Abstract
Upon cell infection, some viruses integrate their genome into the host chromosome, either as part of their life cycle (such as retroviruses), or incidentally. While possibly promoting long-term persistence of the virus into the cell, viral genome integration may also lead to drastic consequences for the host cell, including gene disruption, insertional mutagenesis and cell death, as well as contributing to species evolution. This review summarizes the current knowledge on viruses integrating their genome into the host genome and the consequences for the host cell.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Long Terminal Repeat
- Viral Integration
- Host Chromatin
- Viral Genome Integration
- Integration Site Selection
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Upon host infection, viruses hijack multiple cellular functions in order to promote their replication and favor viral particle progeny. To ensure this, some viruses evolved the ability to integrate their genome into the host chromosomes, yielding to various consequences for the host cell, including gene disruption, oncogenesis or premature cell death, and may ultimately contribute to species evolution through inheritable genome inclusions. Although viral genome integration into the host genome is an obligatory step for viruses such as retroviruses, it may also occur incidentally for some other viruses (Table 1). This review will summarize the current knowledge on viruses integrating into the host genome and the consequences for the host cell.
2 RNA Viruses
By definition, RNA viruses are not able to integrate their genome into the host chromosome, as their genetic information resides in RNA molecules and not DNA. The only exception to this are retroviruses, which are characterized by the reverse transcription of their viral RNA genome into a linear double-stranded DNA molecule (viral DNA intermediate), and thus the substrate for subsequent viral genome integration into the host genome. For retroviruses, integration is a mandatory step for productive infection. Apart from retroviruses, the genome of other RNA viruses has been recently identified in the host genome. However, in these cases, integration seems to have occurred incidentally, as demonstrated for lymphocytic choriomeningitis virus (LCMV), an arenavirus. This section will cover the integration process of retroviruses including endogenous retroviruses and the incidental integration of LCMV.
2.1 Retroviruses
The life cycle of retroviruses, including the prototypic and well studied human immunodeficiency virus type 1 (HIV-1), can be divided in several crucial steps (Fig. 1a): viral entry through host cell-specific receptors dictating viral tropism, core penetration, uncoating, reverse transcription of the viral RNA genome, nuclear translocation and integration of the viral cDNA genome into the host chromosomes, transcription of the integrated provirus*, translation, virion assembly, budding and release (Friedrich et al. 2011).
Integration is a mandatory step of productive retroviral infection. (a) Overview of the HIV-1 life cyle. (See text for details). (b) Viral integration mechanism is divided in three essential steps: (1) 3′ processing, (2) strand transfer, and (3) gap filling. IN: integrase ( yellow oval ). LTR: long terminal repeats. Filled red and green circles indicate 5′ phosphate and 3′OH ends respectively. Arrows indicate the actions performed by the host DNA repair machinery. Black arrows: cleavage of 5′ protruding viral ends. Grey arrows: gap filling of single-strand DNA. (See text for details)
Viral genome integration into the host genome is a hallmark of retroviruses, as it is a mandatory step in the retroviral life cycle and a prerequisite for productive infection. Upon integration, the retrovirus will persist in the infected cell for its entire lifespan, and will affect host gene expression depending on the integration site. Furthermore, if retroviral infection and integration occurs in the germline, the provirus will be transmitted to the progeny, and will thus contribute shaping the genome of future generations. This is the case of the so called “endogenized” retroviruses or endogenous retroviruses (ERV).
2.1.1 Integration Mechanism
After completion of reverse transcription, the linear double-stranded cDNA flanked by the long terminal repeats (LTR) is part of a nucleoprotein complex called preintegration complex (PIC). The PIC contains multiple viral and cellular proteins – including in the case of HIV-1: viral integrase (IN), matrix (MA), Vpr, and cellular barrier-to-autointegration factor (BAF), high-mobility group chromosomal protein A1 (HMGA1), integrase interactor 1 (Ini1), lens epithelium-derived growth factor (LEDGF/p75) – that may contribute to nuclear translocation, integration of the viral genome, and subsequent immediate transcription, and which composition may vary along the way to the host genome (Belshan et al. 2009; Farnet and Haseltine 1991; Fassati and Goff 2001; Lin and Engelman 2003; Miller et al. 1997; Raghavendra et al. 2010). To cross the nuclear membrane and reach the nucleus, retroviruses have evolved different strategies. Simple retroviruses (alpharetroviruses, betaretroviruses, gammaretroviruses and epsilonretroviruses) are able to reach the nucleus only upon nuclear membrane disruption occurring at the time of mitosis, providing a coherent explanation on why these retroviruses infect dividing cells but are unable to infect non-dividing cells (Lewis and Emerman 1994; Roe et al. 1993). In contrast, spumaviruses and lentiviruses have the capacity to infect both dividing and non-dividing cells, entering the nucleus through an active, yet poorly elucidated, mechanism (Suzuki and Craigie 2007). The current model for HIV-1 proposes that a PIC containing minimally the viral integrase and the viral cDNA crosses the nuclear membrane through the nuclear pore complex (NPC), a superstructure mediating the transport of macromolecules between the cytoplasm and the nucleus, via specific interactions with NPC proteins, including importin α3, importin 7, NUP153*, RANBP2* and Transportin-SR2/TNPO3 (Ao et al. 2010; Christ et al. 2008; Levin et al. 2010; Ocwieja et al. 2011; Woodward et al. 2009).
