Abstract
As a ubiquitous, wide-spread bacterium, Pseudomonas aeruginosa thrives in complex environments, forming often multi-species communities called biofilms. Competition for nutrients and space in these conditions may be crucial for bacterial fitness and survival. In this context bacteria employ different strategies to slow down the growth or kill the neighbour or its host. One of these strategies makes use of the complex macromolecular nanomachine that is embedded in the bacterial envelope and serves to export effector proteins from the attacking bacterium towards the prey. This secretion system, being the most recently discovered in Gram-negative bacteria, has been named Type VI Secretion System (T6SS). It is widespread amongst Gram-negative bacteria and is built up of structures homologous to bacteriophage T4-like tail tube and sheath, on one hand, and of two proteins similar to DotU and IcmF, components of the membrane-anchoring complex of the Type IV Secretion System (T4SS), on the other hand. The main function of T6SSs is killing of a target prokaryote or eukaryote cell, but they have been proposed also to be implicated in communication between the members of the multispecies community. Thus, the T6SS constitutes an original membrane-spanning complex that allows Gram-negative bacteria to communicate with other cells in many contexts and with diverse outputs. This book chapter deals with genetic organization of its components, regulatory circuits controlling its expression and activity, as well as with function and structure of the apparatus itself with emphasises on P. aeruginosa systems. More advanced findings on the T6SS machinery in other bacterial species, such as in Escherichia coli and Vibrio cholera, but that may be analogous to P. aeruginosa, have been included.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
As a ubiquitous, wide-spread bacterium, Pseudomonas aeruginosa thrives in complex environments, forming often multi-species communities called biofilms. Competition for nutrients and space in these conditions may be crucial for bacterial fitness and survival. In this context bacteria employ different strategies to slow down the growth or kill the neighbour or its host. One of these strategies makes use of the complex macromolecular nanomachine that is embedded in the bacterial envelope and serves to export effector proteins from the attacking bacterium towards the prey. This secretion system, being the most recently discovered in Gram-negative bacteria, has been named Type VI Secretion System (T6SS). It is widespread amongst Gram-negative bacteria and is built up of structures homologous to bacteriophage T4-like tail tube and sheath, on one hand, and of two proteins similar to DotU and IcmF, components of the membrane-anchoring complex of the Type IV Secretion System (T4SS), on the other hand. The main function of T6SSs is killing of a target prokaryote or eukaryote cell, but they have been proposed also to be implicated in communication between the members of the multispecies community. Thus, the T6SS constitutes an original membrane-spanning complex that allows Gram-negative bacteria to communicate with other cells in many contexts and with diverse outputs. This book chapter deals with genetic organization of its components, regulatory circuits controlling its expression and activity, as well as with function and structure of the apparatus itself with emphasises on P. aeruginosa systems. More advanced findings on the T6SS machinery in other bacterial species, such as in Escherichia coli and Vibrio cholera, but that may be analogous to P. aeruginosa, have been included Fig. 4.1.
a T6SSs target both prokaryotic (left) and eukaryotic (right) cells. The machinery, composed of the Hcp tube, baseplate and sheath structure, is reminiscent of bacteriophage injection structure. T6SS effectors are injected within the target cell as individual proteins or by being fused to other proteins of the machinery. N nucleus, C actin cytoskeleton. b Plate assays showing the killing activity of the P. aeruginosa T6SS towards DH5αEscherichia coli. The method and mutants are described in (Casabona et al. 2013; Hachani 2013). Briefly, genetically modified E. coli (in blue) are challenged by different P. aeruginosa strains. After 4 h of incubation on solid medium, the competition is scrapped off the plate and serial dilutions are made and spotted on selective medium. E. coli survivals will appear blue and P. aeruginosa survivals, white
T6SS in the Microbial World and in P. aeruginosa
First large-scale genome screening gained insights into the phylogenetic distribution, gene content, organization and evolution of the T6SSs (Boyer 2009; Das and Chaudhuri 2003). Using as bait sixteen conserved T6-genes, 176 loci from 92 different bacteria were identified as containing at least five bait genes. These results were in accordance with another study that determined putative T6SS-encoding gene clusters in over one fourth of sequenced Gram-negative bacteria (Bingle et al. 2008). Interestingly, T6SSs are largely confined to proteobacteria, even though they also occur in planctomycetes and acidobacteria (Boyer et al. 2009; Bingle et al. 2008; Shrivastava and Mande 2008; Persson et al. 2009). Originally this secretion system was thought to have a role essentially in pathogenesis; however this point of view changed since the list of T6SS-encoding microbes, along with known human pathogens, included symbionts, pathobionts, non-pathogenic bacteria, commensals and mutualists (Boyer et al. 2009; Jani and Cotter 2010).
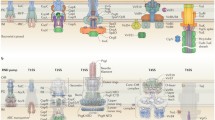
The presence of multiple copies of T6SS loci was found in one third of the genomes (Boyer et al. 2009). It has been proposed that different loci in one genome have distinct evolutionary history, which means that they have been likely acquired by horizontal transfer and not by duplication (Bingle et al. 2008; Shrivastava and Mande 2008). T6SS gene loci can be grouped into five different phylogenetic clusters (Boyer et al. 2009; Barret et al. 2011). Genome of P. aeruginosa PAO1 harbours three T6SS-encoding clusters. These clusters were named Hcp Secretion Islands (HSI-I, -II and -III) upon the Haemolysin-coregulated protein (Hcp) that was the first secreted “core” protein identified (Mougous et al. 2006). Outside of the three HSI clusters and dispersed on the genome, P. aeruginosa harbours genes encoding VgrG proteins, Hcp and other components functionally related with T6SS machineries (Fig. 4.2).
Genetic organization of T6SS-encoding genes. Three clusters HSI-I, -II and –III carry the genes encoding the components of secretion machineries H1-, H2- and H3-T6SS, respectively, and some regulatory elements. Some VgrG and Hcp proteins are encoded out of the loci. The genes are annotated by PA numbers (pseudomonas genome project; http://www.pseudomonas.com) and using nomenclature from (Hachani et al. 2011; Shalom et al. 2007). Colour codes are respected between three clusters for homologous and related genes
These genetic elements of up to 20 kb encode 13 conserved proteins, and frequently additional regulatory elements. Based on the nomenclature proposed by (Shalom et al. 2007), the genes that code for the conserved core components of the T6SS have been named tss, for type six secretion genes. Interestingly, and further supporting the idea of specialized T6SSs with distinct functions, some clusters contain genes that are not conserved and encode accessory components of T6SS. In P. aeruginosa, the accessory genes were named tag (for type six associated genes) and code for proteins with diverse functions, indispensable for the T6 assembly, function and/or regulation.
