Abstract
The study of the organization and dynamics of biological membranes has greatly benefited from the considerable technical advances achieved in the photonics field. Breaking the diffraction limit in optical microscopy to further increase spatial resolution has allowed to define the ultra-structure of the different proteolipid complexes within cellular membranes. Furthermore, the improvements in fluorescence sensitivity have prompted to the analysis of molecular dynamics up to single-molecule level with high temporal resolution. Thanks to all these advances, the concept of tetraspanin-enriched microdomains has been revisited in recent studies that shed new light on tetraspanin dynamics and interactions.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Mean Square Displacement
- Primary Human Endothelial Cell
- Membrane Platform
- Single Molecule Tracking
- High Numerical Aperture Objective
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
4.1 Introduction
Molecular partitioning on cellular membranes and, in particular, the plasma membrane constitutes an essential requirement for the optimal function of an enormous variety of biological processes (Jacobson et al. 2007). Lipids, proteins and carbohydrates spatially coexist in diverse specialized microdomains or nanodomains of the membrane that are characterized by a specific molecular repertoire either in the outer or in the inner leaflets although coupling of the two leaflets is a matter of debate. Thus, receptors and related structural and signaling adaptors are confined within organized membrane units which allow cellular communication with the environment and signal transduction (Dehmelt and Bastiaens 2010). In this regard, the membrane polarity found in some tissues constitutes a further specialized cellular mechanism for compartmentalization of membrane bioactivity such as controlled endocytosis, secretion or proteolysis (Yanez-Mo et al. 2009).
The organizational diversity of various membrane microdomains is supported by a multitude of specific lipid-lipid, lipid-protein and protein-protein interactions. For example, both coalescence of glycosphingolipids and cholesterol (Chl) and their interaction with a number of membrane associated structural and signalling proteins (such as caveolin, flotillins, glycosylphosphatidylinositol (GPI)-linked proteins and the others) are necessary for the assembly of lipid rafts (Lingwood and Simons 2010). These microdomains have been proposed to function as platforms for sorting of GPI-anchored proteins (Mayor and Riezman 2004), the assembly of cytoplasmic signaling complexes (Lasserre et al. 2008; Parton and Hancock 2004) and caveolae (caveolin, cavin, flotillin)-mediated endocytosis (Hansen and Nichols 2010). Microdomains based on tetraspanin-mediated interactions (the so called tetraspanin-enriched microdomains TERMs (Berditchevski et al. 2002) or TEMs (Hemler 2003)) or tetraspanin web (Boucheix and Rubinstein 2001)) relies on the ability of tetraspanin proteins to simultaneously interact with each other and with molecular partners in cis. In this manner, a plethora of transmembrane receptors can be found embedded in a tetraspanin microenvironment which confers them an adequate molecular density to efficiently exert their physiological functions. Furthermore, TEMs spatially confine function-related receptors, which cooperate in certain cellular processes such as adhesion or migration (Barreiro et al. 2008). At the same time, tetraspanins promote local enrichment of actin-anchoring proteins and mediators which act downstream of tetraspanin partners and facilitate amplification of signal transduction (Charrin et al. 2009). TEMs exhibit a specific composition and stoichiometry depending on the cell type and, although a certain degree of redundancy can be found within the tetraspanin family, full restoration of a specific tetraspanin-dependent function cannot normally be achieved with related members of the family (Barreiro et al. 2005).
The initial evidence for existence of tetraspanin microdomains came from biochemical and proteomic approaches involving solubilisation of membranes with a wide range of detergents (Le Naour et al. 2006; Hemler 2005), from structural analysis of tetraspanin dimerization (Drummer et al. 2005) and from images of fixed cells and tissues obtained with conventional fluorescence and electron microscopy equipments (Berditchevski and Odintsova 1999; Abitorabi et.al. 1997). However, all these approaches do not reflect the dynamic nature of these membrane structures. The need for a formal demonstration of TEM existence in living cells led us to study in depth the complex and specific interactions among different tetraspanins and their partners as well as their biophysical properties in native conditions of the plasma membrane even at a single-molecule level (Barreiro et al. 2008; Espenel et al. 2008). Thus, analytical microscopy and spectroscopy techniques are able to “break” the diffraction limit allowed us to produce conclusive data on the differential and unique nature of TEMs in comparison with GPI protein-containing classical raft domains.
4.2 How to Probe Membrane Dynamics
During the last 10 years, important technological improvements in fluorescence microscopy have been done allowing analysis of biological molecules at the single molecule level. This includes the development of highly sensitive camera, high numerical aperture objectives and stable and bright probes. Two techniques in particular have been successfully used to probe the dynamics of biological membranes: fluorescence fluctuation spectroscopy (FFS) and its derivatives, and fluorescence single molecule tracking (SMT). Membrane dynamics can also be probed with FRAP (Fluorescence Recovery After Photobleaching), a technique that renders information about the average diffusion coefficient of a molecular ensemble as well as the percentage of mobile and immobile fractions out of the overall molecular population analyzed. However, the spatial resolution is poor because the size of the bleached area often exceeds the diffraction limit. In addition, it is impossible to determine different modes of motion of molecules within membranes.