Retroviral genome integration occurs in three steps, the first two being catalyzed by the retroviral integrase (IN) protein (Fig. 1b, the example of HIV-1) (Li et al. 2011). IN is bound to the LTR and requires approximately the 32 terminal nucleotides (Bera et al. 2009). First, when the PIC is still in the cytoplasm (Miller et al. 1997), IN hydrolyzes a dinucleotide at each 3′ end, a process called 3′ processing. Second, IN catalyzes the strand transfer reaction, consisting in simultaneously breaking the host DNA asymmetrically and joining it to the recessed viral DNA 3′-OH ends. The IN-mediated asymmetric DNA breaks in the host genome are determined by the retroviral protein structure and vary between 4 and 6 nucleotides (5 in the case of HIV-1). Finally, to stabilize the proviral insertion, the host DNA repair machinery – involving the DNA-dependent kinase (DNA-PK) comprising a DNA-PK catalytic subunit and a DNA binding Ku80/Ku70 complex, and the ligase IV/XRRC4 complex of the non-homologous end joining pathway (NHEJ) – cleaves the viral protruding 5′ nucleotides and fills in the 4–6 bp gap, resulting in the duplication of the gap nucleotide sequence surrounding the provirus.
The retroviral IN enzyme belongs to the family of polynucleotidyl transferases. It contains between 280 and 450 amino acids depending on the retrovirus (for example, HIV-1 IN: 288 amino acids), that are divided in three protein domains (Li et al. 2011). The N-terminal domain (residues 1–50 in HIV-1 IN), containing an HHCC zinc-binding motif, is involved mostly in viral DNA binding, and IN multimerization. The C-terminal domain (residues 212–288 in HIV-1 IN) is also involved in DNA binding and IN multimerization. And most importantly, the catalytic core domain (residues 50–212 in HIV-1 IN), carrying a typical signature with the D,D(35)E acidic triad in the active site, is essential for metal (Mg2+) binding and IN enzymatic activity, and is involved in viral DNA binding as well as host cellular target DNA binding. The catalytic core domain has also been shown to contribute to IN multimerization.
In vitro, purified recombinant IN alone is able to perform 3′ processing and strand transfer. Initial experiments showed that IN was able to catalyze half site integration (one LTR end integrated in the acceptor DNA) using 21-mer oligonucleotides mimicking the U3 or U5 ends of the LTR. However, the use of longer DNA substrates mixed with IN allowed to reconstitute concerted full-site integration (integration of both LTR ends) (Sinha and Grandgenett 2005; Sinha et al. 2002), thereby mimicking the in vivo situation more faithfully and suggesting that other genomic regions in addition to LTR extremities contribute to integration efficiency (Li and Craigie 2005). Although IN is sufficient to perform the first two steps of integration in vitro, multiple PIC components, including LEDGF/p75, were shown to improve the efficiency of this process, both in vitro and in vivo (Van Maele et al. 2006).
The current and commonly accepted model, supported by crystallography, implies that IN activity is linked to its oligomeric state: IN dimers bound to LTR termini catalyze the 3′ processing whereas concerted integration requires IN tetramers (Cherepanov et al. 2011; Delelis et al. 2007; Diamond and Bushman 2005; Faure et al. 2005; Guiot et al. 2006; Hare et al. 2010; Jaskolski et al. 2009).
2.1.2 Integration Site Selection
As mentioned in the previous section, purified IN alone is able to catalyze the first two steps of integration in vitro at any phosphodiester bond of the DNA target, suggesting that IN does not have any DNA sequence preference at the level of the DNA recipient molecule.
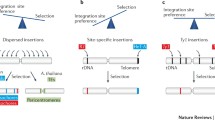
However, a pioneering study by Schroder et al. took advantage of the published human genome sequence and showed that in vivo, the sites of HIV-1 integration were not random but rather favored specific chromosomal features, such as transcription units (Schroder et al. 2002). Since then and thanks to the development of high-throughput sequencing technologies and the availability of the genomic sequence of multiple species, a more complete picture of retroviral integration preferences emerged (Fig. 2a) (Bushman et al. 2005; Ciuffi and Bushman 2006; Lewinski et al. 2005; Lewinski and Bushman 2005; Delelis et al. 2010; Desfarges and Ciuffi 2010).
Retroviral integration site distribution. (a) Host chromosomal preferences in integration site selection diverge among retroviral genera. (+, blue arrows) Gammaretroviruses (MLV) favors integration in promoters and in CpG islands, close to transcription start sites (TSS). (◈, red arrows) Lentiviruses (HIV-1) integrate preferentially into active transcription units. (✯, green arrows) Betaretroviruses (MMTV) integrate randomly. (b) Schematic overview of the tethering model for HIV-1 (left) and MLV (right) (See text for details)
All retroviruses do not display the same integration site preferences. Indeed gammaretroviruses, spumaretroviruses and endogenous retroviruses favor promoters and transcription start sites of active genes, characterized by high CpG islands and DNaseI hypersensitive sites (Mitchell et al. 2004; Wu et al. 2003; Trobridge et al. 2006; Brady et al. 2009; Kim et al. 2008, 2011). Integration of alpharetroviruses and deltaretrovirues is also, although weakly, favored in transcription units and CpG islands (Derse et al. 2007; Mitchell et al. 2004). In contrast, lentiviruses prefer integrating in active genes, along the transcription unit, in both introns and exons, and are often associated with epigenetic marks characterizing active transcription, including H3Ac, H4Ac, H3K4me3, H3K36me3, while disfavoring epigenetic marks associated with repressed transcription such as H3K9me3, H3K27me3, H3K79me3, H4K20me3 and DNA methylation (Brady et al. 2011; Derse et al. 2007; Mitchell et al. 2004; Roth et al. 2011; Schroder et al. 2002; Wang et al. 2007, 2009). Finally, the MMTV betaretrovirus is the only one considered to integrate randomly, with no statistically significant preference for chromosomal features (Faschinger et al. 2008), nevertheless some common integration sites near cellular oncogenes belonging to Wnt and Fgf families have been reported (Callahan and Smith 2000, 2008).
