Abstract
Advances in genetic analysis are fundamentally changing our understanding of the causes of epilepsy, and promise to add more precision to diagnosis and management of the clinical disorder. Single gene mutations that appear among more complex patterns of genomic variation can now be readily defined. As each mutation is identified, its predicted effects can now be validated in neurons derived from the patient’s own stem cells, allowing a more precise understanding of the cellular defect. Parallel breakthroughs in genetic engineering now allow the creation of developmental experimental models bearing mutations identical to the human disorder. These models enable investigators to carry out detailed exploration of the downstream effects of the defective gene on the developing nervous system, and a framework for pursuing new therapeutic target discovery. Once these genetic strategies are combined with interdisciplinary technological advances in bioinformatics, imaging, and drug development, the promise of delivering clinical cures for some genetic epilepsies will be within our reach.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The last decade has witnessed several revolutions in our ability to understand the genetic basis of the epilepsies and its role in diagnosis and treatment. Ten years ago, only a few genes, mostly for ion channels, had been linked to the appearance of epilepsy in large, multigenerational pedigrees. The general belief was that such families were rare, that the numbers of causative genes for epilepsy were few, and that each of the clinically defined Mendelian syndromes was the exclusive product of one gene alone. Furthermore, the absence of a positive family history in a patient with epilepsy suggested that a purely genetic etiology was unlikely. Following this logic, it was anticipated that detecting a mutation in one of these genes would be highly predictive of a specific seizure syndrome, and that only a rare, inherited mutation causes disease. Understandably, most investigators concluded that knowledge of the gene defect could lead shortly to dramatically improved treatments, if not a cure, for the disorder.
In fact, none of these pioneering assumptions has proven to be entirely correct. However, as in the field of DNA sequencing, we have entered the ‘next generation’ of epilepsy genetics, and what began as a search for a few inherited gene errors that could explain why some epilepsies are familial has expanded into a set of powerful research tools and discoveries that have immeasurably accelerated our ability to correctly diagnose and, in some cases, treat the disease. Major strides in clinical phenotyping and classification of epilepsy syndromes have been driven by, and contribute to the identification of, new monogenic epilepsies, both inherited and de novo in origin. Advances in neuroimaging have proven critical to the discovery of genes leading to malformations of cortical development. New methods in molecular genetics and gene sequencing have allowed rapid identification of candidate genes for an increasing number of epilepsy syndromes and potential comorbidities, including sudden unexpected death. Advances in genetic epidemiology, genome-wide association studies and whole exome candidate gene profiling have stimulated the analysis of complex genetic traits. The mathematical aspects of these analyses, as well as the emergence of mutation and polymorphism databases and genotype-phenotype correlations, are now included in the growing new field of epilepsy bioinformatics.
In the neurobiology laboratory, identified genes arising from both human and experimental genetic studies now offer an unparalleled opportunity to examine basic mechanisms of the disease. Genetically engineered models enable the electrophysiological validation of a candidate epilepsy gene using in vivo and in vitro approaches, and are essential to pinpoint the specific brain networks involved. Stem cells derived from patient’s fibroblasts can now be reliably transformed into neurons to evaluate the effects of the mutation on cell biology and signaling within the affected nervous system. Contemporary experimental mouse models not only give investigators the ability to selectively express a predefined human gene mutation in the brain at different stages of brain development, but also to reverse its effects with drugs and other genes. High resolution, chronic imaging techniques using fluorescent reporters of gene expression permit the study of the pathophysiology of a genetic lesion over time, tracking the ‘downstream’ molecular biology of the seizure pathways. Seizures typically arise after prolonged periods of abnormal neural development, and in these cases where the damage is already done, correcting the actual gene defect may come too late to reverse the epileptic condition. However careful analysis of these secondary changes in the physiology and anatomy of the affected neural circuits may offer a second opportunity to discover a novel target for therapy, fulfilling the promise of a cure.
