on 7 infertile and 17 fertile individuals where increased P1/P2 ratio in six of the seven patients was observed (Balhorn et al. 1988). Thereafter, a good number of studies have indicated that fertile men express P1 and P2 in same amount, while alteration in this ratio correlates with male infertility; infertile men show either decreased or increased P1/P2 ratio (Balhorn 2007; Balhorn et al. 1988; Belokopytova et al. 1993; de Yebra et al. 1993; Mengual et al. 2003; Aoki et al. 2005a). Balhorn’s group has shown that the percentage of protamines is different in the patients with abnormal seminal parameters compared to patients with normal parameters. Also within the heterogeneous population of spermatozoa, round-headed spermatozoa from patients contain less protamines and more histones and intermediate proteins than normal spermatozoa. Further, protamine levels vary between individual sperm of infertile males and correlate with viability and DNA integrity (Aoki et al. 2006). Interestingly, studies employing Percoll separation for fractionating sperm have shown that P1/P2 ratio and total protamine from different Percoll fractions within the same sample were not significantly different. However, there were significant differences in P1/P2 ratios in the oligozoospermic and asthenozoospermic groups as compared to normozoospermic indicating that P1/P2 and amount of protamine retention were independent of morphology and motility of sperm cells (Mengual et al. 2003).
This alteration in P1/P2 ratio might be due to alteration in expression of either of two protamines or both. Several studies have indicated lower P2 and increased P2 precursor in infertile men, indicating abnormality in the processing of precursor protein (de Yebra et al. 1993, 1998; Carrell and Liu 2001; Torregrosa et al. 2006). Aoki et al. observed a P1/P2 ratio around 1 in fertile donors; in infertile group, the P1/P2 ratios were either less than 0.8, between 0.8 and 1.2, or greater than 1.2 in 13.6%, 46.7%, and 39.7% of the patients, respectively. P1 and P2 were both under-expressed in patients with a normal P1/P2 ratio. In patients with a high P1/P2 ratio, P1 was normally expressed and P2 was under-expressed. They also reported that patients with abnormal P1/P2 ratios displayed significantly reduced semen quality and sperm penetration ability (Aoki et al. 2005a).
Several studies have also reported the presence of protamine transcript in sperm. Significantly aberrant protamine mRNA ratio was found in infertile individuals, and it correlates with DNA fragmentation and IVF success (Steger et al. 2001; Rogenhofer et al. 2013; Ni et al. 2014a). Significantly higher PRM1 and PRM2 mRNA copy numbers have been observed in normozoospermic versus teratozoospermic samples (Savadi-Shiraz et al. 2015). In contrast, transition protein 2 (TNP2) transcript abundance was significantly higher in teratozoospermic samples and positively correlated with sperm-head defects.
6 Protamines, DNA Compaction, and Integrity
Protamines are essential for sperm-specific packing of DNA. Compaction of DNA shuts off transcription as the DNA is no more amenable to the transcription factors and RNA polymerase. It also protects the DNA from any damage thus maintaining its integrity. This ensures that postfertilization the paternal genome is delivered in a form that allows developing embryo to accurately express genetic information. DNA compaction during chromatin condensation changes a transcriptionally active chromatin into a transcriptionally silent chromatin, and all the genes that are required for spermatogenesis and sperm function are transcribed prior to this transition, i.e., until the round spermatid stage. Live imaging studies in Drosophila have shown that histone-to-protamine transition starts 50–60 h after completion of meiosis and lasts for 5–6 h (Awe and Renkawitz-Pohl 2010). In mice although there is no direct evidence such as live imaging, indirect evidences suggest that this transition starts approximately 156 h after completion of meiosis and it lasts for 120–126 h, i.e., from step 10 to step 15 of spermiogenesis (reviewed by Rathke et al. 2014). This period is characterized by DNA breaks and repair which allows relief from the torsion stress and facilitates removal of the histones and replacement by transition proteins and subsequently protamines (Marcon and Boissonneault 2004). Thereafter, selective translation of the stored mRNA takes place as per the requirements of the sperm.