Despite a conserved gene organization and general predicted structure of the apparatus, the three P. aeruginosa T6SSs seem to be highly host-specific and regulated in a different manner. Indeed, a recent study demonstrated that TssB/TssC proteins, which form the tail sheath of the secretion apparatus, encoded by different clusters are not exchangeable, further suggesting that there is no redundancy between multiple T6SSs within one organism (Lossi et al. 2013). In addition, the major regulatory circuit controlling the expression of HSI-I is the same for other virulence factors associated with chronic infection. H1-T6SS may play a role in chronic infections, as sputa of cystic fibrosis patients harbour both the Hcp1 protein and antibodies against Hcp1 (Mougous et al. 2006). On the contrary, results obtained from animal and nematode models indicate that H2- and H3-T6SS seem to be employed during more aggressive types of infection (Lesic et al. 2009).
Type VI Secretion System effectors
Although some T6SS-dependent phenotypes are revealed by using eukaryotic cells, many have been characterized as having anti-bacterial activity, either intra- or inter-species, where the T6SS is decisive in the outcome of the bacterium when challenged with a competitor (Schwarz et al. 2010a, b). This can be of high importance in the establishment and persistence of a given pathogen during an infection within the host. T6SS may have evolved as “anti-virulence” mechanisms, where the secretion system would have a role in the regulation of cell density within the host, being a key factor in disease progression and microbe persistence by limiting colonization. Other phenotypes that have been associated to T6SS are stress sensing, biofilm formation and persistence in mixed biofilms. Together, T6SSs are involved in diverse social behaviours and act as mediators in inter-bacterial interactions and virulence (Schwarz et al. 2010b).
One of the best studied examples of anti-bacterial T6SS is the P. aeruginosa H1-T6SS that acts as device injecting effector proteins directly into target cells. The first H1-T6SS effectors were elegantly identified using proteomic approach on P. aeruginosa H1-T6SS regulatory mutants (Mougous et al. 2006). These effectors, named Tse1, Tse2, and Tse3 for Type-6-Secretion-Exported, are encoded outside of the HSI-I locus but rely on the machinery to be injected directly into the recipient after physical contact between the two cell types. Tse1 and Tse3 are lytic enzymes able to degrade peptidoglycan by amidase and muramidase activity, respectively. The third effector, Tse2, is toxic in the cytoplasm of recipient cells by a still unknown mechanism. These effectors play a key role in the outcome of P. aeruginosa in interspecies bacterial interactions in niches competition. P. aeruginosa avoids self-intoxication by synthesizing cognate immunity proteins, named Tsi1, Tsi2 and Tsi3 (Type-6-Secretion-Immunity), which bind to the secreted effectors to neutralize their detrimental activities. In addition of P. aeruginosa toxin/antitoxin Tse/Tsi couples (Hood et al. 2010; LeRoux et al. 2012; Russell et al. 2011), the same classes of proteins have been identified in Burkholderia thailandensis, Pseudomonas fluorescens, Salmonella typhimurium (Russell et al. 2012) and Serratia marcescens (English et al. 2012). Further structural analysis of Tse1 showed that this protein belongs to the N1pC/P60 papain-like cysteine peptidases where the catalytic diad is formed by Cys30 and His91. However, unlike the other members of the N1pC/P60 superfamily, Tse1 is a single domain protein without any putative localization domain (Chou et al. 2012). Inhibition of Tse1 by Tsi1 occurs by formation of a hydrogen bond between Tsi1 and the His91 of the Tse catalytic diad which consequently inhibits the enzyme activity (Benz et al. 2012).
A second class of T6SS effectors encompasses a diverse superfamily of bacterial phospholipase enzymes named Tle, for Type VI Lipase Effectors (Russell et al. 2013). Although some of these phospholipases, such as PldA (Tle5), have been implicated in P. aeruginosa virulence against eukaryotic hosts (Wilderman et al. 2001), it seems that they have also a specific function in intra- and interspecies bacterial interactions. Genetic and phenotypic studies demonstrated that the H2-T6SS is required for Tle5 toxic activity against Pseudomonas putida in mixed population. Indeed, purified PldA/Tle5 protein catalyses the release of choline from phosphatidylcholine, the activity being strictly dependent on the conserved catalytic domain in the protein. Furthermore, Tle5 exhibits high in vivo specificity to phosphatidylethanolamine, the major constituent of bacterial membranes. As for Tse molecules, Tle proteins possess their cognate immune partners that prevent self-killing (Russell et al. 2013).
The third class of T6S effectors is composed of VgrGs (Valine-Glycine Repeat protein G). Some of them, in addition of having essential role in T6S function, harbour catalytic domains with detrimental activities for the victim cell, such as actin remodelling and peptidoglycan disruption (Hachani et al. 2011; Ma and Mekalanos 2010; Durand et al. 2012a; Brooks et al. 2013). Dong et al. (2013) recently identified two V. cholerae effectors that possess both anti-bacterial and anti-eukaryotic activity. TseL harbours lipase domain, suggesting that it may target membrane-associated lipids and VasX has affinity for phospholipids in vitro (Miyata et al. 2011). Interestingly, TseL interacts with VgrG3 and its secretion depends on this interaction. Thus, this class of effectors is likely to be secreted in a complex with VgrGs.
The fourth class of potential T6SS effectors is represented by Rhs proteins. This family of YD-peptide repeat proteins is encoded frequently in the vicinity of T6SS loci or VgrG-encoding genes. Some of Rhs proteins have been shown to be a bona fide virulence factors, while others were involved in inter-bacterial competition. P. aeruginosa RhsT, harboured by the PA9 strain, acts by inducing inflammasome-mediated cell mortality (Kung et al. 2012); however its link with T6SS is still to be established.
Finally, the recent work by Leiman and Mekalanos groups further diversifies the list of T6SS effectors by discovering the so-called PAAR (proline-alanine-alanine-arginine) proteins that associate with VgrG on the tip of the injecting device (Shneider et al. 2013). This class of proteins are described later.
Architecture of the Type VI Secretion Apparatus
The T6S machinery is composed of two major sub-structures (Fig. 4.3), one sharing homology with bacteriophage T4 tail, and the other consisting of membrane-spanning peptidoglycan-bound complex that forms the anchoring complex.
Schematic representation of bacteriophage T4 action (a) and homology with T6SS machines (b). The assembly and attack by T6S may be divided in 4 steps: 1 membrane anchored and bacteriophage-like complexes are built up on defined regions within the bacterial envelope; 2 bacteriophage-like complex is extended, perpendicular to the IM, by addition of TssB/C subunits: 3 upon a signal, the TssB/C tubule contracts leading to the export of effectors; 4 the tubule associated AAA + ATPase, ClpV, participates in recycling of TssB and TssC subunits. The legend of the figure with protein names is given below. Colour code is respected between the bacteriophage and T6SS. OM outer membrane, IM inner membrane
Bacteriophage T4 Tail and Needle-like Structure
The marker of T6S functionality is presence of two conserved proteins, Hcp and VgrG, in the bacterial culture supernatants. Early structural studies revealed the striking similarities between these two proteins and proteins of bacteriophages. In addition to Hcp and VgrG, other bacteriophage-like subunits represent core components of the nanomachinery (Fig. 4.3).