2 The Emerging Picture of Epilepsy Genetics
2.1 Gene and Mutation Diversity
It is now estimated that genetic factors contribute to at least 40 % of all epilepsies. While there has been considerable progress in identifying genes for Mendelian epilepsies, the extent of genetic susceptibility to more common sporadic epilepsy syndromes remains unknown. Although limited evidence, in both animal models and human disease, has been gathered that susceptibility to epilepsy conferred by specific mutations might be influenced by non-pathogenic alleles at other genetic loci [1, 15] the characterization and validation of susceptibility variants appears particularly complex and requires large-scale collaborative efforts. Moreover, our understanding genetic susceptibility to a major heterogeneous disorder such as epilepsy would likely be incomplete without reference to a specific syndrome. Over 600 entries for pedigrees showing Mendelian phenotypic inheritance patterns can be found in the Online Mendelian Inheritance in Man (OMIM) database, and genetic loci have been identified in over 160 of these cases. These genes arise not only among the >400 members of the ion channel gene family, but across an extraordinarily diverse group of molecular pathways that also regulate membrane excitability, synaptic plasticity, and rhythmic network firing behavior. Causative genes also include those for presynaptic neurotransmitter release, postsynaptic receptors, transporters, cell metabolism, and importantly, many formative steps in early brain development, such as the proliferation and migration of neuronal precursors, dendritogenesis, synaptogenesis, and glial biology. However, inherited mutations in these known epilepsy genes currently only account for a small fraction of patients. Thus, many additional genes causing seizures are likely to be identified. Within each of these genes, the molecular rearrangements themselves are typically novel, or occur with a very low allele frequency within the epilepsy population. Thus monogenic epilepsies are disorders of many, individually rare, errors in an increasingly broad spectrum of biological pathways.
Most of the idiopathic epilepsies arise sporadically among unaffected family members, or do not appear to follow single gene inheritance patterns. From a purely genetic perspective, this finding may be explained by an inadequate family size, or an underlying complex pattern of multigenic inheritance, or even genetic mosaicism, three possibilities which have long bedeviled the analysis of inherited disease. However a new alternative has arisen from an important insight made over the past decade, and promises a steep increase in our ability to isolate genetic risk of epilepsy in individuals, even in small families – namely, the detection of de novo mutation of single genes or copy number variants of even larger chromosomal regions that encompass them. De novo splice site or nonsense mutations that impair function by removing critical portions of the encoded protein have been identified at convincingly high frequency within specific epilepsy phenotypes, in particular the SCN1A sodium channel linked to the severe myoclonic epilepsy known as Dravet Syndrome. This realization, along with the recent ability to rapidly sequence and assess gene variation in a large list of candidate genes, will greatly contribute to the personal identification of causative genes in the epilepsy clinic.
2.2 Phenotype Complexity
Large scale genotype-phenotype studies within monogenic populations have determined that the simple correspondence between genotype and phenotype can break down, resulting in different ages of onset and clinical seizure severity (phenotypic heterogeneity) within those bearing mutations in the same gene. This poor correlation may be due to the many possible structural alterations in the mutant protein leading to either gain or loss of function. However, even in families with single gene inheritance of an identical gene mutation, a degree of complexity remains, as evidenced by ‘unaffected carriers’ of the ‘causative’ mutation. In these cases, the phenotypic variability in such families can be attributed to the presence of polymorphisms in modifier genes influencing the phenotypic expression or, alternatively, to environmental factors.
Conversely, identical clinical phenotypes may be due to different underlying genotypes (genetic heterogeneity). Most of the broad phenotypic categories of seizure disorders are now recognized to arise from mutations in more than a single gene. In some cases, the different genes for a clinical epilepsy syndrome all contribute a single functional heteromeric unit, such as the different receptor subunits (α,β,γ,δ) contributing to a functional GABAa receptor in generalized epilepsy, the pore forming (α) and regulatory (β1) subunits of the sodium channel in Dravet Syndrome, the different pore forming subunits (KCQ2/3) of the M-current in Benign Neonatal Infantile Epilepsy, or the nicotinic cholinergic receptor subunits (α2,β2,β4) in ADNLFE. In other cases, entirely separate gene pathways may contribute to a very similar phenotype. This property may ultimately explain not only clinical differences in the seizures attached to each gene and their neurological severity in affected patients, but also their pharmacoresistance in clinical subsets of the disorder.