First used to study molecules in solution, FCS (Fluorescence Correlation Spectroscopy) is a single-molecule sensitive technique that analyzes fluctuations in the fluorescence emission of small molecular ensembles in a sub-femtoliter volume. Fluorescence fluctuation analysis thus provides information about a multitude of parameters such as stoichiometry, concentration and molecular diffusion of fluorescently labeled molecules with a very high temporal resolution (reviewed in (Schwille et al. 1999)). In addition, FCS is less invasive than FRAP since it does not require heavy-loaded labeling and, for this reason, molecular species can be observed in a more preserved environment. In order to limit the observation volume, classical FCS setups combine high numerical aperture objective and pinholes (one can also use two-photon excitation as an alternative to pinholes). When compared to one-photon confocal FCS, two-Photon FCS requires an extremely high photon flux density but increases the signal to noise ratio and stability for long-term measurement (Chen et al. 2001), two parameters that are important for measuring diffusion in membranes. Moreover, two-photon FCS allows simultaneous excitation of two or more spectrally distinct dyes with a single line simplifying cross-correlation experiments and enables the detection of co-diffusion of two distinct molecular species (Fig. 4.1). The FCS approach has contributed significantly to the field of membrane organisation and dynamics, especially to studies of lipid rafts using giant unilamellar vesicles or model lipid bilayers (see the reviews (Garcia-Saez and Schwille 2008; Marguet et al. 2006)). A significant progress in the understanding of membrane organization has been achieved with the development by Marguet and colleagues of a new method which allows identification of the motion mode of membrane components by analyzing the FCS diffusion law of τd (i.e. the average time a fluorescent molecule stays within the illuminated area) versus the beam area (Wawrezinieck et al. 2005). More recently, a combination of STED (Stimulated Emission Depletion) microscopy with FCS has been used to probe the dynamics of membrane lipids in living cells (Eggeling et al. 2009). The advantage of this technique is the possibility to tune the diameter of the probed area up to 70-fold below the diffraction barrier. Different approaches facilitating FCS on membranes have also been developed focusing on the problem of photobleaching phenomenon occurring during FCS measurement. It mainly concerns scanning FCS (SFCS) in which the detection volume is scanned through the sample. This includes the continuous wave excitation SFCS (Ries and Schwille 2006) and raster image correlation spectroscopy (RICS), a version of FCS in which involves the spatio-temporal correlation analysis of fluorescence fluctuations in the intensity of a given pixel in an image obtained with a confocal laser-scanning microscope (see an example of diffusion analysis in cell membrane in Digman and Gratton 2009). The measurement of diffusion coefficient of molecules diffusing slowly in the membrane is possible with these two latter techniques (this is generally impossible to achieve with classical FCS).
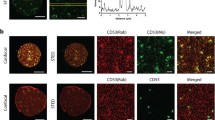
Detection of CD9-CD151 complexes on the plasma membrane of living human primary endothelial cells by fluorescence cross-correlation spectroscopy (FCCS). Representative FCCS measurement at the plasma membrane of transiently transfected primary endothelial cells co-expressing very low levels of mRFP-tagged CD9 and mEGFP-tagged CD151. The figure shows the fluorescence intensity image (kCPS, kilo counts per second), the autocorrelation function (ACF) (black line) derived from the fluorescence intensity trace acquired at the point marked with a grey cross, the best-fitted curve using an anomalous diffusion model (red line), and the diffusion coefficient (D). In the FCS autocorrelation curves, the x axis (τ) represents the delay time in seconds, and the y axis (G(τ)) is the autocorrelation amplitude as a function of delay time. The detection of cross-correlation of green and red fluorescence fluctuations indicates that both fluorescently tagged molecular species co-diffuse in the same supramolecular complexes over time. In this particular example, the diffusion coefficient obtained in the cross-correlation seems to correspond more to EAP diffusion rather than to small tetraspanin complexes exchangeable among platforms which exhibit faster dynamics
SMT within membranes allows the direct visualization of the diffusion of a membrane component. It requires the labeling of only a few molecules in order to track them individually. Precursor studies have investigated the membrane behavior using latex or colloidal gold beads bound to the molecule of interest. The main drawback of this so-called single particle tracking (SPT) technique concerns the size of the bead, which is about two orders of magnitude larger that the size of the tracked membrane component, and can constrain its diffusion (reviewed in Marguet et al. 2006). With the development of very sensitive detectors and organic fluorescent probes that are very stable and bright, it is now possible to track membrane components for long time (within 1 min range). This technique, often referred to as single dye tracking (SDT), requires the nanometer-ranged determination of the localization of the fluorescence spot before linking the different position of the molecules to construct the trajectory (details in Fig. 4.2). This trajectory can give information about the behavior of a membrane component by calculating the mean square displacement (MSD) of the molecules (Saxton and Jacobson 1997). The most popular fluorescent probes to track molecules are the cyanin derivatives such as the cationic Atto647N (Eggeling et al. 2009; Espenel et al. 2008) and quantum dots (Qdots). These Qdots are very bright and the most stable probes available and can be considered as a tool of choice for tracking proteins within membranes, even though the control of the valency of labeling, an important parameter for the tracking of single molecule, still requires further improvement (see Pinaud et al. 2010).
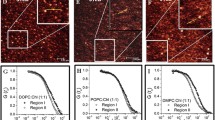
Single molecule tracking of fluorescent molecules. Setup: In order to record single molecule fluorescence signal (a), a setup equipped with a high numerical aperture objective (~1.4) and an EM-CCD camera is necessary. Alternating-laser excitation (ALEX) prevents crosstalk in multicolor experiments (Margeat et al. 2006). Most commonly used probes are cyanin derivatives or quantum dots. (b) is the DIC image corresponding to the area shown in (a). Analysis and processing of single molecule traces: Trajectories are constructed using the individual diffraction limited signal of each molecule (a). The center of each fluorescence peak is determined with sub-pixel resolution by fitting a two-dimensional elliptical Gaussian function (c). The accuracy of the position measurement ∂ is dependent on the wavelength λ and the number of photons N collected. The two-dimensional trajectories of single molecules were constructed frame per frame. The mean square displacement (MSD) of each molecule is plotted as a function of the time interval ∂t and the shape of the curve determine the type of motion (d). For Brownian trajectories, the MSD varies linearly with ∂t and the value of the slope is 4D (brown line in d). If the MSD-∂t (or τ) plot shows positive or negative deviation from a straight line, the MSD is respectively adjusted with a quadratic curve (4Dt + ν 2 t 2)(directed diffusion, red line in d) or with an exponential curve \( \frac{{L}^{2}}{3}[1-\mathrm{exp}(\frac{-12Dt}{{L}^{2}})]\) (confined diffusion, purple line in d) where L is the side of a square domain (Kusumi et al. 1993). The apparent diffusion coefficient values are determined from a linear fit to the MSD-τ plots between the second and the fourth points (D2-4) according to the equation MSD(t) = 4Dt. Thanks to a dual view imaging, it is possible to superimpose trajectories in the context of an ensemble labeling (see a Brownian trajectory in e and a mixed trajectory in f; a mixed trajectory is a combination of Brownian and confined (orange circle) motion modes). All the movies were analyzed using a homemade software (named “PaTrack”) implemented in visual C++ and motion modes within trajectories were automatically attributed using a neural network (manuscript in preparation). Scale bars are 10 μm in b and 1 μm in e and f
It is worth mentioning that labeling with fluorescent proteins (e.g. Green Fluorescent Protein (GFP)) only allows tracking for up to a few seconds because of their weak stability. Therefore, labeling is generally performed with antibodies labeled with the probes mentioned above. Fab fragments should be preferred to full antibodies because of cross-linking ability of the latter reagents (this drawback of using a bivalent antibody was observed in SMT experiments which studied the dynamics of tetraspanin CD9 (Espenel et al. 2008)). On the other hand, both approaches (cell labeling with antibodies and the use of GFP-derived tags) may occlude epitopes important for interactions of a target protein with its ligands and membrane or cytoplasmic partners.