The Tail Sheath
The bacteriophage T4 sheath consists of more than a hundred oligomerized gp18 subunits arranged as rings that form a hollow tube, able to accommodate the bacteriophage tail (Leiman et al. 2004). TssB and TssC subunits have been reported as forming a hollow tubular cogwheel-like complex of several hundreds of angström (Å) long. The internal diameter of the TssB/TssC complex is approximately 100 Å, big enough to accommodate the Hcp tube, estimated to have an external diameter of 85 Å (Lossi et al. 2013; Bonemann et al. 2009; Basler et al. 2012). Although TssB/TssC do not share sequence similarity with the bacteriophage sheath-forming subunit gp18, the modelling of a cross section of TssB/TssC tubules indicated that they shared same structural organization (Cascales and Cambillau 2012). Hcp co-fractionates with TssB and TssC homologues in Francisella novicida, further supporting the hypothesis that TssB and TssC form a tube-like structure able to engulf the Hcp filament (de Bruin et al. 2011). Direct interaction between TssB and TssC has been shown to be mediated by the N-terminus of TssC and was found highly specific as no cross interactions occurred between multiple T6SSs within one organism (Lossi et al. 2013). Assembled cytoplasmic structures appear in a perpendicular fashion to the cell envelope (Bonemann et al. 2009). It has been proposed that the formation of the TssB/TssC complex might be TssC-driven, since the TssB homologue in V. cholerae (VipA) does not form tubules in vitro in the absence of its partner, and in P. aeruginosa the C-terminus of TssC is required for tubule formation. Furthermore, TssC homologue (VipB) is needed for the interaction of the complex with ClpV in V. cholerae, a conserved T6S-ATPase (Lossi et al. 2013; Bonemann et al. 2009). In an effort to elucidate which proteins associate with TssB/TssC sheaths in V. cholerae, mass spectrometry analysis for protein identification was carried out on purified TssB/TssC structures. Four T6S proteins were identified: ClpV, VCA0109, VCA0111 and VCA0114 (Basler et al. 2012). ClpV had already been shown as directly interacting with TssB (Bonemann et al. 2009; Pietrosiuk et al. 2011). VCA0109 is a gp25 homologue, a protein that probably forms part of the baseplate and that is localized in the cytoplasm of P. aeruginosa. Finally, VCA0111 and VCA0114 (TssG and TssK, respectively) are conserved T6SS components, whose exact function is still unknown (see also below).
Recently, it has been shown that TagJ1 and TssB1 form a subcomplex of the sheath by using bacterial two-hybrid assays and NMR peptide mapping (Lossi et al. 2012). This interaction is mediated by the N-terminus of TssB1, conserved in other microorganisms encoding TagJ, such as S. marcescens (Lossi et al. 2012). Interestingly, TagJ1 is present in H1-T6SS in P. aeruginosa and absent in both H2- and H3-T6SS. Moreover, the interaction between TagJ1 and TssB1 is specific, since the authors demonstrated that TagJ1 does not form any complex with TssB2. The exact role of this subcomplex remains to be elucidated.
Hcp: Not Only the Tail Tube
Hcp is a ~ 17 kDa protein that arranges in hexameric rings with an internal diameter of approximately 40 Å (Mougous et al. 2006; Jobichen et al. 2010). Hcp is able to form structured non-helical, nanotube-like assemblies in vitro, which can be stabilized by the introduction of disulfide bonds (Ballister et al. 2008). Hcp shares homology with the protein gp19 that forms the tail tube of phage T4 (Pell et al. 2009; Leiman et al. 2009), further supporting the idea that Hcp may form a tubular structure that could traverse the cell envelope for substrate secretion. First structural studies on T6S effectors revealed that the size of the fully folded Tse1 is compatible with the internal diameter of the Hcp hexamer, thus proposing the Hcp as an effector conduit (Benz et al. 2012). The observation that P. aeruginosa Hcp1 stabilizes Tse effectors posttranslationally prompted Silverman et al. to examine direct interaction between Hcp1 and Tse2 (Silverman et al. 2013). These analyses allowed proposing the Hcp as a multipurpose protein playing a role of both a chaperon and a substrate receptor. Furthermore, as Hcp is known to be a T6S-secreted component, Hcp1-Tse complex is likely to traffic together across the bacterial envelope. However, the final destination of the complex, the way of the effectors’ release and the final fate of the Hcp protein are unknown.
VgrG and PAARs: Not Only a Cell-Puncturing Complex
VgrG proteins, found in the bacterial extracellular milieu, are the second marker of the T6SS functionality. Some bacterial genomes, such as P. aeruginosa, encode up to ten different VgrGs (Boyer et al. 2009; Hachani et al. 2011; Pukatzki et al. 2007). Based on structural analysis of VgrGs and their similarity to the T4 puncturing device, it has been proposed that they are localized at the distal tip of the Hcp tube and act as a membrane-puncturing device. However, the direct interaction between these two proteins has not been shown. The N-ter domain of the VgrG protein encoded by E. coli CFT073 showed that VgrG architecture is comparable to the bacteriophage spike subdomain, notably the gp5-gp27 complex, despite their low sequence identity (Leiman et al. 2009).
As is the case for VgrGs of different species, P. aeruginosa VgrGs (VgrG1a and VgrG1c) multimerize in tripartite structure and their export is Hcp-dependent (Hachani et al. 2011; Pukatzki et al. 2007; Zheng and Leung 2007). VgrG spike is decorated with the protein containing proline-alanine-alanine-arginine (PAAR) repeats. The PAAR protein folds into a symmetrical cone-shaped structure with a sharp tip and a triangular base which sits on the VgrGs surface (Shneider et al. 2013). Extensive bioinformatic analysis showed that PAAR proteins carry C- or N-terminus extensions which are diverse in size and predicted function, comprising peptidase, nuclease or lipase activities. However, most of extensions have no predicted activity. Interestingly, some Rhs proteins harbour PAAR domains and may also act as a tip carrying diverse functional groups (Shneider et al. 2013).
In P. aeruginosa, the H2-T6SS targets eukaryotic cells by modifying PI3K/Akt-dependent bacterial entry into epithelial cells (Sana et al. 2012). VgrGs and Rhs proteins, co-regulated by the H2-T6SS locus, have been suggested to be direct actors of this phenotype.