Pharmacoresistance can itself be considered as a phenotypic trait whose intrinsic mechanisms are, at least in part, influenced by genetic variation. Pharmacogenetic studies have attempted to investigate whether drug resistance is influenced by single nucleotide variants in genes for drug targets, or in other genes related to drug uptake and metabolism, which might explain resistance to drugs [16]. These studies, however, are hampered by serious methodological difficulties, since they do not take into account the causal heterogeneity of ‘epilepsy’ in the populations studied. This oversimplification is reflected in the assumption that a single mechanism would influence drug efficacy in relation to different mechanisms of epileptogenicity, which should be replicated across multiple studies. However results from such studies have not been consistent. For example, a single intronic nucleotide polymorphism in the SCN1A gene was associated with higher prescribed doses of phenytoin and carbamazepine in a UK based study [29], but not in subsequent studies in Austria [34] and Italy [20]. Likewise, studies exploring how gene variants may influence AED penetration into the brain have provided conflicting results [2]. Very large studies on etiologically and ethnically homogeneous populations would be necessary to fully explore the real influence of specific genes on pharmacoresistance.
2.3 Discovery of Novel Comorbidity Syndromes
Epilepsy clinicians have long recognized the frequent association of a variety of cognitive and neuropsychiatric symptoms with seizures, and understood that these occur more often than would be predicted by chance. Whether these result as a direct developmental effect of the cause of the epilepsy, the seizures themselves, or their treatment will always be under debate. Since their co-expression is usually incomplete, other genes may also contribute to the relative penetrance of the co-morbidity.
In the laboratory, mouse models of apparently unrelated disorders, such as Alzheimer’s disease, have delivered firm evidence that single genes can produce both epilepsy and cognitive defects unrelated to antiepileptic treatment, and suggest that antiepileptic treatment may be especially neuroprotective in carriers of these genes [24, 25]. Sudden unexpected death (SUDEP) is another important comorbidity, affecting individuals with idiopathic epilepsy. Recent evidence has confirmed the hypothesis that genes underlying cardiac arrhythmias are co-expressed in brain and produce epilepsy [10]. These genes may prove clinically useful in predicting SUDEP risk and exploring treatments to prevent premature lethality in epilepsy patients.
A great deal of interest is now devoted to understanding the developmental causes of epileptic encephalopathies, in relation to autism spectrum disorders (ASD) and intellectual disability. Exome sequencing in a large cohort of individuals with epileptic encephalopathies and subsequent protein-protein interaction analysis revealed a high interconnectivity between genes carrying de novo mutations, with a much greater probability of overlap with ASD and intellectual disability exome sequencing studies [7].
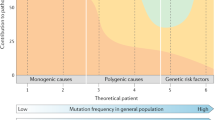
Finally, the ability to probe the full genomic variant profile of unrelated epilepsy patients with and without comorbidities holds enormous promise in understanding the genetic roots of comorbidity. Recently, one such study identified a de novo truncation of the skeletal muscle chloride channel, CLCN1 in a young woman with a childhood writer’s cramp and longstanding pharmacoresistant seizures. This genomic analysis led to the unexpected discovery that CLCN1 is expressed not only in skeletal muscle, but in thalamocortical and cerebellar brain networks, where disruption of chloride-mediated membrane repolarization could lead to hyperexcitability and seizures [5]. This hypothesis-generating study is a harbinger of the kind of novel candidate gene discovery that awaits the widespread use of next generation sequencing (Fig. 25.1).