Although no DNA consensus sequence was identified in vitro, a weak DNA consensus appears in vivo at the host insertion site as well as surrounding the integration site. Furthermore, in the case of HIV-1, a specific nucleosomal DNA architecture, i.e. the outward-facing major groove of the target DNA (possibly consistent with the weak consensus DNA sequence), is favored for integration, presumably due to IN protein structure constraints (Wang et al. 2007).
To date, many hypotheses have been imagined to explain this retroviral-specific integration site selection, including the role of cell cycle, chromatin accessibility and tethering proteins. Although all these models may contribute to integration site selection, only evidence for the tethering model has been identified so far (Fig. 2b). This model suggests that integration site selection is dictated by a protein, directly or indirectly complexed with the retroviral-specific IN, and acting as a tethering protein between the PIC and the host chromatin, thereby promoting integration at a nearby DNA site (Bushman et al. 2005; Ciuffi and Bushman 2006; Desfarges and Ciuffi 2010). Therefore, any PIC component could potentially act as a tethering protein.
Three major lines of evidence argue in favor of this tethering model. The first one takes advantage of chimeric constructs between MLV and HIV-1, and the subsequent analysis of integration site distribution (Lewinski et al. 2006). Swaps between HIV-1 and MLV at the level of Gag and IN highlighted the role of these two viral proteins as major determinants for integration targeting. Indeed, HIV-1 vector containing MLV Gag only displayed specific integration preferences that differed from both HIV-1 and MLV and suggesting that Gag may play a role in integration site selection. In contrast, HIV-1 vector containing MLV IN lost integration preferences for transcription units and acquired preferences for transcription start sites close to MLV phenotype, suggesting that HIV-1 IN is the major determinant for HIV-1 integration site selection. However, an HIV-1 vector containing both MLV Gag and MLV IN preferentially integrated into transcription start sites, completely recapitulating MLV integration site distribution, thereby suggesting that in the case of MLV, both Gag and IN are likely to be major viral determinants of integration site selection.
The second line of evidence resides in the identification of the HIV-1 IN-interacting protein, LEDGF/p75, that was shown to play a key role in integration efficiency as well as integration site distribution (Cherepanov et al. 2005a, b; Ciuffi et al. 2005; Engelman and Cherepanov 2008; Llano et al. 2006; Marshall et al. 2007; Poeschla 2008), thereby providing the proof-of-concept that LEDGF/p75 is acting as the major tethering protein for the HIV-1 PIC. Indeed, cells depleted for LEDGF/p75 do not favor transcription units anymore but rather CpG islands (Ciuffi et al. 2005; Marshall et al. 2007; Schrijvers et al. 2012; Shun et al. 2007). LEDGF/p75 is required for efficient integration and site selection, not only for HIV-1, but for many lentiviruses (SIV, EIAV) (Busschots et al. 2007; Cherepanov 2007; Marshall et al. 2007). In contrast, integration site selection of other retroviruses, such as MLV (a gammaretrovirus), is not affected by LEDGF/p75 depletion, providing additional evidence that LEDGF/p75 is the major tethering factor for lentiviruses only. Of note, Schrijvers et al. recently demonstrated that, in absence of LEDGF/p75, hepatoma-derived growth factor related protein 2 (HRP2) was acting as an alternative tethering protein for HIV-1 PIC, although less efficient than LEDGF/p75 (Schrijvers et al. 2012). Except for Foamy virus (FV), for which H2A/H2B heterodimers were shown to interact with FV Gag, thus tethering FV PIC to chromatin (Tobaly-Tapiero et al. 2008), specific tethering proteins for other retroviral genera remains to be identified.
The third line of evidence originates from experiments using LEDGF/p75 chimera, in which the chromatin binding domain of LEDGF/p75 was substituted with the one of other chromatin binding proteins, including the phage λ repressor protein, H1 histone, KSHV latency-associated nuclear antigen, heterochromatin protein 1-α, inhibitor of growth protein 2 and heterochromatin protein 1-β (Ciuffi et al. 2006; Ferris et al. 2010; Gijsbers et al. 2010, 2011; Meehan and Poeschla 2010; Meehan et al. 2009; Silvers et al. 2010). All these studies showed that, by changing the chromatin binding of LEDGF/p75, integration site selection can be redirected from transcription units to alternative preferential host chromatin sites, dictated by the chromatin binding specificity of the chimeric protein, These data confirm the role of LEDGF/p75 in HIV-1 integration site selection and suggest that integration targeting can be modulated, a feature of great interest for gene therapy studies involving retroviral-based vectors.
Although tethering appears so far to be a major mechanism involved in integration site selection, recent studies demonstrated that integration targeting could also be affected by nuclear import. Indeed, it has been shown that depletion of nuclear pore proteins, such as Transportin-SR2/TNPO3 or resulted in the reduction of HIV-1 integration events in gene dense regions, but has no effect on MLV integration distribution (consistent with the concept that MLV does not enter the nucleus through the nuclear pore). These data provide evidence of a functional coupling between HIV-1 nuclear import and integration, implying a role for proper nuclear trafficking of HIV-1 complexes in integration site distribution (Ocwieja et al. 2011; Schaller et al. 2011).