It is well established that about 5–15% histones are retained in normal human sperm. Elegant studies by Hammoud et al. have shown gene clusters important for embryonic development to be associated with the retained histones in sperm of fertile men (Hammoud et al. 2009). This implies that improper packaging due to higher histone retention as seen in sperm chromatin of infertile men may expose many more gene clusters. A subsequent study by the same group showed that in infertile men, histones retention was random genome-wide, unlike fertile men where the histone retention was seen only at specific gene clusters (Hammoud et al. 2011). The epigenetic marks H3K4me or H3K27me were also reduced on the retained histones in the infertile men. They speculate that these changes may be responsible for the poor reproductive outcome post ICSI/IVF in infertile men.
Any defects in chromatin packaging wreaks havoc with the sperm ability to fertilize or sire a viable offspring either by allowing the untimely transcription of certain genes, allowing certain modifications of histones that may switch the transcription on or off, or increasing the vulnerability of the DNA to drug-induced damage. Observations from the chromodomain helicase DNA-binding protein 5 (CHD5) KO mice are a testimony to the effect of improper condensation on sperm morphology and fertility of the male offspring (Zhuang et al. 2014). H4 hyperacetylation, which is vital for histone replacement during spermiogenesis, is reduced in these mice, and the sperm show deformed nuclei and abnormal head morphology. However, in these mice transcription of important genes, controlling spermatogenesis was not affected. Several groups have shown very lucidly the correlation between protamine compaction, DNA integrity, and sperm quality (Franken et al. 1999; García-Peiró et al. 2011; Manochantr et al. 2012; Utsuno et al. 2014). Chromatin packaging as studied by CMA3 and acidic aniline blue staining negatively correlates with normal sperm morphology (Franken et al. 1999). Utsuno et al. observed abnormal protamination in significantly higher number of spermatozoa with abnormal head morphology compared to those with normal head morphology. DNA fragmentation was also higher in the protamine-deficient spermatozoa. Studies on DNA damage in men undergoing IVF treatment revealed a positive association between DNA damage and abnormal sperm morphology and motility and negative correlation with sperm concentration (Morris et al. 2002). Protamine 2-deficient mice sperm demonstrate a direct correlation between PRM2 haploinsufficiency and frequency of DNA damage as seen from comet assays and ultrastructural analysis (Cho et al. 2003). In studies with human sperm, a positive correlation has been shown between protamine deficiency and sperm DNA damage (De Iuliis et al. 2009; Nili et al. 2009; Tarozzi et al. 2009; Razavi et al. 2010; Manochantr et al. 2012; Utsuno et al. 2014). Several studies have correlated altered P1/P2 ratio with susceptibility to DNA damage (Aoki et al. 2005b, 2006).
7 DNA Integrity and ART Outcomes
DNA integrity also influences sperm penetration and fertilizing ability, IVF and embryo quality, and development in ICSI outcome (Khara et al. 1997; Carrell et al. 1999; Carrell and Liu 2001; Nasr-Esfahani et al. 2004; de Mateo et al. 2009). DNA fragmentation and CMA3 positivity indicative of protamine deficiency negatively correlate with the fertilization rate in ICSI patients; DNA methylation negatively correlated with DNA fragmentation (Tavalaee et al. 2009). However, Tarozzi et al. observed a close relationship between sperm protamination and fertilization and pregnancy only in IVF; in ICSI there was a correlation between DNA fragmentation and pregnancy (Tarozzi et al. 2009). In men enrolled for ICSI, a positive association was seen between sperm damage and impairment of postfertilization embryo cleavage (Morris et al. 2002). In another study of individuals referred for ICSI, CMA3 positivity showed a significant negative correlation with fertilization rate post ICSI (Iranpour 2014). An isolated study using cleavage-stage frozen-thawed embryos from cycles of IVF and ICSI has however observed no significant difference in the biochemical pregnancy, clinical pregnancy, and miscarriage rates between sperm showing DFI <30% and those >30% (Ni et al. 2014b). The group did find some association between DFI and blastocyst formation in the ICSI group. A recent study investigating the influence of sperm DNA fragmentation on the pregnancy outcome and pregnancy loss after ART in couples going for either autologous ICSI, ICSI using donor eggs, or IUI observed that while the pregnancy rates were not significantly different, pregnancy losses correlated positively with the DNA fragmentation which was measured as DNA fragmentation index (DFI). The study indicates that sperm samples showing DFI >27% are associated with an increased risk of early pregnancy loss (Rilcheva Violeta et al. 2016). A similar observation has been reported earlier (Jin et al. 2015). Additionally, this group observed that when the DFI exceeded 27.3%, the live birth and implantation rates were significantly reduced in women with reduced ovarian reserve vis-a-vis women with normal ovarian reserve.