4.3 Current View of the Dynamics of Tetraspanins
Characterization of the membrane dynamics of tetraspanins (TM4) remains an understudied area of research. Two papers, published almost simultaneously, have provided the first description of TEMs as physical entities in the plasma membrane of living cells and demonstrated that TM4, especially CD9, are very dynamic proteins (Barreiro et al. 2008; Espenel et al. 2008). Despite the fact that different biophysical approaches have been used in these studies, the authors have reached a similar general conclusion—although most of the TM4 molecules are dynamic, freely diffusing in the plasma membrane, they can also be confined in protein assemblies containing TM4 aggregates.
The work by Barreiro et al. have combined FRAP and FCS experiments to evaluate the dynamics of GFP-tagged CD9 and CD151 in living primary human endothelial cells in the context of cell adhesion. In FRAP experiments, CD9 diffusion coefficient was evaluated to be 0.47 μm2/s with a mobile fraction representing up to 88%; the diffusion coefficient of CD151 was ~0.41 μm2/s and its mobile fraction was 84%. The median values obtained with FCS although lower (0.17 μm2/s for CD9 and 0.14 μm2/s for CD151), were in the range similar to those obtained using FRAP. These results are in agreement with the differential size of the molecular samples measured by the two techniques (FRAP renders the average apparent diffusion of a huge molecular population versus a small number of molecules measured by FCS, whose average diffusion is much similar to single-molecule diffusion). FCS autocorrelation curves were describable by an anomalous diffusion model with an anomalous coefficient of 0.77 for CD9 and 0.71 for CD151. This parameter was interpreted as a transient interaction or confinement of TM4 diffusing molecules within endothelial adhesive platforms (EAPs) that also contain receptors such as ICAM-1 and VCAM-1. This interpretation is supported by an increase of the anomalous coefficient (and a corresponding decrease of the diffusion coefficient) when receptor-mediated docking structures were formed (i.e. increasing engagement of TM4 molecules in these structures). The difference found in the anomalous coefficients for these TM4 could suggest that CD151 is slightly more confined within EAP than CD9. This result can be related to the preferential interaction of CD151 with VCAM-1 that diffuses slower and is more confined than ICAM-1, a preferable partner for CD9 in endothelial cells. It underlines the specificity of interaction within TEMs and dependence of dynamics of individual TM4 on their specific partners. Furthermore, molecular complexes containing both CD9 and CD151 were also found by fluorescence cross-correlation analysis (Fig. 4.1; Barreiro and Zamai unpublished data). Interestingly, even in a different cell type and measuring by SMT at the basal instead of the apical membrane, the diffusion coefficient of CD9 molecules was in the same range (Espenel et al. 2008). Specifically, using SMT, the diffusion coefficient of CD9 in the prostate cancer cells PC3 was found to be 0.23 μm2/s. Furthermore, three different types of trajectories were identified from MSD analysis: Brownian, pure confined and mixed (a mixture of alternated confined and Brownian motion). Pure confined trajectories (23% of the total trajectories) can be paralleled with the percentage of immobile fraction in FRAP experiments in endothelial cells and, in both cell types, proteins displaying immobile and confined behavior represent the smaller fraction of the molecules. Using dual-view SMT setup, the authors were able to show that transient confinement of CD9 described above occurred in TM4 assemblies that behave like interactive platforms in permanent exchange with the rest of the membrane (Fig. 4.3). These platforms are somewhat similar to the specialized EAPs found in activated endothelial cells that contain adhesion receptors such as VCAM-1 and ICAM-1. In more recent experiments we extended these observations to other cell lines (such as HeLa and CHO cells) to show that the dynamic behaviour of CD9 (i.e. diffusion coefficient, type of motion and percentage of confinement in these cells) is similar to that observed in PC3 (Rassam and Milhiet, unpublished results). Altogether these results suggest that CD9 is a very dynamic membrane protein but a considerable fraction of CD9 molecules can be transiently trapped in TEMs. A tiny percentage of molecules also seem to be immobile in stable membrane platforms, at least for a few minutes. The relationship between the confinement of TM4 in membrane platforms and their functions deserves further investigation. All these results obviously raises a question about the membrane behavior of other TM4. CD81 molecules have been shown to be part of TEMs in primary endothelial cells (EAPs) (Barreiro et al. 2005) and PC3 cells and is considered to be one of the preferable partners of CD9 in TM4 assemblies (Charrin et al. 2009). Study of its dynamics should further extend our understanding of TM4 membrane behavior.