Detection of heterozygous de novo nonsense mutation in CLCN1, encoding a premature stop codon in the CLC-1 chloride channel protein in a proband with generalized tonic clonic and absence seizures with a subtle myotonic phenotype. (a). PCR amplification of the final coding exon (exon 23) in the trio yielded a 550 bp product. (b) Sequence chromatograms for the trio shows the heterozygous base pair substitution encoding a premature stop codon in the proband, but not in either parent. (c) Schematic diagram of a single alpha-subunit of the CLC-1 channel protein showing the location of the C-terminal truncation R976X mutation in the proband. (d) Typical absence seizure in the proband (From Chen et al. [5])
2.4 Gene Testing
Recent identification of causative genes for a number of early-onset severe epilepsies has created the opportunity for diagnostic genetic testing in this population. Some examples include brain malformations and epileptic encephalopathies of infancy. At present, the clinical impact of genetic testing in these syndromes is by itself limited, due to the small percentage of patients in whom a single, causative gene mutation can be identified and the lack of specific, gene-directed, treatment options. However, genetic counseling can certainly be improved by recognizing a specific etiology. It also sets the stage for further research advances in understanding how each of the genes give rise to epileptogenic defects, and discovering which of these may be reversible. It has been claimed that, in some cases, discovery of a single causative gene defect may reduce the need for further diagnostic investigation at the biochemical level. However, from a practical standpoint, since genomic variants require time to analyze, this information typically arrives after reversible causes have been clinically excluded. It is also essential to understand that recent profiling studies of whole genomes and large sets of candidate exomes such as ion channel genes have determined that patients with sporadic epilepsy often carry more than one potentially causative mutation [17], complicating the interpretation. Furthermore, all individuals carry, on average, 50–100 loss of function variants in disease genes that for the most part produce no apparent clinical effects [30], signifying that the mere presence of a variant does not predict clinical status. This is likely explained by the presence of other ‘protective’ modifier genes. Thus, as we gain access to a broader view of the genetic landscape in individuals with epilepsy, we expect to routinely encounter patients with a complex genetic basis for their seizure phenotype.
The next steps toward increasing the power of genetic testing in epilepsy include identifying more genes for monogenic epilepsies, and learning to understand the contribution of specific genes in epilepsies with complex inheritance. This will require continued genotype-phenotype correlations, coupled with functional studies of the abnormal proteins to more accurately understand the pathophysiological implications of each new mutation and how they combine to create neural excitability phenotypes. While bioinformatic analysis offers an increasingly powerful way of categorizing the potential damage a gene variant may inflict on protein function, it cannot conclusively predict its actual effect upon a neuron, and indeed, despite being expressed in multiple cell types, it may not affect them all equally. However we are now entering the era of ‘personalized’ mutation analysis, where the mutant functional defect can be determined directly in the patient’s own cells, sometimes finding that it is counter to the expected result. For example, a currently held hypothesis for the mechanism of epilepsy in Dravet Syndrome, as studied in a mouse model of Scn1a haploinsufficiency, is based on the failure of interneurons to fire adequately in the face of reduced sodium current through Scn1a ion channels [31]. Analysis of membrane excitability in stem-cell derived neurons transformed from a Dravet Syndrome patient showed that the mutation, predicted to reduce the density of functional sodium channels, resulted in increased sodium current and hyperexcitability in cells classified as both excitatory and inhibitory, implying a distinctly different pathogenic mechanism [19]. These studies may have practical implications for diagnosis, genetic counseling and possible treatment, as well as increasing our knowledge of normal brain function and mechanisms of epileptogenesis.