The Baseplate Assembly
The T4 bacteriophage baseplate, a macromolecular complex composed of at least 130 proteins of 14 different families, is required for initiation of tail tube and sheath assembly. It is assembled around the “hub”, a central three-fold-symmetric cylindrical structure (Rossmann et al. 2004). In T4, gp25 forms a wedge-shaped complex with gp6 and gp53 that is localized around the centre of the baseplate and has a key role in the connection of its central and peripheral parts (Kostyuchenko et al. 2003; Yap et al. 2010).
TssE shares approximately 40 % sequence similarity with gp25 of T4 bacteriophage (Leiman et al. 2009; Lossi et al. 2011). Moreover, it shows also remarkable similarity with gp25 secondary and tertiary structures as revealed by prediction tools. P. aeruginosa TssE is localized in the cytoplasm and has no lysozyme activity, unlike T4 gp25 protein family (Lossi et al. 2011). It is conceivable to propose that TssE might form part of the baseplate of the T6SS, that would assemble in the inner leaflet of the IM of the bacterium prior to secretion, and that the baseplate would be the linker structure between the tail and spike complex and the membrane-anchored complex.
Several questions remain unanswered. Does TssE form part of a larger IM baseplate-like structure? Does TssE interact with TssM and/or TssL in the inner leaflet of the IM? How is TssE recruited, if so, to the IM machinery complex?
Interestingly, it has been observed by electron microscopy that the T6S machinery presents a bell-shaped platform that connects the sheath to the IM (Basler et al. 2012). TssB/TssC elongated tube assembly is TssE-dependent in V. cholerae and they can be co-purified. Gp25 interacts with gp5-gp27 trimer in T4 bacteriophages, suggesting that TssE may have a similar role in the T6SS (Basler et al. 2012, Basler and Mekalanos 2012; Kapitein et al. 2013).
Membrane-Associated Components
The T6S machinery is anchored to the envelope of Gram-negative bacteria by several membrane-associated proteins forming a complex bound to peptidoglycan. This membrane subcomplex has been isolated by Cascales and co-workers from E. coli (Aschtgen et al. 2010a), and is composed at least by TssJ, TssM and TssL (Fig. 4.3). All these proteins are indispensable for T6SS activity.
TssJ is an OM lipoprotein able to homodimerize (Aschtgen et al. 2008; Rao et al. 2011b). Its X-ray crystal structure showed its arrangement in parallel β-sheets connected by a protruding loop that in E. coli is required for interaction with the periplasmic domain of TssM (Rao et al. 2011b; Felisberto-Rodrigues et al. 2011; Robb et al. 2013). The structural conservation of this loop across all species suggests that the interaction with TssM may also be conserved (Robb et al. 2013).
TssL is an IcmH-like integral IM protein (Aschtgen et al. 2012). IcmH (DotU) is needed for the correct functioning of T4bSS in Legionella pneumophila, since knockout mutants are impeded in intracellular growth (Zusman et al. 2004). The C-terminus of TssL homologue in EAEC is oriented towards the periplasm (Aschtgen et al. 2012). Its N-terminus folds as a hook-like structure composed of two three-helix bundles and is necessary for TssL autoassociation (Durand et al. 2012b). Autoassociation has been proposed to be mediated by the trans-membrane domain of TssL, since overproduced cytoplasmic domains are monomeric in solution and they only interact transiently and with low affinity in bacterial two-hybrid assays. Mutation of crucial residues of the trans-membrane segments completely abrogates TssL-TssL interaction, further supporting the hypothesis. Autoassociation has been suggested to be crucial for TssL function, maybe by increasing local concentration of the protein and allowing recruitment of other T6S components (Durand et al. 2012b). The cytoplasmic domain of TssL behaves as a soluble globular protein and does not present hydrophobic patches. It does not present any apparent catalytic domain or catalytic residues, which further suggests that TssL is a structural component of the apparatus. TssL proteins can bear a peptidoglycan-binding (PGB) domain thought to stabilize the apparatus in the cell envelope by anchoring it to the cell wall. In the case of some T6SSs, TssL lacking the recognized PGB domain co-occurs with TagL, another integral IM protein able to bind peptidoglycan in vivo and in vitro (Aschtgen et al. 2010a, b). In P. aeruginosa, curiously, the HSI-II and HSI-III loci do not harbour any protein with identified PGB domains.
TssM is an integral IM protein with a large periplasmic domain (744 residues in EAEC (Felisberto-Rodrigues et al. 2011)). In Agrobacterium tumefaciens TssM has ATP-binding and hydrolysis activity, the latter being crucial for Hcp recruitment (Ma et al. 2009, 2012). On the other hand, TssM of Edwardsiella tarda does not require its NTP-binding domain for Hcp secretion (Zheng and Leung 2007). Moreover, some TssM do not possess any NTP-binding domains. These findings argue that TssM-associated ATPase activity is not widespread through T6SSs and that it might be related to specific needs of each bacterium. In P. aeruginosa, TssM1 harbours only one predicted transmembrane helix and no predicted NTP-binding domain.
TssM, in addition of interacting with TssJ, is able to form a complex with the C-terminal periplasmic domain of TssL. TssJ and TssL are both able to homodimerize, in the OM and in the IM, respectively. It is plausible that these three proteins form a membrane-spanning complex that is bound to peptidoglycan and has a ring shape-like structure. Of note, ring shape-like structures seem to be a common feature of different membrane-spanning machineries, such as T3SS and T4SS (Schraidt and Marlovits 2011; Fronzes et al. 2009).
Does the membrane-spanning complex function as a channel and support for the syringe-like complex? It has been suggested that the TssM-L-J complex can accommodate the phage-like injection machinery. The interaction of TssM via its periplasmic domain both with Hcp and TssB supports this model, where the membrane-spanning complex would stabilize the needle-like complex.
Recently, the IM proteome of P. aeruginosa PAO1 has been explored by mass spectrometry (Casabona et al. 2013). Among the detected proteins, 23 components encoded by the HSI-I locus have been identified. Notably, two predicted cytoplasmic proteins, TssE and TssK, were found associated with the IM, further suggesting their role in syringe formation. Interestingly, E. coli TssK homologue is a trimer able to interact with several components of the T6SS (TssM, TssA, TssB and Hcp), suggesting that it may be the missing glue between the two complexes (Zoued et al. 2013). Another protein, of yet unknown function, TagJ, forms a complex with TssB. Curiously TagJ is one of the “accessory” components found only in approximately 30 % of T6SSs, which suggests the subtle adjustments in T6SS assemblies (Lossi et al. 2011).
Mechanism of Injection, a Dynamic Story
As discussed in the previous sections, bioinformatic, structural and biochemical analyses suggest that T6SSs are contractile injection systems reminiscent of tailed phages that employ a syringe-like macromolecular nanomachine to puncture host cell membrane.