Conclusions and Future Directions
DNA integrity and its proper compaction in sperm are vital to its fertilizing ability as well as for early embryonic development in the preimplantation stage. Poor DNA compaction in sperm severely hampers its fertilizing ability and further development that accounts for fertility loss in natural conception or poor success of IVF/ICSI procedures. While literature is replete with evidences on histone retention and protamine deficiency in infertile cases, our knowledge on impact of several histone modifications on the fertility of male is limited and needs attention. Further research in this direction may identify sperm chromatin tests that may predict the success of ARTs. At the same time, further studies are needed to understand the significance of the retained histones in sperm maturation and their contribution toward the fertilizing ability of sperm.
References
Akama K, Sato H, Furihata-Yamauchi M, Komatsu Y, Tobita T, Nakano M (1996) Interaction of nucleosome core DNA with transition proteins 1 and 3 from boar late spermatid nuclei. J Biochem 119:448–455
Aoki VW, Liu L, Carrell DT (2005a) Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod 20:1298–1306
Aoki VW, Moskovtsev SI, Willis J, Liu L, Mullen JB, Carrell DT (2005b) DNA integrity is compromised in protamine-deficient human sperm. J Androl 26:741–748
Aoki VW, Emery BR, Liu L, Carrell DT (2006) Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl 27:890–898
Awe S, Renkawitz-Pohl R (2010) Histone H4 acetylation is essential to proceed from a histone- to a protamine-based chromatin structure in spermatid nuclei of Drosophila melanogaster. Syst Biol Reprod Med 56:44–61
Balhorn R (2007) The protamine family of sperm nuclear proteins. Genome Biol 8:227
Balhorn R, Reed S, Tanphaichitr N (1988) Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia 44:52–55
Belokopytova IA, Kostyleva EI, Tomilin AN, Vorob’ev VI (1993) Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol Reprod Dev 34:53–57
Belva F, Henriet S, Liebaers I, Van Steirteghem A, Celestin-Westreich S, Bonduelle M (2007) Medical outcome of 8-year-old singleton ICSI children (born >or =32 weeks’ gestation) and a spontaneously conceived comparison group. Hum Reprod 22:506–515
Bonduelle M, Bergh C, Niklasson A, Palermo GD, Wennerholm UB (2004) Medical follow-up study of 5-year-old ICSI children. Reprod Biomed Online 9:91–101
Brunner AM, Nanni P, Mansuy IM (2014) Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin 7:2
Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH (2010) Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17:679–687
Carrell DT, Liu L (2001) Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J Androl 22:604–610
Carrell DT, Emery BR, Liu L (1999) Characterization of aneuploidy rates, protamine levels, ultrastructure, and functional ability of round-headed sperm from two siblings and implications for intracytoplasmic sperm injection. Fertil Steril 71:511–516
Catena R, Ronfani L, Sassone-Corsi P, Davidson I (2006) Changes in intranuclear chromatin architecture induce bipolar nuclear localization of histone variant H1T2 in male haploid spermatids. Dev Biol 296:231–238
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ et al (2002) Genomic instability in mice lacking histone H2AX. Science 296:922–927
Chevaillier P, Chirat F, Sautiere P (1998) The amino acid sequence of the ram spermatidal protein 3--a transition protein TP3 or TP4? Eur J biochem 258:460–464
Cho C, Jung-Ha H, Willis WD, Goulding EH, Stein P, Xu Z, Schultz RM, Hecht NB, Eddy EM (2003) Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod 69:211–217
De Iuliis GN, Thomson LK, Mitchell LA, Finnie JM, Koppers AJ, Hedges A, Nixon B, Aitken RJ (2009) DNA damage in human spermatozoa is highly correlated with the efficiency of chromatin remodeling and the formation of 8-hydroxy-2′-deoxyguanosine, a marker of oxidative stress. Biol Reprod 81:517–524
de la Barre AE, Angelov D, Molla A, Dimitrov S (2001) The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J 20:6383–6393