2.2 Incidental Integration of Non-retroviral RNA Viruses
As mentioned at the beginning of this section, RNA viruses normally do not integrate. However, the genomic sequence of lymphocytic choriomeningitis virus (LCMV), an arenavirus, has been identified in genome of infected mice and is seemingly the result of an incidental event that will be described hereafter.
Arenaviruses are the etiologic agents of hemorrhagic fever disease in humans. Arenaviruses are enveloped viruses containing a bisegmented negative single stranded RNA genome coding for four viral proteins: an RNA-dependent RNA polymerase, the nucleocapsid, the glycoprotein and a RING-domain containing protein. The replication of arenaviruses is completely different from retroviruses, with a broader cell tropism (Emonet et al. 2011). Viral replication takes place exclusively in the cytoplasm in which RNA synthesis is performed by the virally encoded RNA-dependent RNA polymerase (RdRp). Although RdRps belong to the reverse transcriptase-like superfamily, no reverse transcriptase activity has been detected so far. Therefore, these viruses normally do not integrate into the host chromosomes. However, studies aiming at characterizing LCMV persistence in infected mice were able to detect LCMV DNA sequences by PCR in ∼60% of mice 200 days post-infection (long after LCMV blood clearance), at a frequency of about 1 LCMV DNA copy in 104–105 splenocytes (Klenerman et al. 1997). LCMV DNA was also detected in murine and hamster cell lines (which are considered as the natural hosts for LCMV), but not in non-natural host cell lines (human, monkey, dog, cow). Further analysis highlighted a role for retrotransposons*, encoding a reverse transcriptase (RT), in the generation of LCMV DNA and subsequent integration. Interestingly, murine and hamster cells display a high level of endogenous RT activity, consistent, in part, with the natural host restriction observed. Recently, Geuking et al. showed that RT from endogenous retrotransposons can illegitimately recombine with the exogenous LCMV RNA genome by template switching, providing additional data pointing towards the role of retrotransposons in reverse transcribing and integrating LCMV genomic sequences (Geuking et al. 2009).
Totiviridae and Partitiviridae are superfamilies containing a broad range of RNA viruses infecting fungi, protozoa, nematods, arthropods and plants. Similarly to arenaviruses, neither reverse transcriptase activity, nor integration activity have been reported for these viruses. However, sequences of the capsid and the RdRp genes have been identified in many eukaryotic genomes, suggesting that integration of these viral sequences can occur more frequently as initially expected (Liu et al. 2010). Based on these observations, the question remains: how can these viral sequences integrate in the host genome? Liu and coworkers proposed two models (Liu et al. 2010): (i) an illegitimate and incidental recombination with retrotransposons may occur, leading to the integration of viral sequences, as described for LCMV (Geuking et al. 2009; Tanne and Sela 2005) or (ii) the double-strand-break repair machinery of the host cell may capture nearby viral DNA sequences and insert them in some instable regions of the genome, as described in yeast (Frank and Wolfe 2009; Puchta 2005). Although both models can each contribute, only the first model enacting a role for retrotransposons can explain the prior appearance of a viral DNA intermediate, essential for being considered as a substrate of host genome insertion.
3 DNA Viruses
Unlike RNA viruses, the genome of DNA viruses is already a potential substrate for host genome integration, without the need for prior processing. In general, the genome of DNA viruses is translocated to the nucleus, where it remains as an episome to ensure viral persistence. However, the genome of some DNA viruses can be found inserted in the host genome. The mechanisms underlying these integration events, incidental or non-incidental, are still poorly characterized, and the potential advantages for these DNA viruses to integrate are still obscure. Understanding these mechanisms should help elucidate the role of DNA virus integration in the viral life cycle. This section will summarize the current knowledge on integration of some prototypic DNA viruses as well as highlighting some mechanisms involved in this process.
3.1 Adeno-Associated Virus Type 2 (AAV-2)
The adeno-associated virus (AAV) is a widespread virus classified among the parvoviridae family. The relationship between AAV and the host remains obscure due partially to the absence of associated pathology. Replication of AAV is strictly conditioned by the presence in the same infected cell of helper viruses such as adenoviruses (Ad), human papillomaviruses (HPV) or herpes simplex viruses (HSV). In absence of helper viruses, AAV integrates its genome in a site-specific way. The molecular mechanism involved in AAV integration has only been investigated for the type 2 serotype (AAV-2). The genome organization of AAV-2 consists of two major open reading frames coding for the non-structural proteins Rep (Rep78, Rep68, Rep52 and Rep40) and structural proteins Cap (VP1, VP2 and VP3), flanked by inverted terminal repeats (ITR). The site-specific integration of AAV-2 is located in a non-repetitive element at the position 19q13,42 corresponding to the long arm of the chromosome 19, in a gene-dense region named AAVS1 (for AAV integration site 1) (Fig. 3a) (Kotin et al. 1991). Analysis of AAVS1 host sequence revealed two cis-acting sequences involved in AAV-2 integration: the terminal resolution site (TRS) corresponding to the Rep-specific endonuclease site and the Rep binding site (RBS) (Brister and Muzyczka 1999; McCarty et al. 1994a, b). Interestingly, this TRS-RBS motif is also present in the ITR of the viral genome, suggesting that the sequence homology between AAV-2 ITR and the host genome site – TRS and RBS sequences – plays a role in AAV-2 integration. Recently, two new AAV-2 integration sites have been reported in chromosomes 5 (5p13.3) and 3 (3p24.3), named AAVS2 and AAVS3 respectively, that also carry a RBS motif (Hüser et al. 2010).