We can now consult lists of epilepsy syndromes in which a chromosomal locus or loci have been mapped, and those in which one or more gene mutations or variants have been identified. These lists are constantly expanding as new loci and genes are identified. Our view on the correlations between phenotype and genotype in genetic epilepsies is also rapidly changing in relation to new findings emerging from exome sequencing and the use of diagnostic panels as unexpected phenotypes become associated with mutations of specific genes and vice-versa. A constantly updated database will be essential to establish all the known gene mutations and polymorphisms and their clinical correlates, so that genotype-phenotype correlations can be determined. This is the objective of the Human Variome Project [13] and an achievable goal for epilepsy genetics. For example, over 700 mutations in SCN1A have now been reported in the SCN1A Variant Database [14] and offer the possibility of predicting the onset, if not the severity, of the related phenotype with a reasonable likelihood.
3 Does Knowledge of a Specific Genetic Cause Influence Treatment?
The appropriateness or inadvisability of a given treatment in a specific condition has in some instances been acquired in clinical practice and then scientifically justified by genetic knowledge. For example, the potential for lamotrigine, a sodium channel blocker, to aggravate seizures in Dravet syndrome was initially reported well before the discovery that loss of function SCN1A mutations were the cause of the syndrome [11]. However from a clinical perspective, a number of specific conditions provide evidence that improved understanding of epilepsy genetics, together with enhanced knowledge of molecular pathology and electroclinical characteristics, substantiate more rational and effective treatment choices resulting in better patient management. In cases where genotype-phenotype correlations suggest that the epilepsy may have a benign course, gene testing may support the decision to withhold antiepileptic drug therapy during critical periods of brain maturation. Examples of this are mainly related to benign familial epilepsies starting in the first years of life due to PRRT2, KCNQ2 and SCN2A gene mutations [33].
Only in very rare conditions, however, do the treatment choices specifically target the inherited pathophysiological mechanism. An interesting example is represented by autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) due to mutation of neuronal nicotinic acid acetylcholine receptor alpha subunit in which the therapeutic effect of nicotine patch treatment on refractory seizures was elegantly albeit anecdotally demonstrated using an N-of-1 trial [32]. Other studies have confirmed that transdermal nicotine administration may be a suitable treatment option for patients with ADNFLE and severe seizures [4]. However, the translational dimension of these observations is limited as nicotine is highly addictive and may cause cardiovascular effects. More specific examples derive from clinical conditions in which a rationale therapeutic approach is prompted by administering a substance that can correct a metabolic defect that causes epilepsy. Pyridoxine-dependent epilepsy, for example, is an autosomal recessive disorder in which seizures manifesting in the neonatal period or in infancy can only be controlled after administration of high doses of pyridoxine [12]. If untreated, the disorder can lead to life threatening status epilepticus. Affected patients require lifelong pyridoxine supplementation but antiepileptic medication is usually unnecessary. While prognosis for seizure control is excellent in most patients, neurodevelopmental impairment is often present and although it has been suggested that children who are treated early have a better outcome, this is not always the case [9]. Pyridoxine-dependent epilepsy is likely underdiagnosed and for this reason in many centers pyridoxine administration is part of a treatment protocol for neonatal seizures. Pyridoxine dependent epilepsy is caused by mutations in the ALDH7A1 gene, which encodes for an aldehyde dehydrogenase (antiquitin) acting in the cerebral lysine catabolism pathway [21]. Affected individuals have α-aminoadipic semialdehyde (AASA) levels, which cause an intracellular reduction in the active vitamin B6 co-factor pyridoxal-5′ -phosphate (PLP) and a concomitant imbalance of glutamic acid and γ-aminobutyric acid (GABA). Folinic acid-responsive seizures are very similar to PDE [8]. Early seizures can also be caused by deficient pyridox(am)ine 5′ -phosphate oxidase (PNPO), which respond to pyridoxal-5′ -phosphate supplementation [9].