de Mateo S, Gazquez C, Guimera M, Balasch J, Meistrich ML, Ballesca JL, Oliva R (2009) Protamine 2 precursors (Pre-P2), protamine 1 to protamine 2 ratio (P1/P2), and assisted reproduction outcome. Fertil Steril 91:715–722
de Yebra L, Ballesca JL, Vanrell JA, Bassas L, Oliva R (1993) Complete selective absence of protamine P2 in humans. J Biol Chem 268:10553–10557
de Yebra L, Ballesca JL, Vanrell JA, Corzett M, Balhorn R, Oliva R (1998) Detection of P2 precursors in the sperm cells of infertile patients who have reduced protamine P2 levels. Fertil Steril 69:755–759
Drabent B, Bode C, Bramlage B, Doenecke D (1996) Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem Cell Biol 106:247–251
Drabent B, Benavente R, Hoyer-Fender S (2003) Histone H1t is not replaced by H1.1 or H1.2 in pachytene spermatocytes or spermatids of H1t-deficient mice. Cytogenet Genome Res 103:307–313
Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I (2001) Histone variant H2A.Z is required for early mammalian development. Curr Biol 11:1183–1187
Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A (2003) H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4:497–508
Franken DR, Franken CJ, De La Guerre H, De Villiers A (1999) Normal sperm morphology and chromatin packaging: comparison between aniline blue and chromomycin A3 staining. Andrologia 31:361–366
García-Peiró A, Martínez-Heredia J, Oliver-Bonet M, Abad C, Amengual MJ, Navarro J, Jones C, Coward K, Gosálvez J, Benet J (2011) Protamine 1 to protamine 2 ratio correlates with dynamic aspects of DNA fragmentation in human sperm. Fertil Steril 95:105–109
Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW (1987) Sequence-specific packaging of DNA in human sperm chromatin. Science 236:962–964
Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F, Seibler J, Roellig D, Kranz A, Anastassiadis K et al (2009) The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin 2:5
Grimes SR Jr, Platz RD, Meistrich ML, Hnilica LS (1975) Partial characterization of a new basic nuclear protein from rat testis elongated spermatids. Biochem Biophys Res Commun 67:182–189
Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478
Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT (2011) Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod 26:2558–2569
Heidaran MA, Showman RM, Kistler WS (1988) A cytochemical study of the transcriptional and translational regulation of nuclear transition protein 1 (TP1), a major chromosomal protein of mammalian spermatids. J Cell Biol 106:1427–1433
Ichijima Y, Sin HS, Namekawa SH (2012) Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell Mol Life Sci 69:2559–2572
Iguchi N, Tanaka H, Yomogida K, Nishimune Y (2003) Isolation and characterization of a novel cDNA encoding a DNA-binding protein (Hils1) specifically expressed in testicular haploid germ cells. Int J Androl 26:354–365
Iguchi N, Tanaka H, Yamada S, Nishimura H, Nishimune Y (2004) Control of mouse hils1 gene expression during spermatogenesis: identification of regulatory element by transgenic mouse. Biol Reprod 70:1239–1245
Ioannou D, Miller D, Griffin DK, Tempest HG (2016) Impact of sperm DNA chromatin in the clinic. J Assist Reprod Genet 33:157–166
Iranpour FG (2014) Impact of sperm chromatin evaluation on fertilization rate in intracytoplasmic sperm injection. Adv Biomed Res 3:229
Irez T, Sahmay S, Ocal P, Goymen A, Senol H, Erol N, Kaleli S, Guralp O (2015) Investigation of the association between the outcomes of sperm chromatin condensation and decondensation tests, and assisted reproduction techniques. Andrologia 47:438–447
Jin J, Pan C, Fei Q, Ni W, Yang X, Zhang L, Huang X (2015) Effect of sperm DNA fragmentation on the clinical outcomes for in vitro fertilization and intracytoplasmic sperm injection in women with different ovarian reserves. Fertil Steril 103:910–916
Khara KK, Vlad M, Griffiths M, Kennedy CR (1997) Human protamines and male infertility. J Assist Reprod Genet 14:282–290
Kistler WS, Geroch ME, Williams-Ashman HG (1975) A highly basic small protein associated with spermatogenesis in the human testis. Investig Urol 12:346–350