Integration site distribution of DNA viruses. (a) Host chromosomal preferences in integration site selection of some DNA viruses. (+, brown arrows) MDV/HHV-6 viruses favor integration in telomeres. (▲, red arrow) AAV-2 integrates preferentially at the AAVS1 site, (◈, purple arrows) Ad integrates preferentially in gene loci, (✯, orange arrows) EBV integrates in heterochromatin. (b) Schematic overview of the integration mechanism potentially involved in some DNA viruses, AAV, EBV, KHSV and Ad (from left to right). TRS terminal resolution site, RBS Rep binding site, ITR inverted terminal repeat, oriP origin of replication, HMGB2 high mobility group protein 2, MeCP2 methyl-CpG-binding protein 2, MBD methyl-CpG-binding domain, SYREC symmetric recombinant, NHEJ non homologous end joining repair machinery, HR homologous recombination repair machinery (See text for details)
Biochemical characterization of the proteins Rep68 and Rep78 revealed several activities, including DNA binding, ATPase, helicase and endonuclease activities, essential to direct site-specific integration of AAV-2 genome (Surosky et al. 1997). All together, these data point to a molecular model of AAV-2 integration in which the viral genome is tethered to a specific AAVS locus via concomitant binding of Rep68/78 on both cellular and viral RBS (Weitzman et al. 1994). More particularly, AAV-2 integration starts with Rep68/Rep78 complex introducing a nick at the adjacent cellular TRS that may induce the non homologous end-joining pathway (NHEJ) repair machinery. Non homologous recombination between the viral ITRs and the host DNA results in the viral insertion of AAV-2 in the host genome and the partial duplication of the integration site (Henckaerts and Linden 2010; Lamartina et al. 2000; Urcelay et al. 1995).
In conclusion, AAV long persistence, the absence of pathogenicity and the site-specific integration at AAVS loci render AAV a very attractive candidate for gene therapy. However, to date, nothing is known about the long-term effect of AAV integration at the AAVS locus, which is close to a gene-dense region, containing among others the myosin light chain phosphatase MBS85, an enzyme important for smooth muscle contraction.
3.2 Herpes Viruses
Herpes viruses are DNA enveloped viruses, classified in three families based on their sequence phylogeny: α, β and γ herpes viruses. They contain a linear double- stranded DNA that is delivered in the nucleus upon viral entry and circularized. It usually remains episomal, i.e. as an extrachromosomal circular DNA. However, some herpes viruses can integrate their genome into the host chromosomes, although these observations are considered as exceptions of the herpesvirus life cycle. In this part, we will highlight the features concerning integration of the γ-herpesvirus Epstein-Barr virus (EBV) and the β-herpesvirus Human Herpes Virus 6 (HHV-6) into the host chromosomes.
3.2.1 Epstein-Barr Virus (EBV)
EBV is the prototypical member of the γ-herpesvirus family and is known to establish a long persistent infection in B-lymphocytes as well as in epithelial cells. EBV is associated with several proliferative disorders and cancers, including Burkitt’s lymphoma, Hodgkin’s lymphoma and nasopharyngeal carcinoma (Epstein et al. 1964; Gutensohn and Cole 1980; Zur Hausen and Schulte-Holthausen 1970). Two stages of EBV infection exist: (i) the lytic or productive cycle, in which the infected cell is actively releasing new infectious viral particles, and (ii) the latent cycle, in which only a few viral proteins are expressed, some of which are directly linked with cell proliferation and thus cancer. During latent infection, the EBV genome persists as an episome. However, the presence of linearized EBV genome in the host genome has been identified and confirmed using different approaches, including cytological hybridization, FISH*, PCR*, genomic library screening and sequencing. The presence of integrated EBV genome suggests an alternative way for EBV to establish long term infection (Gao et al. 2006; Hurley et al. 1991; Lestou et al. 1993). However, the question whether integration site selection occurs randomly or not is still a matter of debate, mainly due to the technical difficulties to isolate EBV integration events from EBV episomes (Gao et al. 2006; Takakuwa et al. 2004). Nevertheless, data so far suggest that EBV integration is not random and occurs preferentially in regions corresponding to heterochromatin (Gao et al. 2006; Lestou et al. 1993) (Fig. 3a). However, EBV integration has also been identified in genes, including MACF1*, BACH-2* (putative tumor suppressor gene), REL* and BCL-11A* (proto-oncogenes), thereby revealing a potential impact of EBV integration in disrupting the expression of some cellular genes (Takakuwa et al. 2004).
The EBV episome maintenance is ensured by the viral Epstein-Barr nuclear antigen 1 (EBNA-1) protein, attaching the episome to the host chromatin via AT-hook motifs (Fig. 3b). The interaction of EBNA-1 with the cellular EBNA-1 Binding Protein 2 (EBP2) and high-mobility group protein 2 (HMGB2) may also play a role in attaching the EBV episome to the host chromatin during interphase and mitosis (Jourdan et al. 2012). This chromatin attachment process could be enlarged to other family members, including the Kaposi’s sarcoma herpes virus (KSHV). Indeed, it was shown that KSHV episomal genome was attached to the host chromatin via the cellular histones 2A and 2B, the methyl-CpG-binding protein 2 (MeCP2) and the LANA (latency associated nuclear antigen) viral protein (Fig. 3b) (Barbera et al. 2006; Matsumura et al. 2010; Verma and Robertson 2003). Although the mechanisms involved in EBV and KHSV genome integration into the host chromatin remains to be elucidated, it is tempting to hypothesize, based on the retroviral tethering model, that viral DNA episome integration requires initially these docking proteins (EBNA-1 complex, LANA complex), thereby creating an opportunity for the subsequent incidental recombination and insertion into the host DNA, probably mediated by the cellular DNA repair machinery.