T6SSs are highly dynamic structures that follow assembly/disassembly contact-dependent cycles that can be monitored by time-lapse fluorescence microscopy (LeRoux et al. 2012; Basler and Mekalanos 2012; Brunet et al. 2012). For example, P. aeruginosa cells can respond to T6SS activity in a neighbouring sister cell with an increase in their own T6SS dynamics. This phenomenon has been baptized bacterial dueling (Basler et al. 2013), and has also been documented for pathogenic E. coli EAEC cells. Interestingly, the propagation of T6SS activities was shown to increase over time, spreading through the bacterial lawn, suggesting the rapid genesis of the signal within cooperative microorganisms which could contribute to coordination in a spatial and temporal fashion of a bacterial community to eliminate competing bacteria.
Indeed, recent data obtained by time-lapse fluorescence microscopy showed that T6SS dueling also occurred between heterologous species (Basler et al. 2013). More precisely, the dueling response of P. aeruginosa was triggered by the T6SS activity of a heterologous microorganism which had attacked first whereas P. aeruginosa T6SS dynamic was absent in mixture containing species devoid of T6S activity. This ability to respond to a T6SS-mediated attack and efficiently kill the competitors was lost when the TagQRST-PpkA-Fha1-PppA regulatory cascade (see parts on regulation) was inactivated. This suggested that this cascade may form a regulatory module involved in direct sensing of envelope perturbations such as OM breach, peptidoglycan disruption/modification and/or IM perforation upon exogenous T6SS attack. Envelope perturbations will then be the point where the new T6SS will be assembled and “fired”.
Finally, two reports describe inter-bacterial intoxication via T6SS in a quantitative manner in vivo (LeRoux et al. 2012; Brunet et al. 2012). By monitoring prey lysis over time, they demonstrated that cell-cell contact is needed for T6SS-mediated intoxication but it is not the cue for T6SS activation. Moreover, the T6SS is not an extended extracellular appendage, since immediate cell-contact (less than 200 nm) was needed for prey lysis. The authors proposed that T6SS-associated attacks may act as a cue to the donor that the susceptible cell is in its vicinity, conceivable given the broad distribution of T6SS among Gram-negative bacteria. In bacteriophages, host recognition occurs through a reversible interaction of the conserved tip of the long fibres with the OM protein OmpC or with LPS (Yu and Mizushima 1982). This triggers conformational changes in the short fibres that extend and bind to the outer core of LPS in an irreversible fashion. The binding is followed by contraction of the outer tail sheath, penetration of the bacterial membrane by the hollow inner tail tube (Leiman et al. 2004, 2009). Interestingly, structural studies revealed that the aromatic and positive residues on the surface of the fibre tips are most likely receptor-binding determinants with the receptor in the bacterial surface (Bartual et al. 2010). Up-to-date, homologues of these fibre tip components have not been found in T6SS.
In the case of bacteriophages, the syringe-like complex is used only once to inject DNA into the host. On the contrary, T6SS injection device can be reused. It has been suggested that ClpV, a conserved Hsp100/Clp family of AAA + of the T6S, is responsible for the recycling of the TssB/TssC tubules. ClpV is able to disassemble and remodel the tubules in vitro and interacts directly with TssC in a specific manner. Finally, it has also been demonstrated that ClpV prevents the formation of non-productive TssB/TssC tubules formed spontaneously in the cell (Bonemann et al. 2009; Pietrosiuk et al. 2011; Kapitein et al. 2013). This would assure the presence of a pool of non-assembled TssB and TssC units, needed for T6S assembly upon cue sensing.
Regulation of Type VI Secretion Systems in P. aeruginosa
As for most of the complex secretion machineries embedded within bacterial membranes, the three T6SSs are highly regulated at different levels, including post-translational, transcriptional and post-transcriptional levels. Few signals and some molecular actors involved in these types of regulation are starting to be revealed and are summarized in Fig. 4.4.
Regulation of the P. aeruginosa HSI loci. The principal regulators and pathways involved in transcriptional, post-transcriptional and post-translational control are indicated. The positive effects are depicted by green arrows, the negative ones by red bars. For the regulation of orphan elements and the link between c-di-GMP and RsmZ/Y pathway, see the text. The direct binding of RsmA on HSI-I mRNAs was demonstrated by RNA mobility shift assays (Brencic and Lory 2009), that of LasR on PA1656-59 locus (HSI-II) by electrophoretic mobility shift assay (Gilbert et al. 2009) and of MvaT on PA1656 (HSI-II) and PA2371-73 (HSI-III) by ChIP-on-Chip (Castang et al. 2008)
Post-Translational Regulation
H1-T6SS activity depends on additional proteins. Seven proteins, PpkA, PppA, TagR, TagS, TagT, TagQ and Fha1, form part of the so-called threonine phosphorylation pathway (TPP) (Hsu et al. 2009), and TagF is a negative regulator acting independently of the TPP (Silverman et al. 2011).
The TPP pathway relies on a transmembrane serine-threonine kinase, PpkA, which dimerizes and autophosphorylates to then phosphorylate Fha1, a protein that contains a Forkhead-Associated domain. Once phosphorylated, Fha1 recruits the ATPase ClpV1 to the IM and to TssB/C tubes, triggering both syringe assembly and effector export. The activity of PpkA is antagonized by its cognate phosphatase, PppA, which de-phosphorylates Fha1. TagR, a periplasmic protein that acts upstream of PpkA, is essential for the activation of the system. The modulation of the phosphorylation status of Fha1 on Thr362 determines the triggering of H1-T6SS activation (Hood et al. 2010; Hsu et al. 2009; Mougous et al. 2007). A recent study has described TagT, TagS and TagQ as additional players in the TPP by acting upstream of the PpkA/PppA phosphorylation checkpoint (Casabona et al. 2013). TagT and TagS form a membrane-bound ABC transporter with ATPase activity and TagQ is an OM lipoprotein that faces the periplasm. It was shown that TagR, under some conditions, associates to the OM in a TagQ-dependent manner. Thus, TagTSRQ form a novel signalling module in charge of sensing exogenous T6SS attacks, most probably by sensing cell envelope disruption or interacting with exogenous T6SS effectors. The sensing by TagTSRQ module promotes local Fha1 phosphorylation that leads to T6S machinery assembly and firing (Basler et al. 2013; Casabona et al. 2013).
Independent of the TPP, another posttranslational pathway has been described in which Fha1 phosphorylation is negatively regulated by TagF. The environmental cues that govern the two pathways are different, since surface growth induces TPP activation, but not the TagF-dependent pathway. Even though the mechanism by which TagF represses the H1-T6SS activation is still unknown, the authors have shown that both TPP and TagF pathways converge in the recruitment of ClpV1 (Silverman et al. 2011).