Single molecule tracking of CD9 in PC3 cells. Time lapse of a mixed trajectory of CD9 obtained by superposition of ensemble CD9 labeling achieved with anti-CD9 Cy3B-labeled antibodies (green) and a fluorescent single CD9 molecule (red spot) labeled with Atto647N-conjugated Fab fragments of the anti-CD9 SYB-1 (trajectories of the single molecule is indicated by the white thin line). The single CD9 molecule enters and exits from a tetraspanin-enriched domain (green) and is transiently confined when its position overlaps with the ensemble labeling (Adapted from (Espenel et al. 2008))
In addition to protein-protein interactions that could affect TM4 membrane behavior, lipids also influence CD9 dynamics and partitioning. In this regard, it has been observed that Chl depletion led to a decrease in the association of CD9 with TEMs and, consequently, affected dynamic behaviour of the tetraspanin in the plasma membrane. In addition, the experiments using a palmitoylation-deficient mutant have shown that CD9 Brownian diffusion is slowed down by palmitoylation, a phenomenon that can be explained by the preferential interaction of palmitoylated CD9 with more ordered lipid phase (as has already been suggested for classical raft microdomains (Brown and London 2000)). In addition, palmitoylation is also involved in the interaction of CD9 with TEMs. In agreement with the effects on CD9 dynamics observed upon Chl depletion, saturated fatty acid chains such as palmitate could favor association of CD9 with TEMs that are enriched in Chl. However, because Chl depletion does not alter TEMs in terms of shape and position, it is tempting to speculate that different pools of Chl co-exist on the plasma membrane. The first pool will include molecules mostly involved in the general physical properties of membrane by interacting with fatty acid chains of lipids. Another pool could be more involved in organizing the architecture of protein assemblies. This second pool will be less accessible to the drugs used for Chl depletion and weakly exchanged with the rest of the membrane explaining the preservation of TEMs upon depletion of this lipid. The existence of the second pool of Chl is in agreement with the ability of this lipid to be chemically cross-linked with CD9 (Charrin et al. 2003b).
Because of the development of the dual-view SMT mentioned above, it has been possible to perform dynamic co-localization experiments using Fab fragments of anti-CD9 mAb (Charrin et al. 2003a), labeled with spectrally distinct fluorophores (for more details, see (Espenel et al. 2008)). We have been able to characterize a complex of two molecules of CD9 diffusing in the membrane, both exhibiting a Brownian motion, before and during their association. CD9-CD9 dynamic co-localization that concerned 15% of the Brownian trajectories was always transient. The diffusion coefficient of both molecules was not modified when interacting with each other, suggesting that CD9 molecules were more likely diffusing in a cluster of molecules than as individual transmembrane protein. This cluster or nanodomain could accommodate several proteins and lipids including TM4, their partners and Chl. This hypothesis is supported by FCS experiments performed on endothelial cells since photon-counting histograms revealed that more than one fluorescent protein was present in diffusing entities (Barreiro et al. 2008). Association of proteins within clusters appears to be dependent on Chl and CD9 palmitoylation.
Several properties of TEMs can be related to so-called lipid rafts, especially the involvement of Chl in their formation as well as their resistance to the extraction by detergent. In addition, our SMT experiments demonstrate that CD55, often used as a raft marker (Charrin et al. 2002), exhibits a membrane behavior similar to that of CD9 in terms of motion mode (Brownian, confined, and mixed diffusion). It is important to mention that a similar behavior has been described for other membrane proteins such as EGFR and the GPI-anchored proteins CD59 (Suzuki et al. 2007; Sergé et al. 2008). This behavior is likely more related to the general organization of the plasma membrane in eukaryotic cells (a mosaic of nano or microdomains) than to the properties of rafts or TEMs. However, our results provide compelling evidence that rafts and TEMs are different physical entities at the plasma membrane. The SMT approach demonstrates that CD55 was never confined in TEMs whereas most of the CD9 trajectories passed through these areas. Moreover, the dynamics of a raft marker (a GPI-anchored protein coupled to EGFP) in EAP of endothelial cells evaluated by FRAP was clearly different from those of the tetraspanins CD9 and CD151. In addition, classical detergent-based biochemical experiments with endothelial cells indicate that the two types of domains were not located in the same lipid environment (Barreiro et al. 2008).
4.4 Dynamics and Functions
Although there are no clear biochemical functions directly ascribed to tetraspanin proteins (except for their role as membrane organizers), it is well established that tetraspanins act as modulators of the function of their lateral partners regulating a multitude of processes such as cell-cell or cell-matrix adhesion and migration (Yanez-Mo et al. 2009), protein trafficking (Berditchevski and Odintsova 2007), fusion (Rubinstein et al. 2006), cancer invasion (Stipp 2010), pathogen infection (Thali 2009; Tham et al. 2010) and others. Thus, the insertion in TEMs bestows some transmembrane receptors with the local molecular density (clustering) thus resulting in fully functional proteins. Hence, the spatial organization of receptors at the plasma membrane is as critical as their appropriate expression level in order to accomplish their function. In most cases, receptor clustering is induced upon certain stimuli by dynamic coalescence of receptor-containing TEMs. This is the case for endothelial adhesion receptors (Barreiro et al. 2008). We have investigated in depth the spatial organization and the functional regulation of endothelial receptors which seem to be governed by their inclusion in TEMs. Inflammatory conditions induce the expression of a variety of adhesion receptors (e.g. VCAM-1, ICAM-1, E- and P-selectins) at the luminal surface of endothelium to allow leukocyte-endothelial cell contacts and promote leukocyte extravasation (Barreiro et al. 2007). These endothelial adhesion receptors are not evenly distributed in the plasma membrane, but show a patterned distribution in submicron-sized domains, as observed with scanning electron microscopy of immunogold-labelled samples (Barreiro et al. 2008). The treatment with the exogenous large extracellular loop of CD9 coupled to GST disturbs this spatial organization, decrease receptor clustering, alter the molecular dynamics of the clusters and, consequently, negatively affect the receptor adhesive functions (Barreiro et al. 2005, 2008). This argues in favour of TEMs acting as cell-cell adhesion modulators. Once a leukocyte contacts with the endothelium in order to extravasate towards the inflammatory focus, the tetraspanin nanoclusters containing endothelial adhesion receptors that we have coined as EAPs rapidly coalesce around the leukocyte forming the so-called endothelial docking structure (Barreiro et al. 2002). The insertion of a variety of adhesion receptors in this specialized kind of TEMs implies the co-recruitment of all these receptors with complementary adhesive functions towards the leukocyte-endothelial contact area. In this manner, endothelial adhesion receptors are exposed to be available in case of leukocyte transendothelial migration. Noteworthy, most of the endothelial receptors embedded within EAPs are driven to the vicinity of the adhered leukocytes without recognizing any ligand on the leukocyte side and even without the participation of the actin cytoskeleton, but only due to their interaction with the tetraspanins (Barreiro et al. 2008). In this regard, the complexity of the molecular inter-relationships within EAPs has been dissected by using FRET-FLIM (fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy) and discovered the preferential binding of ICAM-1 to CD9, that of VCAM-1 to CD151, and the interaction of CD9 and CD151 with themselves and each other in living primary human endothelial cells. Thus, an array of receptors with related functions becomes spatially congregated to ensure the progression of the stepwise process of extravasation (Barreiro et al. 2008).