Krejci J, Stixova L, Pagacova E, Legartova S, Kozubek S, Lochmanova G, Zdrahal Z, Sehnalova P, Dabravolski S, Hejatko J et al (2015) Post-translational modifications of histones in human sperm. J Cell Biochem 116:2195–2209
Kremling H, Luerssen H, Adham IM, Klemm U, Tsaousidou S, Engel W (1989) Nucleotide sequences and expression of cDNA clones for boar and bull transition protein 1 and its evolutionary conservation in mammals. Differentiation 40:184–190
Lee J, Park HS, Kim HH, Yun YJ, Lee DR, Lee S (2009) Functional polymorphism in H2BFWT-5′UTR is associated with susceptibility to male infertility. J Cell Mol Med 13:1942–1951
Li W, Wu J, Kim SY, Zhao M, Hearn SA, Zhang MQ, Meistrich ML, Mills AA (2014) Chd5 orchestrates chromatin remodelling during sperm development. Nat Commun 5:3812
Lu S, Xie YM, Li X, Luo J, Shi XQ, Hong X, Pan YH, Ma X (2009) Mass spectrometry analysis of dynamic post-translational modifications of TH2B during spermatogenesis. Mol Hum Reprod 15:373–378
Luense LJ, Wang X, Schon SB, Weller AH, Lin Shiao E, Bryant JM, Bartolomei MS, Coutifaris C, Garcia BA, Berger SL (2016) Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin 9:24
Manochantr S, Chiamchanya C, Sobhon P (2012) Relationship between chromatin condensation, DNA integrity and quality of ejaculated spermatozoa from infertile men. Andrologia 44:187–199
Marcon L, Boissonneault G (2004) Transient DNA strand breaks during mouse and human spermiogenesis:new insights in stage specificity and link to chromatin remodeling. Biol Reprod 70:910–918
Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I (2005) Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A 102:2808–2813
Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA (2012) National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med 9:e1001356
Meetei AR, Ullas KS, Rao MR (2000) Identification of two novel zinc finger modules and nuclear localization signal in rat spermatidal protein TP2 by site-directed mutagenesis. J Biol Chem 275:38500–38507
Mengual L, Ballesca JL, Ascaso C, Oliva R (2003) Marked differences in protamine content and P1/P2 ratios in sperm cells from percoll fractions between patients and controls. J Androl 24:438–447
Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, Fenaille F, Shiota H, Debernardi A, Hery P, Curtet S et al (2013) Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev 27:1680–1692
Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ et al (2009) Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461:664–668
Morris ID, Ilott S, Dixon L, Brison DR (2002) The spectrum of DNA damage in human sperm assessed by single cell gel electrophoresis (Comet assay) and its relationship to fertilization and embryo development. Hum Reprod 17:990–998
Mylonis I, Drosou V, Brancorsini S, Nikolakaki E, Sassone-Corsi P, Giannakouros T (2004) Temporal association of protamine 1 with the inner nuclear membrane protein lamin B receptor during spermiogenesis. J Biol Chem 279:11626–11631
Nasr-Esfahani MH, Salehi M, Razavi S, Mardani M, Bahramian H, Steger K, Oreizi F (2004) Effect of protamine-2 deficiency on ICSI outcome. Reprod Biomed Online 9:652–658
Nekrasov M, Soboleva TA, Jack C, Tremethick DJ (2013) Histone variant selectivity at the transcription start site: H2A.Z or H2A.Lap1. Nucleus 4:431–438
Ni K, Steger K, Yang H, Wang H, Hu K, Chen B (2014a) Sperm protamine mRNA ratio and DNA fragmentation index represent reliable clinical biomarkers for men with varicocele after microsurgical varicocele ligation. J Urol 192:170–176
Ni W, Xiao S, Qiu X, Jin J, Pan C, Li Y, Fei Q, Yang X, Zhang L, Huang X (2014b) Effect of sperm DNA fragmentation on clinical outcome of frozen-thawed embryo transfer and on blastocyst formation. PLoS One 9:e94956
Nili HA, Mozdarani H, Aleyasin A (2009) Correlation of sperm DNA damage with protamine deficiency in Iranian subfertile men. Reprod Biomed Online 18:479–485
Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y (2007) Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450:119–123
Oliva R, Bazett-Jones D, Mezquita C, Dixon GH (1987) Factors affecting nucleosome disassembly by protamines in vitro. Histone hyperacetylation and chromatin structure, time dependence, and the size of the sperm nuclear proteins. J Biol Chem 262:17016–17025