Transcriptional and Post-Transcriptional Regulations
HSI-I was discovered through microarray analysis revealing a new locus negatively controlled by RetS, a key component of the RetS/LadS/Gac/Rsm pathway known to orchestrate the chronic/acute virulence switch (Mougous et al. 2006; Goodman et al. 2004; Ventre et al. 2006; Coggan and Wolfgang 2012). Sensor kinases RetS and LadS control in opposite manner HSI-I expression that is inhibited by RetS and activated by LadS, thus promoting expression during chronic stage of infection (Mougous et al. 2006). The involvement of the other players of the cascade, namely GacA, RsmA and RsmYZ, was further confirmed by microarray analyses (Brencic et al. 2009). Furthermore, a direct binding of RsmA to the leader sequence of transcripts encoding PA0081 and PA0082 was shown (Brencic and Lory 2009). Interestingly genes located in the HSI-III are also under regulation of the pathway as transcript levels were downregulated in gacA and rsmYZ mutants (Brencic et al. 2009). In addition, H1-T6SS is inversely controlled with T3SS by c-di-GMP; indeed, an increased level of this second messenger, either triggered by retS mutation, ladS overexpression, or WspR adenylate cyclase activity, was shown to activate H1-T6SS activity and inhibit T3SS synthesis (Moscoso et al. 2011). This effect requires the RsmY and RsmZ small regulatory RNAs but the exact mechanism connecting the two pathways is still unknown. The LadS/Gac/Rsm pathway, but not c-di-GMP, was recently shown to be required for the induction of both biofilm formation and HSI-I gene expression, along with the repression of T3SS genes, in response to sub-inhibitory concentration of kanamycin (Jones et al. 2013). This might be of importance for the survival within polymicrobial niche, even if no increase in bacterial killing was observed, probably linked to absence of Tse3 effector induction (Jones et al. 2013).
An inverse relationship between mucoidy and T6SS expression has been underlined in several studies. Mutation in mucA, encoding the anti-sigma factor of AlgU, leads to the release of the ExtraCytoplasmic Function (ECF) sigma factor; then AlgU is free to activate transcription of genes involved in alginate synthesis. This is the common mechanism for transition to mucoid phenotype (Damron and Goldberg 2012). Transcriptome analysis indicated that AlgU exerts a negative effect on HSI-I and HSI-II genes, as well as on vgrG2a (PA1511) and hcpC-vgrG2b (PA0263–62) (Tart et al. 2005). This suggests that mucoid conversion, the hallmark of chronic infection, should also decrease H1-T6SS activity, an idea contradicting the observed Hcp1 production in CF sputum (Mougous et al. 2006). In the same line, mucA mutants isolated from CF individuals were shown to exhibit reduced expression of virulence factors, such as T3SS but also H1-T6SS genes, compared to the non-mucoid strains (Rau et al. 2010). Furthermore, transcriptome and proteome analyses of two isogenic strains isolated from a CF patient pointed out a net difference in T6SS expression between the non-mucoid and mucoid strains (Rao et al. 2011a, 2008). Indeed, three proteins encoded within HSI-I, namely TssB1 (PA0083), TssC1 (PA0084) and Hcp1 (PA0085), were less abundant in the mucoid strain, and the transcriptome analysis indicated a decreased expression in the mucoid strain of most of HSI-I operons and few HSI-II genes, namely tssA2 (PA1656), tssC2 (PA1658) and tssE2 (PA1659). The authors proposed that the non-mucoid bacteria during initial phases of infection could require T6SS to outcompete other bacteria, but then bacteria convert to mucoid phenotype and do not require any more their T6SS (Rao et al. 2011a).
Quorum sensing (QS) coordinates expression of the 3 HSI loci, as reported by numerous studies using approaches such as transcriptome analysis, RT-qPCR, or ChIP-on-chip ((Bernard et al. 2010); and references herein). Both HSL-based (Rhl and Las) and HAQ-based (MvfR) systems affect and control differentially the three loci, with HSI-I being negatively controlled by LasR and MvfR, while HSI-II and HSI-III are positively controlled by Las/Rhl and MvfR (Lesic et al. 2009; Sana et al. 2012; Schuster et al. 2003). A direct binding of LasR was even observed on PA1656–59 region (HSI-II) (Gilbert et al. 2009). Hence, and as confirmed by transcriptional fusion analyses (Sana et al. 2013), H2-T6SS and H3-T6SS synthesis occurs at high cell density at the same timing as numerous other virulence determinants. The loci encoding these two T6SSs also belong to the MvaT regulon, as PA1656 (HSI-II), PA2371–73 (3 HSI-III-encoded genes) as well as the two T6SS related genes PA1512–11 (hcpA-vgrG2a), are direct targets of MvaT (Castang et al. 2008). MvaT is an H-NS family member functioning as a repressor of gene expression and proposed to be a transcriptional silencer of AT-rich foreign DNA (Castang et al. 2008). HSI-II is also induced upon iron limitation, as the repressor Fur controls its expression, probably directly, 2 putative Fur boxes being located in the tssA2 promoter (Sana et al. 2012). HSI-II locus (PA1657–70) as well as hcpC (PA0263), vgrG2a (PA1511) and vgrG6 (PA5266), are repressed by TetR family repressor, PsrA, able to bind and respond to long-chain fatty acid signals, that controls fatty acid metabolism (Kang et al. 2008). PsrA also regulates expression of Tfp and rpoS, the latter being potentially of importance for stationary growth adaptations (see (Potvin et al. 2008), and references herein).
Based on transcriptional fusion and mutant analyses, a recent study has reported the control of HSI-II and HSI-III by the alternative sigma factor 54 (σ54), known to be crucial for virulence in P. aeruginosa (Sana et al. 2013). Whereas σ54 (RpoN) activates one of the two HSI-III operons, PA2364–59, it affects negatively the HSI-II locus and the PA2365–74 HSI-III operon probably through an indirect mechanism. The transcriptional activation by RNAP-σ54, in general, requires bacterial Enhancer-Binding Proteins (EBPs) to provide energy through ATP-hydrolysis and direct molecular interaction. Both the identification of a consensus sequence for binding of σ54 in HSI-III locus by bioinformatics analyses (Bernard et al. 2010) and the positive control of RpoN exerted on PA2364–59 suggest that this HSI-III operon might require such an EBP activator. Interestingly, two EBPs are potentially encoded within the two loci, the sigma factor activators Sfa2 (PA1663) and Sfa3 (PA2359) (Filloux et al. 2008). However, none of the two proteins was found involved in HSI-III operon expression, and Sfa2 participates with RpoN to the HSI-II repression (Sana et al. 2013). A crosstalk was previously reported between HSI-II and HSI-III, as HSI-II deletion triggered a reduced hcp3 transcription (Lesic et al. 2009); as it does not rely on a positive effect of Sfa2 on HSI-III as it was suggested, the mechanism is still unknown.