3.2.2 Human Herpes Virus-6 (HHV-6)
HHV-6 is the causal agent of the roseola infantum occurring during the first years of life and characterized by an intense fever for a few days. After the primary infection, the virus is able to establish latency in some monocytes and macrophages. Viruses may be reactivated from latency, particularly in immunosuppressed patients, thereby causing secondary infections with severe complications such as encephalitis (Kondo et al. 1991, 2002; Vu et al. 2007). Integration of HHV-6 (also named chromosomally integrated human herpes virus 6, ciHHV-6) into the host chromosomes is well defined and remains one of the most consistent observations of DNA virus integration, with at least 34 published examples (Pellett et al. 2011). Although the molecular mechanism involved in this process is still not fully understood, a few hints are starting to emerge.
The HHV-6 genome architecture is organized in two main regions: (i) the unique long region (UL) containing several gene blocks responsible for viral replication, and (ii) direct repeats (DR) flanking the genome. The right DR (DRR) and the left DR (DRL) contain a perfect [TAACCC]58 repeated sequence arrangement identical to the human telomeric repeat sequence, as well as an imperfect telomeric repeat sequence arrangement referred to the het region (Gompels and Macaulay 1995). To date, all integration sites reported were localized in the telomeric regions with no preference for a given chromosome (Fig. 3a), suggesting that HHV-6 integrates its genome via homologous recombination between the viral and cellular telomeric sequences (Arbuckle et al. 2010; Nacheva et al. 2008). Recently, a role for the still poorly characterized HHV-6 U94 protein in HHV-6 integration was proposed, based on its strong homology with AAV-2 Rep68/78, particularly at the level of single-stranded DNA binding activity (Dhepakson et al. 2002).
The HHV-6 closely related Marek’s disease virus (MDV) was shown to have also viral telomeric sequences that facilitate MDV integration into host telomeres. Minimal changes in these sequences not only strongly reduced integration efficiency but also modified the integration site selection to regions outside the telomeres (Kaufer et al. 2011), providing additional evidence that the viral DR sequence is essential for integration targeting.
3.3 Hepatitis B Virus (HBV)
The hepatitis B virus is one of the most common human pathogen responsible for the development of hepatocellular carcinoma (Neuveut et al. 2010). During acute infection, HBV can integrate its genome into the host chromosomes and present several similarities with retroviral integration. Although initial analyses of several HBV integration sites revealed random integration events in all chromosomes (Tokino and Matsubara 1991; Yaginuma et al. 1987), a recent large-scale analysis identified favored HBV integration events in transcriptionally active regions (Murakami et al. 2005). Furthermore, HBV integration target genes (including hTERT*, PDGF receptor*, the mixed lineage leukemia 2 or the 60 S ribosomal protein) were preferentially involved in cell proliferation, survival and oncogenesis (Ferber et al. 2003; Murakami et al. 2005; Tamori et al. 2005). Future studies are needed to further unveil the molecular mechanism of HBV integration the exact role of HBV integration in the establishment of hepatocellular carcinoma.
3.4 Adenoviruses (Ad)
Adenoviruses are double stranded DNA viruses, usually perceived as non-integrating viruses with a genome persisting under episomal form. However, in hamster cells, the complete genome of Ad12 was found to be stably integrated into the host chromosomes, with a few nucleotide modifications at the viral junctions. Similarly, Stephen et al. infected hamster immortalized (HT-1080 and C32) and primary fibroblasts (FF-92) with an Ad5-derived vector and identified 59 integration sites: 29 were found in active transcription units in all chromosomes and 15 out of the 30 integration sites identified outside genes were located near genes, suggesting preferential integration of Ad in gene loci (Fig. 3a) (Stephen et al. 2008, 2010). The current model suggests that Ad ITR contains specific symmetric recombinant (SYREC) sequences, which have stretches homologous to cellular repetitive elements, and that could thus allow Ad host genome insertion through patchy nucleotide homology (Fig. 3b) (Deuring and Doerfler 1983; Deuring et al. 1981; Doerfler 2009; Stabel and Doerfler 1982; Wronka et al. 2002). Further analysis of Ad integration events in vitro and in vivo revealed that both homologous recombination and heterologous recombination (non homologous end joining pathway) were involved in this SYREC-mediated integration process (Hoglund et al. 1992; Stephen et al. 2008, 2010; Wronka et al. 2002). Adenovirus-based vectors are currently the most used vectors in gene therapy, representing 24.2% of the clinical trials (source http://www.wiley.com//legacy/wileychi/genmed/clinical). Understanding the frequency and the mechanisms of Ad integration and recombination should help render these vectors safer for gene therapy trials.
4 Consequences of Viral Integration on the Host Cell
The site of the viral integration event can have multiple consequences for the host, as well as for the virus itself. Indeed, viral integration can lead to cell death or proliferation as a result of insertional mutagenesis. However, integration can also lead to consequences for the virus, i.e. active production or transcriptional silencing, a process also called latency that is key to establish viral persistence. Finally, integration in the germline can contribute shaping the host genome and participate in species evolution. Each of these effects will be further discussed below.