A third important example of a direct link between genetic diagnosis and effective treatment choice is represented by the use of the ketogenic diet in the treatment of the GLUT1 deficiency syndrome, an autosomal dominant disorder due to a mutation in the SLC2A1 gene. Brain glucose transport occurs by facilitated transport, predominantly via GLUT1, located on the blood–brain barrier endothelium, SLC2A1 mutations result in insufficient transport of glucose into the brain. Patients with GLUT1 deficiency had originally been described as exhibiting a severe neurological syndrome with early intractable seizures, followed by developmental delay, microcephaly and paroxysmal or continuous dyskinesia. This condition was initially identified and subsequently diagnosed in clinical practice, based on low glucose levels in the CSF (hypoglycorrhachia) in the setting of normal serum glucose or of abnormal CSF/serum glucose ratios [6]. However, a wide range of variants have been described, resulting in variable degrees of impairment of glucose transport [23], complicating the utility of the genetic information in clinical practice [18]. Neurologic consequences of GLUT1 deficiency presumably arise from disordered brain energy metabolism, secondary to reduced transport. D -glucose is the main fuel for the brain, although alternative fuels such as ketone bodies can be used. The treatment of choice for GLUT1 deficiency syndrome is a diet that mimics the metabolic state of fasting and provides ketones as an alternative fuel for the brain, effectively restoring brain energy metabolism. The ketogenic diet is a high-fat, adequate protein, low carbohydrate diet that provides 87–90 % of daily calories as fats and is used in the treatment of drug resistant childhood epilepsy. As the developing brain requires substantially more energy in young children, patients with GLUT1 deficiency syndrome should be started on the diet as early as possible and should remain on the diet at least until adolescence. Although some patients with milder seizure disorders may respond to antiepileptic medication, most do not and seizure response to the ketogenic diet is remarkable. Also, some pharmacological agents such as phenobarbital and diazepam, impair GLUT1 function and should be avoided [3]. In the past few years, the range of clinical epilepsy phenotypes where GLUT1 mutations and a positive response to the ketogenic diet have been identified is expanding [22, 26–28], raising the possibility that the gene acts as a modifier of other coexisting abnormalities. The usefulness of the ketogenic diet in GLUT1 deficiency syndrome was demonstrated before the syndrome was linked to mutations of the GLUT1 gene, based on the expected pathophysiological consequences of low levels of glycorrachia, however, the possibility of uncovering GLUT1 mutations in patients with atypical clinical presentations of GLUT1 deficiency and even borderline or normal levels of glycorrachia, has brought about invaluable advantages for the diagnosis and treatment of this disorder.
4 Conclusion
In the epilepsy clinic, genetic analysis has revealed not only the presence of more monogenic epilepsy syndromes, but, thanks to continually emerging genome-phenome correlations, is also pointing the way to earlier and more accurate diagnosis. Gene-specific classification of patients will aid clinical stratification to tailor more relevant diagnostic testing and better characterization of the natural history of the disease, enabling improved outcome prediction and genetic counseling for family planning. Future clinical treatment trials will almost certainly include genomic characterization to enhance the detection of a drug response signal as well as any gene-linked adverse effects. The relative contributions of major categories of genetic influence, including inherited monogenic epilepsies, de novo mutations, and sporadic individuals with complex multigenic inheritance are under exploration in many different seizure types and are beginning to inform genetic counseling in the neuropediatric setting. The definitions of classical epilepsy syndromes have been enlarged to account for multiple genetic etiologies, and novel comorbidity syndromes.
In the epilepsy neurobiology laboratory, genetics continues to reveal mechanistic insight into the rich biological diversity of gene defects leading to epilepsy phenotypes. Genes linked to epilepsy have opened the door to understanding the neurobiology and pathology of epilepsies and localizing the vulnerable neural pathways at the molecular, cellular, and functional levels. Defects in ion channels and a broad range of other cellular signaling pathways including receptors, transporters, and proteins for exocytotic release of neurotransmitters now constitute primary classes of epileptogenic mechanisms. A second major category of epilepsy genes involves transcription factors regulating the early migration and maturation of interneurons, and a third group controls metabolic functions within the cell and neuron-glia relationships.