A transcriptome study indicated a strong increase in PAO1 HSI-III (PA2365–74 operon) gene expression as well as hcpC (PA0263) upregulation upon contact with respiratory epithelia but the underlying mechanism is not known (Chugani and Greenberg 2007).That might be relevant for the virulence in eukaryotes, as well as the observation that expression of HSI-III operons is maximal at 37 ℃ (Sana et al. 2013). However, lowering growth temperature from 37 to 25 ℃ strongly induced expression of hcpB (PA5267) encoding an orphan T6SS element in PAO1, as well as vgrG2b (PA0262) and tssB1, a HSI-I-encoded gene, but at a lower level (Termine and Michel 2009). The temperature decrease conducts also to upregulation of hcpD (PA14_43070) and hcpB (PA14_69560) in the PA14 isolate (Wurtzel et al. 2012). Furthermore, secretome analysis indicated that suboptimal temperature influences secretion of Hcp, whose production was increased at 25 ℃. The temperature effect might give a competitive advantage over environmental bacteria and be of high importance for the persistence in hospitals (Termine and Michel 2009).
Deciphering all the intertwined regulatory pathways, the cross-talks, the sensed signals controlling expression of the T6SSs will help understanding their physiological roles and relevance. To complete the overall picture of T6SSs regulation, the reader can consult the following reviews (Bernard et al. 2010; Silverman et al. 2012; Leung et al. 2011; Miyata et al. 2013).
Concluding Remarks and Future Directions
T6SSs are the last protein machineries discovered in Gram-negative bacteria. Their structural and regulatory complexity, role in bacterial physiology and in host-bacteria interactions made it an extremely attractive field of investigations. P. aeruginosa, together with EPEC and V. cholera, became especially appropriate model for studying all of these aspects of T6SS biology. The homology with the T4 bacteriophage allowed the strong impact of structural studies and revealed tridimensional organization of T6SS sub-assemblies. However, the whole apparatus has not been yet isolated and visualized. In our point of view, some other challenges are confronting us. Although some membrane signalling modules have been identified, the environmental signals that trigger T6SS assembly and effector firing are still unknown. The first pieces of evidence for “membrane perturbation signals” came from the recent paper of Ho et al. (Ho et al. 2013), in which the authors identified mating pair formation system by T4SS as one of the natural membrane-disturbing agents that may induce T6SS response to delimit the acquisition of foreign DNA. The second challenge concerns the T6SS effectors; the list of T6SS-associated effectors is being rapidly increasing and up to date encompasses five effector families. Studies of how these effectors are recruited and recognized by the given syringe and what is the timing of their ejection will certainly require new technological and methodological developments. Finally, the activity of T6SSs being essential for bacterial fitness in different settings, including multi-species infections, may be considered in the future as target for novel anti-bacterial therapy.
References
Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E (2008) SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol 190:7523–7531
Aschtgen MS, Gavioli M, Dessen A, Lloubes R, Cascales E (2010a) The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol Microbiol 75:886–899
Aschtgen MS, Thomas MS, Cascales E (2010b) Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence 1:535–540
Aschtgen MS, Zoued A, Lloubes R, Journet L, Cascales E (2012) The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 Type VI secretion system, is inserted by YidC. Microbiologyopen 1:71–82
Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD (2008) In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci U S A 105:3733–3738
Barret M, Egan F, Fargier E, Morrissey JP, O’Gara F (2011) Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology 157:1726–1739
Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ (2010) Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci U S A 107:20287–20292
Basler M, Mekalanos JJ (2012) Type 6 secretion dynamics within and between bacterial cells. Science 337:815
Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186
Basler M, Ho BT, Mekalanos JJ (2013) Tit-for-Tat: Type VI secretion system counterattack during bacterial cell–cell interactions. Cell 152:884–894
Benz J, Sendlmeier C, Barends TR, Meinhart A (2012) Structural insights into the effector-immunity system Tse1/Tsi1 from Pseudomonas aeruginosa. PLoS ONE 7:e40453
Bernard CS, Brunet YR, Gueguen E, Cascales E (2010) Nooks and crannies in type VI secretion regulation. J Bacteriol 192:3850–3860
Bingle LE, Bailey CM, Pallen MJ (2008) Type VI secretion: a beginner’s guide. Curr Opin Microbiol 11:3–8
Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A (2009) Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. Embo J 28:315–325
Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104
Brencic A, Lory S (2009) Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632
Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S (2009) The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445
Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S (2013) Lytic activity of the Vibrio cholerae Type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem 288:7618–7625
Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E (2012) Imaging type VI secretion-mediated bacterial killing. Cell Rep 3:36–41
Casabona MG, Silverman JM, Sall KM, Boyer F, Coute Y, Poirel J, Grunwald D, Mougous JD, Elsen S, Attree I (2013) An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ Microbiol 15:471–486
Cascales E, Cambillau C (2012) Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci 367:1102–1111
Castang S, McManus HR, Turner KH, Dove SL (2008) H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A 105:18947–18952
Chou S, Bui NK, Russell AB, Lexa KW, Gardiner TE, LeRoux M, Vollmer W, Mougous JD (2012) Structure of a peptidoglycan amidase effector targeted to Gram-negative bacteria by the type VI secretion system. Cell Rep 1:656–664
Chugani S, Greenberg EP (2007) The influence of human respiratory epithelia on Pseudomonas aeruginosa gene expression. Microb Pathog 42:29–35
Coggan KA, Wolfgang MC (2012) Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr issues mol biol 14:47–70
Damron FH, Goldberg JB (2012) Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Molecular microbiology 84:595–607
Das S, Chaudhuri K (2003) Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. Silico Biol 3:287–300
de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE (2011) The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology 157:3483–3491
Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ (2013) Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci USA 110:2623–2628
Durand E, Derrez E, Audoly G, Spinelli S, Ortiz-Lombardia M, Raoult D, Cascales E, Cambillau C (2012a) Crystal structure of the VgrG1 actin cross-linking domain of the Vibrio cholerae type VI secretion system. J Biol Chem 287:38190–38199
Durand E, Zoued A, Spinelli S, Watson PJ, Aschtgen MS, Journet L, Cambillau C, Cascales E (2012b) Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J Biol Chem 287:14157–14168
English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ (2012) New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol 84:921–936
Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E (2011) Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog 7:e1002386
Filloux A, Hachani A, Bleves S (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570–1583
Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G (2009) Structure of a type IV secretion system core complex. Science 323:266–268
Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M (2009) Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol 73:1072–1085
Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S (2004) A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754
Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jove D, Filloux A (2011) Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem 286:12317–12327
Hachani A, Lossi NS, Filloux A (2013) A visual assay to monitor T6SS-mediated bacterial competition. J Vis Exp. 2013 Mar 20;(73):e50103. doi: 10.3791/50103.