Integrins also constitute another representative example of molecules which can be regulated by avidity (clustering), although they also exhibit conformational changes to enhance their affinity for ligands (Carman and Springer 2003; Luo et al. 2007). Different integrin conformations have distinct diffusion profiles which control integrin clustering, vary according to the activation state of the cell and seem to be governed by the binding of the integrin to the actin cytoskeleton (Das et al. 2009). In this context, CD81 has been involved in the regulation of VLA-4 (α4β1) and VLA-5 (α5β1) adhesion strengthening to multivalent ligands, in particular during the interaction of VLA-4 to its endothelial ligand VCAM-1 under physiological flow conditions (Feigelson et al. 2003). Moreover, an anti-CD81 antibody has been shown to induce high avidity of the integrin LFA-1, the counter-receptor of the endothelial ICAM-1 (VanCompernolle et al. 2001). Therefore, the leukocyte integrins and their endothelial ligands participating in the leukocyte-endothelial adhesive interactions appear to be regulated in terms of avidity by their inclusion in TEMs. This regulatory mechanism could modulate the adhesion threshold of these transmembrane receptors to avoid undesired interactions at non-inflammatory sites of the vasculature but to promote a robust adhesion under stringent conditions as high shear flow stress at proper inflammatory scenarios. Same arguments could be applied to integrins functioning in cell-extracellular matrix interactions, a key process for cell adhesion, migration and invasion.
More recently, TM4 has been suggested to be involved in the sequestration of VAMP7, a membrane protein mediating exocytosis during neuritogenesis, phagocytosis and lysosomal secretion, within lipid microdomains (Danglot et al. 2010). Using Qdot-based single molecule tracking, it was shown that depletion in the tetraspanin CD82 in HeLa cells restrain EGFR diffusion. Effects upon CD82 depletion was explained by a modification of the membrane content in Chl and gangliosides, because this TM4 can regulate the expression of these lipids (Charrin et al. 2003b; Delaguillaumie et al. 2004; Le Naour et al. 2006; Odintsova et al. 2006; Regina Todeschini and Hakomori 2008), favouring the exit of EGFR molecules from lipid microdomains, and thus their dimerization and endocytosis. In this case, CD82 would be more involved in the homeostasis of the plasma membrane in terms of lipid composition than in the formation of a network of protein-protein interactions including EGFR.
The involvement of TEMs in receptor compartmentalization in other cellular contexts such as enzymatic activity (Yanez-Mo et al. 2008), antigen presentation at immune synapse (Mittelbrunn et al. 2002; Unternaehrer et al. 2007), viral and bacterial infection (Nydegger et al. 2006; Silvie et al. 2006; Tham et al. 2010), as well as gamete fusion (Ziyyat et al. 2006) has been also described. However, no detailed biophysical approaches have been applied on these topics to date, except in the case of HIV-1 assembly (Krementsov et al 2010). The employment of powerful analytical microscopy and spectroscopy techniques will help in solving the contribution of TEMs to these crucial physiopathological processes. The performance of biophysical studies will also favour the development of specific new therapeutic agents directed against TM4, which we hypothesize could have a general and more potent effect than the single targeting and inhibition of one of the receptors involved in cancer invasion, inflammation or pathogen infection. However, due to the ubiquitous nature of TM4, it will also be critical to develop new strategies for the administration, specific targeting and local delivery of TM4-based therapies (Barreiro et al. 2009).
4.5 Perspectives and Future Developments
The studies described in this chapter have demonstrated for the first time that TM4 are dynamic molecules that can exhibit a Brownian motion within plasma membrane or be transiently confined in TEMs (Fig. 4.4). Further works are now required to have a more precise view of the dynamic network of protein-protein interactions. Specifically, we need to uncover how TM4 interact with each other and with their membrane partners, how dynamic these TEMs are and what are precise molecular composition and stoichiometry of these membrane platforms. The relationship of these membrane platforms with the cytoskeleton needs also further clarification since some TM4 interact with proteins such as EWI-2, ICAM-1, VCAM-1 or CD44 which in turn bind to the actin cytoskeleton linkers ezrin–radixin–moesin (ERM) proteins. ERM could also act as a linker between TEMs and the cytoskeleton (Sala-Valdes et al. 2006). Whether TM4 could remain only transiently bound to the actin cytoskeleton (and this binding could be induced upon certain stimuli) is still a matter of investigation. It is noteworthy to mention that, even if TM4 are most of the time related to membrane assemblies, it is likely that isolated molecules have a key role in their cellular functions and single molecule analysis is a prerequisite for better understanding the molecular mechanisms underlying TM4 function.