Mouse models bearing mutations in each of these gene-delineated pathways allow us to examine the fine details of how they alter developmental plasticity in the epileptic brain, to learn when and where cellular pathology arises, and how it spreads to alter excitability in cortical networks through remodeling of gene expression and synaptic reorganization. Mice engineered to conditionally express gene mutations in specific circuits provide information on which circuits are necessary or sufficient to produce the seizure phenotype. Finally we can learn whether there are critical developmental stages for correcting or reversing the gene defect, exactly what the desired drug effect should be at the cellular level, and which molecular targets are most effective in preventing the epileptic disorder. Taken together, these advances hold great promise for improving the clinical management of seizure disorders.
References
Bergren SK, Rutter ED, Kearney JA (2009) Fine mapping of an epilepsy modifier gene on mouse Chromosome 19. Mamm Genome 20:359–366
Bournissen FG, Moretti ME, Juurlink DN, Koren G, Walker M, Finkelstein Y (2009) Polymorphism of the MDR1/ABCB1 C3435T drug-transporter and resistance to anticonvulsant drugs: a meta-analysis. Epilepsia 50:898–903
Brockmann K (2009) The expanding phenotype of GLUT1-deficiency syndrome. Brain Dev 31:545–552
Brodtkorb E, Picard F (2006) Tobacco habits modulate autosomal dominant nocturnal frontal lobe epilepsy. Epilepsy Behav 9:515–520
Chen TT, Klassen TL, Goldman AM, Marini C, Guerrini R, Noebels JL (2013) Novel brain expression of ClC-1 chloride channels and enrichment of CLCN1 variants in epilepsy. Neurology 80:1078–1085
De Vivo DC, Trifiletti RR, Jacobson RI, Ronen GM, Behmand RA, Harik SI (1991) Defective glucose transport across the bloodbrain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N Engl J Med 325:703–709
Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS, Berkovic SF et al (2013) De novo mutations in epileptic encephalopathies. Nature 501:217–221
Gallagher RC, Van Hove JL, Scharer G (2009) Folinic acid-responsive seizures are identical to pyridoxine-dependent epilepsy. Ann Neurol 65:550–556
Gospe SM (2011) Pyridoxine-dependent epilepsy. In: Shorvon S, Andermann F, Guerrini R (eds) The causes of epilepsy. Cambridge University Press, Cambridge, pp 237–241
Goldman AM, Glasscock E, Yoo J, Chen TT, Klassen TL, Noebels JL (2009) Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained death. Sci Transl Med 1:2ra6
Guerrini R, Dravet C, Genton P, Belmonte A, Kaminska A, Dulac O (1998) Lamotrigine and seizure aggravation in severe myoclonic epilepsy. Epilepsia 39:508–512
Hoffmann GF, Schmitt B, Windfuhr M (2007) Pyridoxal 50-phosphate may be curative in early-onset epileptic encephalopathy. J Inherit Metab Dis 30:96–99
The Human Variome Project: http://www.humanvariomeproject.org/
SCN1A variant database: http://www.molgen.ua.ac.be/SCN1AMutations/
Jorge BS, Campbell CM, Miller AR et al (2011) Voltage-gated potassium channel KCNV2 (Kv8.2) contributes to epilepsy susceptibility. Proc Natl Acad Sci U S A 108:5443–5448
Kasperaviciute D, Sisodiya SM (2009) Epilepsy pharmacogenetics. Pharmacogenomics 10:817–836
Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, McPherson J, Bourquin T, Lewis L, Villasana D, Morgan M, Muzny D, Gibbs R, Noebels J (2011) Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell 145:1036–1048
Klepper J (2012) GLUT1 deficiency syndrome in clinical practice. Epilepsy Res 100:272–277