Padavattan S, Shinagawa T, Hasegawa K, Kumasaka T, Ishii S, Kumarevel T (2015) Structural and functional analyses of nucleosome complexes with mouse histone variants TH2a and TH2b, involved in reprogramming. Biochem Biophys Res Commun 464:929–935
Parte PP, Rao P, Redij S, Lobo V, D'Souza SJ, Gajbhiye R, Kulkarni V (2012) Sperm phosphoproteome profiling by ultra performance liquid chromatography followed by data independent analysis (LC-MS(E)) reveals altered proteomic signatures in asthenozoospermia. J Proteome 75:5861–5871
Pentakota SK, Sandhya SP, Sikarwar A, Chandra N, Satyanarayana Rao MR (2014) Mapping post-translational modifications of mammalian testicular specific histone variant TH2B in tetraploid and haploid germ cells and their implications on the dynamics of nucleosome structure. J Proteome Res 13:5603–5617
Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S (2003) Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol 23:5354–5365
Pradeepa MM, Rao MR (2007) Chromatin remodeling during mammalian spermatogenesis: role of testis specific histone variants and transition proteins. Soc Reprod Fertil Suppl 63:1–10
Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R (2014) Chromatin dynamics during spermiogenesis. Biochim Biophys Acta 1839:155–168
Razavi SH, Nasr-Esfahani MH, Deemeh MR, Shayesteh M, Tavalaee M (2010) Evaluation of zeta and HA-binding methods for selection of spermatozoa with normal morphology, protamine content and DNA integrity. Andrologia 42:13–19
Reeves RH, Gearhart JD, Hecht NB, Yelick P, Johnson P, O'Brien SJ (1989) Mapping of PRM1 to human chromosome 16 and tight linkage of Prm-1 and Prm-2 on mouse chromosome 16. J Hered 80:442–446
Rilcheva Violeta S, Ayvazova Nina P, Ilieva Lyubomira O, Ivanova Svetlana P, Konova EI (2016) Sperm DNA integrity test and assisted reproductive technology (art) outcome. J Biomed Clin Res 9(1):21–29
Rogenhofer N, Dansranjavin T, Schorsch M, Spiess A, Wang H, von Schonfeldt V, Cappallo-Obermann H, Baukloh V, Yang H, Paradowska A et al (2013) The sperm protamine mRNA ratio as a clinical parameter to estimate the fertilizing potential of men taking part in an ART programme. Hum Reprod 28:969–978
Savadi-Shiraz E, Edalatkhah H, Talebi S, Heidari-Vala H, Zandemami M, Pahlavan S, Modarressi MH, Akhondi MM, Paradowska-Dogan A, Sadeghi MR (2015) Quantification of sperm specific mRNA transcripts (PRM1, PRM2, and TNP2) in teratozoospermia and normozoospermia: new correlations between mRNA content and morphology of sperm. Mol Reprod Dev 82:26–35
Schenk R, Jenke A, Zilbauer M, Wirth S, Postberg J (2011) H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120:275–285
Schon SB, Luense LJ, Wang X, Donahue G, Garcia BA, Bartolomei MS, Berger SL (2015) A comprehensive assessment of histone modifications in human sperm. Fertil Steril 104:e296–e2e7
Shinagawa T, Takagi T, Tsukamoto D, Tomaru C, Huynh LM, Sivaraman P, Kumarevel T, Inoue K, Nakato R, Katou Y et al (2014) Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 14:217–227
Shinagawa T, Huynh LM, Takagi T, Tsukamoto D, Tomaru C, Kwak HG, Dohmae N, Noguchi J, Ishii S (2015) Disruption of Th2a and Th2b genes causes defects in spermatogenesis. Development 142:1287–1292
Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M et al (2015) Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 350:aab2006
Silvestroni L, Frajese G, Fabrizio M (1976) Histones instead of protamines in terminal germ cells of infertile, oligospermic men. Fertil Steril 27:1428–1437
Steger K, Klonisch T, Gavenis K, Drabent B, Doenecke D, Bergmann M (1998) Expression of mRNA and protein of nucleoproteins during human spermiogenesis. Mol Hum Reprod 4:939–945
Steger K, Failing K, Klonisch T, Behre HM, Manning M, Weidner W, Hertle L, Bergmann M, Kliesch S (2001) Round spermatids from infertile men exhibit decreased protamine-1 and -2 mRNA. Hum Reprod 16:709–716
Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K et al (2005) HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol 25:7107–7119
Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P (1978) Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp Cell Res 117:347–356
Tarozzi N, Nadalini M, Stronati A, Bizzaro D, Dal Prato L, Coticchio G, Borini A (2009) Anomalies in sperm chromatin packaging: implications for assisted reproduction techniques. Reprod Biomed Online 18:486–495
Tavalaee M, Razavi S, Nasr-Esfahani MH (2009) Influence of sperm chromatin anomalies on assisted reproductive technology outcome. Fertil Steril 91:1119–1126
Torregrosa N, Dominguez-Fandos D, Camejo MI, Shirley CR, Meistrich ML, Ballesca JL, Oliva R (2006) Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum Reprod 21:2084–2089
Trostle-Weige PK, Meistrich ML, Brock WA, Nishioka K, Bremer JW (1982) Isolation and characterization of TH2A, a germ cell-specific variant of histone 2A in rat testis. J Biol Chem 257:5560–5567
Trostle-Weige PK, Meistrich ML, Brock WA, Nishioka K (1984) Isolation and characterization of TH3, a germ cell-specific variant of histone 3 in rat testis. J Biol Chem 259:8769–8776
Unni E, Zhang Y, Kangasniemi M, Saperstein W, Moss SB, Meistrich ML (1995) Stage-specific distribution of the spermatid-specific histone 2B in the rat testis. Biol Reprod 53:820–826
Urahama T, Harada A, Maehara K, Horikoshi N, Sato K, Sato Y, Shiraishi K, Sugino N, Osakabe A, Tachiwana H et al (2016) Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis. Epigenetics Chromatin 9:2
Utsuno H, Miyamoto T, Oka K, Shiozawa T (2014) Morphological alterations in protamine-deficient spermatozoa. Hum Reprod 29:2374–2381
van Roijen HJ, Ooms MP, Spaargaren MC, Baarends WM, Weber RF, Grootegoed JA, Vreeburg JT (1998) Immunoexpression of testis-specific histone 2B in human spermatozoa and testis tissue. Hum Reprod 13:1559–1566
Wennerholm UB, Bergh C, Hamberger L, Lundin K, Nilsson L, Wikland M, Kallen B (2000) Incidence of congenital malformations in children born after ICSI. Hum Reprod 15:944–948
Yan W, Ma L, Burns KH, Matzuk MM (2003) HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc Natl Acad Sci U S A 100:10546–10551
Ying HQ, Scott MB, Zhou-cun A (2012) Relationship of SNP of H2BFWT gene to male infertility in a Chinese population with idiopathic spermatogenesis impairment. Biomarkers 17:402–406
Yuen BT, Bush KM, Barrilleaux BL, Cotterman R, Knoepfler PS (2014) Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 141:3483–3494
Zalensky AO, Siino JS, Gineitis AA, Zalenskaya IA, Tomilin NV, Yau P, Bradbury EM (2002) Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J Biol Chem 277:43474–43480
Zhang X, San Gabriel M, Zini A (2006) Sperm nuclear histone to protamine ratio in fertile and infertile men: evidence of heterogeneous subpopulations of spermatozoa in the ejaculate. J Androl 27:414–420
Zhao M, Shirley CR, Yu YE, Mohapatra B, Zhang Y, Unni E, Deng JM, Arango NA, Terry NH, Weil MM et al (2001) Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol 21:7243–7255
Zhao M, Shirley CR, Mounsey S, Meistrich ML (2004) Nucleoprotein transitions during spermiogenesis in mice with transition nuclear protein Tnp1 and Tnp2 mutations. Biol Reprod 71:1016–1025
Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD, Brodeur GM (2014) CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev 131:35–46
Acknowledgments
We acknowledge with gratitude the research contributions of all the authors whose work has been cited in this review (IR/394/07-2016). We also thank Mr. Vivian Lobo, a graduate student from my laboratory, for his inputs with the figures and Mr. Vaibhav Shinde for his help with the graphic design. We thank the Department of Biotechnology, India for the Junior Research Fellowship to Mr Aniket Patankar.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Patankar, A., Parte, P. (2017). Sperm Chromatin Compaction and Male Infertility. In: SINGH, R., Singh, K. (eds) Male Infertility: Understanding, Causes and Treatment. Springer, Singapore. https://doi.org/10.1007/978-981-10-4017-7_17
Download citation
DOI: https://doi.org/10.1007/978-981-10-4017-7_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-4016-0
Online ISBN: 978-981-10-4017-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)