Model of tetraspanin membrane organization. The scheme represents the dynamic concept of tetraspanin-enriched microdomains (TEMs) obtained in biophysical studies. Tetraspanins (TM4) interact with themselves and their partners at the membrane conforming TEMs, which are supramolecular platforms with variable composition and stoichiometry. These platforms are dynamic, slowly diffusing throughout the membrane and showing a continuous exchange of components (1, bidirectional blue arrows). There are also smaller tetraspanin ensembles which diffuse faster exhibiting a mixture of alternated Brownian and confined motion, i.e., free diffusion in membrane versus retention within platforms (2, coloured traces). Some of these ensembles can coalesce and co-diffuse over time and then diverge (3). Finally, it is also shown the clustering of TEMs into bigger immobilized platforms induced by the binding of tetraspanin partners to ligands and cytoskeleton constraints (4). A multivalent ligand able to bind to several TM4 partners simultaneously is exemplified
Future studies should benefit from recent technological developments in optical fluorescence microscopy that circumvent the diffraction law (Patterson 2009). Among them, super resolution microscopy such PALM (Photo-Activated Localization Microscopy) and STORM (Stochastic Optical Reconstruction Microscopy) should greatly improve our knowledge in the composition of TM4 assemblies. These two technologies that are relatively easy to implement will allow the estimation of the number of molecules per TEMs, their stoichiometry, and their spatial organization in these microdomains with a lateral resolution of 20 nm. Both methods are based on photoswitchable or photoactivable fluorophores. In each imaging cycle, only a fraction of the fluorophores is turned on, allowing their positions to be determined with nanometer accuracy. The fluorophore positions obtained from a series of imaging cycles are used to reconstruct the entire image (Betzig et al. 2006; Rust et al. 2006). With STORM, small fluorescent probes are used and it is easy to perform multicolor experiments that are necessary to make a mapping of several TM4 and partners on the same cell. Interestingly it can now be applied to conventional probes (Heilemann et al. 2008). PALM often required eosFP, a mutant of the Green Fluorescent Protein that can be photoactivated only once. Therefore, it could be used to quantitatively determine the number of a given TM4 within TEMs. Because PALM does not require the fixation of cells, it will be interesting in the future to combine PALM and tracking of activated TM4 proteins in living cells, the so-called sptPALM (Manley et al. 2008). Further development in SMT should also help in deciphering the molecular mechanism underlying the dynamics and partitioning of TM4. Besides developments in the detection of single molecules (new EM-CCD cameras, more stable probes) that will improve time and lateral resolution, tracking in 3D is a crucial step for better understanding the behavior of TM4 because a distinct membrane topology of cells cultured under 3D conditions is likely to influence the dynamic behavior of proteins. More recently, Hell’s group has developed a new optical setup allowing tracking with a very high spatial (20–40 nm) and time (<1 ms) resolution (Sahl et al. 2010). This setup is based on a confocal microscope, where the pinhole is replaced by three separate point detectors arranged in close proximity to obtain the localization of the fluorescent molecule. This technique appears especially well-suited for probing membrane components on a very large time scale. Finally, FRET at the single molecule level in living cells, which is not still available, represents another attractive way to probe direct interaction between two proteins within TEMs.
Abbreviations
- Chl:
-
Cholesterol
- FCCS:
-
Fluorescence Cross-Correlation Spectroscopy
- FCS:
-
Fluorescence Correlation Spectroscopy
- FRAP:
-
Fluorescent Recovery After Photobleaching
- FRET-FLIM:
-
Fluorescence Resonance Energy Transfer-Fluorescence Lifetime Imaging Microscopy
- PALM:
-
Photo-Activated Localization Microscopy
- SMT:
-
Single Molecule Tracking
- STED:
-
Stimulated Emission Depletion
- STORM:
-
Stochastic Optical Reconstruction Microscopy
- TEM:
-
Tetraspanin-Enriched Microdomain
References
Abitorabi MA, Pachynski RK, Ferrando RE, Tidswell M, Erle DJ (1997) Presentation of integrins on leukocyte microvilli: a role for the extracellular domain in determining membrane localization. J Cell Biol 139(2):563–571. doi:10.1083/jcb.139.2.563
Barreiro O, Yanez-Mo M, Serrador JM, Montoya MC, Vicente-Manzanares M, Tejedor R, Furthmayr H, Sanchez-Madrid F (2002) Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J Cell Biol 157(7):1233–1245. doi:10.1083/jcb.200112126
Barreiro O, Yanez-Mo M, Sala-Valdes M, Gutierrez-Lopez MD, Ovalle S, Higginbottom A, Monk PN, Cabanas C, Sanchez-Madrid F (2005) Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood 105(7):2852–2861. doi:10.1182/blood-2004-09-3606
Barreiro O, de la Fuente H, Mittelbrunn M, Sanchez-Madrid F (2007) Functional insights on the polarized redistribution of leukocyte integrins and their ligands during leukocyte migration and immune interactions. Immunol Rev 218:147–164. doi:10.1111/j.1600-065X.2007.00529
Barreiro O, Zamai M, Yanez-Mo M, Tejera E, Lopez-Romero P, Monk PN, Gratton E, Caiolfa VR, Sanchez-Madrid F (2008) Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol 183(3):527–542. doi:10.1083/jcb.200805076
Barreiro O, Aguilar RJ, Tejera E, Megias D, de Torres-Alba F, Evangelista A, Sanchez-Madrid F (2009) Specific targeting of human inflamed endothelium and in situ vascular tissue transfection by the use of ultrasound contrast agents. JACC Cardiovasc Imag 2(8):997–1005. doi:10.1016/j.jcmg.2009.04.012
Berditchevski F, Odintsova E (1999) Characterization of integrin-tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J Cell Biol 146(2):477–492. doi:10.1083/jcb.146.2.477