Liu Y, Lopez-Santiago LF, Yuan Y, Jones JM, Zhang H, O’Malley HA, Patino GA, O’Brien JE, Rusconi R, Gupta A, Thompson RC, Natowicz MR, Meisler MH, Isom LL, Parent JM (2013) Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Ann Neurol 74:128–133
Manna I, Gambardella A, Bianchi A et al (2011) A functional polymorphism in the SCN1A gene does not influence antiepileptic drug responsiveness in Italian patients with focal epilepsy. Epilepsia 52:40–44
Mills PB, Struys E, Jakobs C (2006) Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med 12:307–309
Mullen SA, Marini C, Suls A, Mei D, Della Giustina E, Buti D, Arsov T, Damiano J, Lawrence K, De Jonghe P, Berkovic SF, Scheffer IE, Guerrini R (2011) Glucose transporter 1 deficiency as a treatable cause of myoclonic astatic epilepsy. Arch Neurol 68:1152–1155
Nickels K, Wirrell E (2010) GLUT1-ous maximus epilepticus: the expanding phenotype of GLUT-1 mutations and epilepsy. Neurology 75:390–391
Noebels J (2011) A perfect storm: converging paths of epilepsy and Alzheimer’s dementia intersect in the hippocampal formation. Epilepsia 52(Suppl 1):39–46
Palop JJ, Chin J, Roberson ED et al (2007) Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer’s disease. Neuron 55:697–711
Striano P, Weber YG, Toliat MR, Schubert J, Leu C, Chaimana R, Baulac S, Guerrero R, LeGuern E, Lehesjoki AE, Polvi A, Robbiano A, Serratosa JM, Guerrini R, Nürnberg P, Sander T, Zara F, Lerche H, Marini C, EPICURE Consortium (2012) GLUT1 mutations are a rare cause of familial idiopathic generalized epilepsy. Neurology 78:557–562
Suls A, Dedeken P, Goffin K et al (2008) Paroxysmal exercise induced dyskinesia and epilepsy is due to mutations in SLC2A1, encoding the glucose transporter GLUT1. Brain 131:1831–1844
Suls A, Mullen SA, Weber YG, Verhaert K, Ceulemans B, Guerrini R, Wuttke TV, Salvo-Vargas A, Deprez L, Claes LR, Jordanova A, Berkovic SF, Lerche H, De Jonghe P, Scheffer IE (2009) Early-onset absence epilepsy caused by mutations in the glucose transporter GLUT1. Ann Neurol 66:415–419
Tate SK, Depondt C, Sisodiya SM et al (2005) Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci U S A 102:5507–5512
The 1000 Genomes Project Consortium (2010) A map of human genome variation from population scale sequencing. Nature 467:1061–1073
Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, Catterall WA (2006) Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci 9:1142–1149
Willoughby JO, Pope KJ, Eaton V (2003) Nicotine as an antiepileptic agent in ADNFLE: an N-of-one study. Epilepsia 44:1238–1240
Zara F, Specchio N, Striano P et al (2013) Genetic testing in benign familial epilepsies of the first year of life: clinical and diagnostic significance. Epilepsia 54:425–436
Zimprich F, Stogmann E, Bonelli S, Baumgartner C, Mueller JC, Meitinger T, Zimprich A, Strom TM (2008) A functional polymorphism in the SCN1A gene is not associated with carbamazepine dosages in Austrian patients with epilepsy. Epilepsia 49:1108–1109
Acknowledgements
We extend our gratitude to Phil Schwartzkroin, a careful scientist, wise Editor-in-Chief, and trusted friend who has inspired us both throughout our careers in epilepsy research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Guerrini, R., Noebels, J. (2014). How Can Advances in Epilepsy Genetics Lead to Better Treatments and Cures?. In: Scharfman, H., Buckmaster, P. (eds) Issues in Clinical Epileptology: A View from the Bench. Advances in Experimental Medicine and Biology, vol 813. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-8914-1_25
Download citation
DOI: https://doi.org/10.1007/978-94-017-8914-1_25
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-8913-4
Online ISBN: 978-94-017-8914-1
eBook Packages: MedicineMedicine (R0)