Ho BT, Basler M, Mekalanos JJ (2013) Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science 342:250–253
Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37
Hsu F, Schwarz S, Mougous JD (2009) TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol 72:1111–1125
Jani AJ, Cotter PA (2010) Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8:2–6
Jobichen C, Chakraborty S, Li M, Zheng J, Joseph L, Mok YK, Leung KY, Sivaraman J (2010) Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS ONE 5:e12910
Jones C, Allsopp L, Horlick J, Kulasekara H, Filloux A (2013) Subinhibitory concentration of kanamycin induces the Pseudomonas aeruginosa type VI secretion system. PLoS ONE 8:e81132
Kang Y, Nguyen DT, Son MS, Hoang TT (2008) The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 beta-oxidation operon. Microbiology 154:1584–1598
Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A (2013) ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87:1013–1028
Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG (2003) Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol 10:688–693
Kung VL, Khare S, Stehlik C, Bacon EM, Hughes AJ, Hauser AR (2012) An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc Natl Acad Sci USA 109:1275–1280
Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG (2004) Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118:419–429
Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA 106:4154–4159
LeRoux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD (2012) Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci USA 109:19804–19809
Lesic B, Starkey M, He J, Hazan R, Rahme LG (2009) Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155:2845–2855
Leung KY, Siame BA, Snowball H, Mok YK (2011) Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol 14:9–15
Lossi NS, Dajani R, Freemont P, Filloux A (2011) Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa. Microbiology 157:3292–3305
Lossi NS, Manoli E, Simpson P, Jones C, Hui K, Dajani R, Coulthurst SJ, Freemont P, Filloux A (2012) The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel subcomplex in the bacterial type VI secretion system. Mol Microbiol 86:437–456
Lossi NS, Manoli E, Forster A, Dajani R, Pape T, Freemont P, Filloux A (2013) The HsiB1C1 (TssB/TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheath-like structure. J Biol Chem 288:7536–7548
Ma AT, Mekalanos JJ (2010) In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci USA 107:4365–4370
Ma LS, Lin JS, Lai EM (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol 191:4316–4329
Ma LS, Narberhaus F, Lai EM (2012) IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J Biol Chem 287:15610–15621
Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S (2011) Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun 79:2941–2949
Miyata ST, Bachmann V, Pukatzki S (2013) Type VI secretion system regulation as a consequence of evolutionary pressure. J Med Microbiol 62:663–676
Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A (2011) The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol 13:3128–3138
Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530
Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ (2007) Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 9:797–803
Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR (2009) The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA 106:4160–4165
Persson OP, Pinhassi J, Riemann L, Marklund BI, Rhen M, Normark S, Gonzalez JM, Hagstrom A (2009) High abundance of virulence gene homologues in marine bacteria. Environ Microbiol 11:1348–1357
Pietrosiuk A, Lenherr ED, Falk S, Bonemann G, Kopp J, Zentgraf H, Sinning I, Mogk A (2011) Molecular basis for the unique role of the AAA + chaperone ClpV in type VI protein secretion. J Biol Chem 286:30010–30021
Potvin E, Sanschagrin F, Levesque RC (2008) Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev 32:38–55
Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA 104:15508–15513
Rao J, DiGiandomenico A, Unger J, Bao Y, Polanowska-Grabowska RK, Goldberg JB (2008) A novel oxidized low-density lipoprotein-binding protein from Pseudomonas aeruginosa. Microbiology 154:654–665
Rao J, Damron FH, Basler M, Digiandomenico A, Sherman NE, Fox JW, Mekalanos JJ, Goldberg JB (2011a) Comparisons of two proteomic analyses of non-mucoid and mucoid Pseudomonas aeruginosa clinical isolates from a cystic fibrosis patient. Front Microbiol 2:162
Rao VA, Shepherd SM, English G, Coulthurst SJ, Hunter WN (2011b) The structure of Serratia marcescens Lip, a membrane-bound component of the type VI secretion system. Acta Crystallogr D Biol Crystallogr 67:1065–1072
Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF, Jelsbak L, Hoiby N, Yang L, Molin S (2010) Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol 12:1643–1658
Robb CS, Assmus M, Nano FE, Boraston AB (2013) Structure of the T6SS lipoprotein TssJ1 from Pseudomonas aeruginosa. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:607–610
Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG (2004) The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol 14:171–180
Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347
Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD (2012) A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11:538–549
Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD (2013) Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512
Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S (2012) The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem 287:27095–27105
Sana TG, Soscia C, Tonglet CM, Garvis S, Bleves S (2013) Divergent control of two Type VI secretion systems by RpoN in Pseudomonas aeruginosa. PLoS ONE 8:e76030
Schraidt O, Marlovits TC (2011) Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331:1192–1195
Schuster M, Lostroh CP, Ogi T, Greenberg EP (2003) Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079
Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD (2010a) Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068
Schwarz S, Hood RD, Mougous JD (2010b) What is type VI secretion doing in all those bugs? Trends Microbiol 18:531–537
Shalom G, Shaw JG, Thomas MS (2007) In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699
Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG (2013) PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353
Shrivastava S, Mande SS (2008) Identification and functional characterization of gene components of Type VI secretion system in bacterial genomes. PLoS ONE 3:e2955
Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD (2011) Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol 82:1277–1290
Silverman JM, Brunet YR, Cascales E, Mougous JD (2012) Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66:453–472
Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD (2013) Haemolysin coregulated protein is an exported receptor and chaperone of Type VI secretion substrates. Mol Cell 51:584–593
Tart AH, Wolfgang MC, Wozniak DJ (2005) The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol 187:7955–7962
Termine E, Michel GP (2009) Transcriptome and secretome analyses of the adaptive response of Pseudomonas aeruginosa to suboptimal growth temperature. Int Microbiol 12:7–12
Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A (2006) Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA 103:171–176
Wilderman PJ, Vasil AI, Johnson Z, Vasil ML (2001) Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol Microbiol 39:291–303
Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S (2012) The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945
Yap ML, Mio K, Leiman PG, Kanamaru S, Arisaka F (2010) The baseplate wedges of bacteriophage T4 spontaneously assemble into hubless baseplate-like structure in vitro. J Mol Biol 395:349–360
Yu F, Mizushima S (1982) Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J Bacteriol 151:718–722
Zheng J, Leung KY (2007) Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66:1192–1206
Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, Cascales E, Journet L (2013) TssK Is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type vi secretion system. J Biol Chem 288:27031–27041
Zusman T, Feldman M, Halperin E, Segal G (2004) Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect Immun 72:3398–3409
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Casabona, MG., Elsen, S., Cogoni, V., Attrée, I. (2015). P. aeruginosa Type VI Secretion Machinery: Another Deadly Syringe. In: Ramos, JL., Goldberg, J., Filloux, A. (eds) Pseudomonas. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9555-5_4
Download citation
DOI: https://doi.org/10.1007/978-94-017-9555-5_4
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-9554-8
Online ISBN: 978-94-017-9555-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)