Berditchevski F, Odintsova E (2007) Tetraspanins as regulators of protein trafficking. Traffic 8(2):89–96. doi:10.1111/j.1600-0854.2006.00515.x
Berditchevski F, Odintsova E, Sawada S, Gilbert E (2002) Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J Biol Chem 277(40):36991–37000
Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313(5793):1642–1645. doi:10.1126/science.1127344
Boucheix C, Rubinstein E (2001) Tetraspanins. Cell Mol Life Sci 58:1189–1205
Brown DA, London E (2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem 275(23):17221–17224. doi:10.1074/jbc.R000005200
Carman CV, Springer TA (2003) Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr Opin Cell Biol 15(5):547–556. doi:S0955067403001121 [pii]
Charrin S, Manie S, Oualid M, Billard M, Boucheix C, Rubinstein E (2002) Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett 516:139–144. doi:S001457930202522X [pii]
Charrin S, Manie S, Billard M, Ashman L, Gerlier D, Boucheix C, Rubinstein E (2003a) Multiple levels of interactions within the tetraspanin web. Biochem Biophys Res Commun 304(1):107–112. doi:S0006291X0300545X [pii]
Charrin S, Manie S, Thiele C, Billard M, Gerlier D, Boucheix C, Rubinstein E (2003b) A physical and functional link between cholesterol and tetraspanins. Eur J Immunol 33:2479–2489. doi:10.1002/eji.200323884
Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E (2009) Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J 420(2):133–154. doi:10.1042/BJ20082422
Chen Y, Muller JD, Eid JS, Gratton E (2001) Two-photon fluorescence fluctuation spectroscopy. In: Valeur B, Brochon JC (eds) New trends in fluorescence spectroscopy. Springer, Berlin, pp 277–296
Danglot L, Chaineau M, Dahan M, Gendron MC, Boggetto N, Perez F, Galli T (2010) Role of TI-VAMP and CD82 in EGFR cell-surface dynamics and signaling. J Cell Sci 123(Pt 5):723–735. doi:10.1242/jcs.062497
Das R, Cairo CW, Coombs D (2009) A hidden Markov model for single particle tracks quantifies dynamic interactions between LFA-1 and the actin cytoskeleton. PLoS Comput Biol 5(11):e1000556. doi:10.1371/journal.pcbi.1000556
Dehmelt L, Bastiaens PI (2010) Spatial organization of intracellular communication: insights from imaging. Nat Rev Mol Cell Biol 11(6):440–452. doi:10.1038/nrm2903
Delaguillaumie A, Harriague J, Kohanna S, Bismuth G, Rubinstein E, Seigneuret M, Conjeaud H (2004) Tetraspanin CD82 controls the association of cholesterol-dependent microdomains with the actin cytoskeleton in T lymphocytes: relevance to co-stimulation. J Cell Sci 117(Pt 22):5269–5282. doi:10.1242/jcs.01380
Digman MA, Gratton E (2009) Analysis of diffusion and binding in cells using the RICS approach. Microsc Res Tech 72(4):323–332. doi:10.1002/jemt.20655
Drummer HE, Wilson KA, Poumbourios P (2005) Determinants of CD81 dimerization and interaction with hepatitis C virus glycoprotein E2. Biochem Biophys Res Commun 328(1):251–257. doi:10.1016/j.bbrc.2004.12.160
Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW (2009) Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457(7233):1159–1162. doi:10.1038/nature07596
Espenel C, Margeat E, Dosset P, Arduise C, Le Grimellec C, Royer CA, Boucheix C, Rubinstein E, Milhiet PE (2008) Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J Cell Biol 182(4):765–776. doi:10.1083/jcb.200803010
Feigelson SW, Grabovsky V, Shamri R, Levy S, Alon R (2003) The CD81 tetraspanin facilitates instantaneous leukocyte VLA-4 adhesion strengthening to vascular cell adhesion molecule 1 (VCAM-1) under shear flow. J Biol Chem 278(51):51203–51212. doi:10.1074/jbc.M303601200
Garcia-Saez AJ, Schwille P (2008) Fluorescence correlation spectroscopy for the study of membrane dynamics and protein/lipid interactions. Methods 46(2):116–122. doi:10.1016/j.ymeth.2008.06.011
Hansen CG, Nichols BJ (2010) Exploring the caves: cavins, caveolins and caveolae. Trends Cell Biol 20(4):177–186. doi:10.1016/j.tcb.2010.01.005
Heilemann M, Van De Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M, 33 (2008) Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed 47(33):6172–6176, doi:10.1002/anie.v47:33
Hemler ME (2003) Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol 19:397–422. doi:10.1146/annurev.cellbio.19.111301.153609
Hemler ME (2005) Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6(10):801–811. doi:10.1038/nrm1736
Jacobson K, Mouritsen OG, Anderson RG (2007) Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol 9(1):7–14. doi:10.1038/ncb0107-7
Krementsov DN, Rassam P, Margeat E, Roy NH, Schneider-Schaulies J, Milhiet PE, Thali M (2010) HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 11(11):1401–1414. doi:10.1111/j.1600-0854.2010.01111.x
Kusumi A, Sako Y, Yamamoto N (1993) Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J 65(5):2021–2040. doi: 0006-3495/93/11/2021/20
Lasserre R, Guo XJ, Conchonaud F, Hamon Y, Hawchar O, Bernard AM, Soudja SM, Lenne PF, Rigneault H, Olive D, Bismuth G, Nunes JA, Payrastre B, Marguet D, He HT (2008) Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat Chem Biol 4(9):538–547. doi:10.1038/nchembio.103
Le Naour F, Andre M, Boucheix C, Rubinstein E (2006) Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics 6(24):6447–6454. doi:10.1002/pmic.200600282
Lingwood D, Simons K (2010) Lipid rafts as a membrane-organizing principle. Science 327(5961):46–50. doi:10.1126/science.1174621
Luo BH, Carman CV, Springer TA (2007) Structural basis of integrin regulation and signaling. Annu Rev Immunol 25:619–647. doi:10.1146/annurev.immunol.25.022106.141618
Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J (2008) High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Meth 5(2):155–157. doi:10.1038/nmeth.1176
Margeat E, Kapanidis AN, Tinnefeld P, Wang Y, Mukhopadhyay J, Ebright RH, Weiss S (2006) Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys J 90(4):1419–1431
Marguet D, Lenne PF, Rigneault H, He HT (2006) Dynamics in the plasma membrane: how to combine fluidity and order. EMBO J 25(15):3446–3457. doi:10.1038/sj.emboj.7601204