4.1 Cell Death
Apopotosis is a general mechanism involved in cell homeostasis regulation eliminating aberrant cells, with altered physiological parameters as well as a compromised genome integrity (Roulston et al. 1999). Upon viral invasion, the presence of a linear double-stranded DNA is sensed by the host DNA repair machinery as a DNA break, which will lead to cell apoptosis unless successfully repaired (Daniel et al. 1999; O’Brien 1998). Following the same concept, if the cell is invaded by multiple viral particles, thus multiple DNA genomes, it is likely that the DNA repair machinery will be overwhelmed, and will thus fail in repairing all the DNA molecules, thereby resulting in cell death. Similarly, if too many viral genomes integrate successfully, the integrity of the host genome itself may be compromised, also leading to cell death. In addition, viral integration will eventually lead to gene expression deregulation that may induce cell apoptosis. For instance, it has been reported that integration of HBV in ATP2A1/SERCA-1* gene resulted in gene disruption and in the expression of a chimeric non functional protein HBVx/SERCA-1 (Fig. 4). This chimeric protein lost calcium and ATP binding domains, thereby strongly disturbing the reticulum endoplasmic calcium homeostasis and inducing apoptosis (Chami et al. 2000).
Schematic overview of global consequences of viral integration events. Viral genome (red) insertion into gene exons (yellow) or introns (black line) eventually leads to gene disruption (left). Viral genome insertion into or close to promoters (blue) leads to an influence of viral enhancers on host gene expression regulation, thus overexpression by gene activation (right)
4.2 Tumorigenesis
Many viruses have been characterized based on their ability to induce cellular transformation and thus tumors. However, two mechanisms of virus-induced cellular transformation should be distinguished.
The first one leads to a rapid tumorigenesis process and is exemplified by oncoviruses, i.e. viruses coding for a viral oncogene and thus directly responsible for the cellular proliferation, such as some retroviruses (MMTV*, MLV*, RSV*, HTLV*) and DNA viruses (HPV, EBV, HBV, Ad) (Nevins 2007). Of note, it has been suggested that Adenoviruses are more likely to induce cell death in permissive cells (including human cells), while inducing a tumor in non-permissive cells (hamster cells), often linked to Adenoviral genome integration (Doerfler 2011, 2012).
The second mechanism, which is directly related to viral integration, is called insertional mutagenesis. In this case, tumorigenesis is a slow process directly related to the viral integration site, which disturbs the cell homeostasis. Indeed, viral integration alters and modulates the expression of cellular nearby genes (Fig. 4). A first scenario is the result of gene disruption by the viral integration event. If the disrupted gene is a tumor suppressor gene for example, this may ultimately lead to cellular transformation. Second, viral integration occurring close to cellular oncogenes may result in viral promoter-induced overexpression of the oncogene. The best illustration of this event occurred in a gene therapy trial aiming at correcting the severe combined immunodeficiency-X1 disease (SCID-X1) using a gammaretroviral vector providing a functional IL2RG* gene (Cavazzana-Calvo et al. 2000). Although this trial was successful, restoring an immune function, 4 out of 9 patients developed leukemia in the 5 years following viral transduction (Hacein-Bey-Abina et al. 2008). The analysis of viral integration sites in the transduced cells identified integration events nearby the LMO2* proto-oncogene, yielding to LMO2 overexpression (Hacein-Bey-Abina et al. 2008). The aberrant expression of LMO2 is a major determinant of T cell immortalization as recently demonstrated in vitro after gammaretroviral transduction of the proto-oncogene LMO2 in T cells (Newrzela et al. 2011). Although it was shown that MLV vectors preferentially integrate at promoters and regions close to the transcription start site (Kim et al. 2008, 2011; Mitchell et al. 2004; Wu et al. 2003), exon 1 of LMO2 locus was shown to be a hotspot for MLV integration in T cells, with 1 integration out of 2.125 ´ 105 (Yamada et al. 2009). Nonetheless, new MLV-derived vectors containing chromatin insulator elements from the chicken β-globin have been engineered to block the viral enhancer activity of the promoter, thereby reducing the risk of MLV-induced leukemia (Emery 2011).
To obtain a more global picture of cellular homeostasis alterations upon viral integration events, Soto-Giron and Garcia-Vallejo (2012) recently attempted at predicting the changes due to HIV-1 integration in macrophages, using protein networks interacting directly with HIV-1 or indirectly through regulatory pathways (Balakrishnan et al. 2009; Schroder et al. 2002). They selected a few genes targeted by retroviral integration and compared the interactome of these gene products between non-infected and HIV-1 infected macrophages. By computational analysis, they showed that integration in 5 selected genes induced profound alteration of the global transcription network (Soto-Giron and Garcia-Vallejo 2012). Another illustration of cell homeostasis deregulation upon viral integration, leading to tumor development, resides in HBV infected cells, where multiple pathways involved in cell cycle are deregulated, including Wnt/β-catenin signaling, Ras/MAPK, PTEN/Akt, p14ARF/p53, and TGF-β pathways (Neuveut et al. 2010).
Accumulation of genetic changes, chromosomal rearrangements, alterations of gene expression and cellular pathways as consequences of viral integration contribute incrementally to deregulate cell growth and induce tumor development when apoptosis is not involved. The database named DrVIS has recently been developed in order to report the association between viral integration sites and malignant diseases (Zhao et al. 2012). However, to date, the exact role of viral integration in cancer induction has not been fully clarified for many viruses.
4.3 Viral Persistence
Many viruses can exist in a latent state, thus establishing a persistent infection. During this phase, viruses are transcriptionally silent, either completely or partially, allowing them to escape immune surveillance and establish viral reservoirs. Viral reservoirs represent a major obstacle for therapeutic strategies and virus eradication.