Mayor S, Riezman H (2004) Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol 5(2):110–120. doi:10.1038/nrm1309
Mittelbrunn M, Yanez-Mo M, Sancho D, Ursa A, Sanchez-Madrid F (2002) Cutting edge: dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APC. J Immunol 169(12):6691–6695
Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M (2006) Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol 173(5):795–807. doi:10.1083/jcb.200508165
Odintsova E, Butters TD, Monti E, Sprong H, van Meer G, Berditchevski F (2006) Gangliosides play an important role in the organization of CD82-enriched microdomains. Biochem J 400(2):315–325
Parton RG, Hancock JF (2004) Lipid rafts and plasma membrane microorganization: insights from Ras. Trends Cell Biol 14(3):141–147. doi:10.1016/j.tcb.2004.02.001
Patterson G (2009) Fluorescence microscopy below the diffraction limit. Semin Cell Dev Biol. doi:10.1016/j.semcdb.2009.08.006
Pinaud F, Clarke S, Sittner A, Dahan M (2010) Probing cellular events, one quantum dot at a time. Nat Meth 7(4):275–285. doi:10.1038/nmeth.1444
Regina Todeschini A, Hakomori SI (2008) Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim Biophys Acta 1780(3):421–433
Ries J, Schwille P (2006) Studying slow membrane dynamics with continuous wave scanning fluorescence correlation spectroscopy. Biophys J 91(5):1915–1924. doi:10.1529/biophysj.106.082297
Rubinstein E, Ziyyat A, Wolf JP, Le Naour F, Boucheix C (2006) The molecular players of sperm-egg fusion in mammals. Semin Cell Dev Biol 17(2):254–263. doi:10.1016/j.semcdb.2006.02.012
Rust MJ, Bates M, Zhuang X (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Meth 3(10):793–796. doi:10.1038/nmeth929
Sahl SJ, Leutenegger M, Hilbert M, Hell SW, Eggeling C (2010) Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc Natl Acad Sci USA 107(15):6829–6834. doi:10.1073/pnas.0912894107
Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sanchez-Madrid F, Yanez-Mo M (2006) EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem 281(28):19665–19675. doi:10.1074/jbc.M602116200
Saxton MJ, Jacobson K (1997) Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct 26:373–399. doi:10.1146/annurev.biophys.26.1.373
Schwille P, Korlach J, Webb WW (1999) Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry 36(3):176–182. doi:10.1002/(SICI)1097-0320(19990701)36:3<176::AID-CYTO5>3.0.CO;2-F
Sergé A, Bertaux N, Rigneault H, Marguet D (2008) Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Meth 5(8):687–694. doi:10.1038/nmeth.1233
Silvie O, Charrin S, Billard M, Franetich JF, Clark KL, van Gemert GJ, Sauerwein RW, Dautry F, Boucheix C, Mazier D, Rubinstein E (2006) Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J Cell Sci 119(Pt 10):1992–2002. doi:10.1242/jcs.02911
Stipp CS (2010) Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med 12:e3. doi:10.1017/S1462399409001355
Suzuki KG, Fujiwara TK, Edidin M, Kusumi A (2007) Dynamic recruitment of phospholipase C gamma at transiently immobilized GPI-anchored receptor clusters induces IP3-Ca2+ signaling: single-molecule tracking study 2. J Cell Biol 177(4):731–742. doi:10.1083/jcb.200609175
Thali M (2009) The roles of tetraspanins in HIV-1 replication. Curr Top Microbiol Immunol 339:85–102. doi:10.1007/978-3-642-02175-6_5
Tham TN, Gouin E, Rubinstein E, Boucheix C, Cossart P, Pizarro-Cerda J (2010) Tetraspanin CD81 is required for Listeria monocytogenes invasion. Infect Immun 78(1):204–209. doi:10.1128/IAI.00661-09
Unternaehrer JJ, Chow A, Pypaert M, Inaba K, Mellman I (2007) The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc Natl Acad Sci USA 104(1):234–239. doi:10.1073/pnas.0609665104
VanCompernolle SE, Levy S, Todd SC (2001) Anti-CD81 activates LFA-1 on T cells and promotes T cell-B cell collaboration. Eur J Immunol 31(3):823–831. doi:10.1002/1521-4141(200103)31:3<823::AID-IMMU823>3.0.CO;2-D
Wawrezinieck L, Rigneault H, Marguet D, Lenne PF (2005) Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J 89(6):4029–4042. doi:10.1529/biophysj.105.067959
Yanez-Mo M, Barreiro O, Gonzalo P, Batista A, Megias D, Genis L, Sachs N, Sala-Valdes M, Alonso MA, Montoya MC, Sonnenberg A, Arroyo AG, Sanchez-Madrid F (2008) MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood 112(8):3217–3226. doi:10.1182/blood-2008-02-139394
Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F (2009) Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 19(9):434–446. doi:10.1016/j.tcb.2009.06.004
Ziyyat A, Rubinstein E, Monier-Gavelle F, Barraud V, Kulski O, Prenant M, Boucheix C, Bomsel M, Wolf JP (2006) CD9 controls the formation of clusters that contain tetraspanins and the integrin alpha 6 beta 1, which are involved in human and mouse gamete fusion. J Cell Sci 119(Pt 3):416–424. doi:10.1242/jcs.02730
Acknowledgements
We would like to thank Dr. Moreno Zamai for participating in data acquisition and analysis of Fig. 4.1. This work was supported by the Spanish Ministry of Science and Innovation (SAF2008-06235); and MEICA from Genoma España Foundation, INSINET from Comunidad Autónoma de Madrid [FONCICYT-C002-2009-1ALA/127249], and RECAVA [RD06/0014-0030] from Instituto de Salud Carlos III to F.S-M. This work was also supported by the Association Nationale pour la Recherche (ANR-06-BLAN-0378) to P.E.M. O.B. is a post-doctoral fellow from Spanish Ministry of Science and Innovation ‘Juan de la Cierva Program’(JDC 08). Centro Nacional de Investigaciones Cardiovasculares is supported by the Ministry of Science and Innovation and the Pro CNIC Foundation.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Barreiro, O., Sanchez-Madrid, F., Espenel, C., Milhiet, PE. (2013). Dynamic Partitioning of Tetraspanins Within Plasma Membranes. In: Berditchevski, F., Rubinstein, E. (eds) Tetraspanins. Proteins and Cell Regulation, vol 9. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-6070-7_4
Download citation
DOI: https://doi.org/10.1007/978-94-007-6070-7_4
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-6069-1
Online ISBN: 978-94-007-6070-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)