A well-known example is illustrated by HIV-1, which can persist in resting memory CD4+ T cells (Chomont et al. 2009; Chun et al. 1997a, b, 1995; Finzi et al. 1999, 1997). Indeed, despite a very efficient combination therapy (highly active antiretroviral therapy, HAART), HIV-1 is not eliminated from the organism and rebounds upon HAART interruption. Although the mechanisms underlying virus reactivation, allowing the virus to exit a transcriptionally silent and latent state in favor of a productive state releasing infectious particles, is not yet completely understood, it is nevertheless obvious that this can only be achieved thanks to the presence of the integrated HIV-1 genome in the infected cell (Joos et al. 2008; Zhang et al. 2000). To date, it is thought that the only way to successful HIV-1 eradication resides in purging the viral reservoir, and that this could be achieved by reactivation of viral transcription from latently infected cells (Siliciano 2010).
The molecular mechanisms promoting and maintaining in vivo latency of DNA and RNA viruses have not been completely elucidated and are still the focus of many investigations. In the case of HIV-1, three major players are currently involved in latency: (i) the availability of cellular transcription factors. Indeed, a current model implies that HIV-1 is transcriptionally active in activated infected T cells, and that when the T cells evolve to a resting memory state, many transcription factors become unavailable, thus silencing viral transcription (Coiras et al. 2009). Furthermore, epigenetic modifications implicating de novo methylation of the provirus and chromatin remodeling complexes may also contribute to the transcriptional silencing of the integrated retrovirus (Agbottah et al. 2006; Blazkova et al. 2009; Kauder et al. 2009; Mahmoudi et al. 2006; Treand et al. 2006). (ii) The level of the viral transactivator protein, Tat, which is responsible for efficient viral transcription, and (iii) the site of viral integration. Indeed, it has been shown that infected cells in a latent state were characterized with proviruses in heterochromatin and centromeric regions (Jordan et al. 2003; Lewinski et al. 2005) and were found more often in sense orientation, leading to decreased viral transcription due to RNA interference (Shan et al. 2011).
Although herpes viruses establish latency via persistent episomes, it has been shown that HHV-6 integration was also able to promote latency. Indeed, by a mechanism similar to HIV-1, HHV-6 integration into telomeric heterochromatin, which are transcriptionally inactive regions may affect viral transcriptional activity, thereby favoring latency (Arbuckle et al. 2010; Arbuckle and Medveczky 2011; Nacheva et al. 2008). This latent HHV-6 is non cytopathic as completely or partially silent. However, the reactivation of integrated HHV-6 by HDAC inhibitors, such as trichostatin-A, induces efficient viral production, as well as cytopathic effects (cell death and syncytium formation), which are eventually deleterious for the host (Arbuckle et al. 2010; Duelli and Lazebnik 2007).
4.4 Species Evolution
The integrating virus can be persistent not only at the level of the cell but also at the level of the organism. Indeed, viral integration may have a significant impact on the organism and its progeny if the virus succeeds in infecting the germ line.
Retroviruses are the only viral group that has remnants in the form of integrated endogenous elements (ERV for Endogenous Retrovirus), accumulating over time in the human genome, and reaching to date approximately 8% of the total genome (Jern and Coffin 2008). In humans, HERVs resemble to exogenous retroviruses, however, due to accumulated mutations, they lost their ability to replicate and can thus be considered as defective endogenous retroviruses. Even if retroviruses usually infect somatic cells, infection of a germ line cell can sometimes occur. In this way, HERVs were fixed in the human genome and could be transmitted through generations as a classical human gene driven by Mendelien’s rules.
Integration of viral elements followed by endogenization can lead to profound consequences for the host, ultimately shaping its genome. The proof of concept of this is illustrated by syncytin genes that are expressed in trophoblasts. Syncytins display fusogenic activities that contribute to the formation of multinucleated syncytiotrophoblast cells, and are thus essential for placenta morphogenesis (Rawn and Cross 2008). It has been shown that the syncytin-1 gene corresponds to the env gene of an endogenous retrovirus belonging to the HERV-W family that was fixed in the human genome 45 million years ago (Mi et al. 2000). Similarly, another fusogenic protein named Syncytin-2 has been identified, corresponding to the env gene of HERV-FRD (Blaise et al. 2003). During primate evolution, these genes were conserved, and thus “captured” by the host as they provided a benefit for the host. In contrast, gag and pol genes accumulated inactivating mutations, leading to a replication-incompetent retrovirus that could be otherwise detrimental to the host.
As mentioned earlier, 8% of the human genome is composed of ERV remnants. Further investigations on these retroviral sequences should provide additional information about retroviral genes that are functional, like env-derived syncytins, and therefore likely to play a role in host cellular processes.
5 General Conclusions
Integration of viral genome into host chromosomes results from (i) an essential step of life cycle, such as for retroviruses, or (ii) an incident, for some RNA viruses and DNA viruses. However, the high integration frequency of some DNA viruses (i.e. HHV-6) and its role in establishing beneficial latency may challenge the view of incidental integration. Nevertheless, incidental or not, genome integration of DNA and RNA viruses have profound consequences for the host, including premature cell death and tumorigenesis, and that will in turn affect the rate of viral expression, thereby guiding the virus in a productive or latent cycle. In addition, viral integration events in the germ line may contribute to shaping the host genome, eventually providing selective advantages for the host, and contributing to species evolution.
A better understanding of viral integration mechanisms, integration frequency, integration site selection and the impact of viral integration on the virus-associated disease outcome should help designing new strategies aiming at eradicating persistent viral infections, as well as improving virus-derived delivery vectors for gene therapy.
References
Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F (2006) Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 3:48
Ao Z, Danappa Jayappa K, Wang B et al (2010) Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J Virol 84:8650–8663
Arbuckle JH, Medveczky PG (2011) The molecular biology of human herpesvirus-6 latency and telomere integration. Microbes Infect 13:731–741
Arbuckle JH, Medveczky MM, Luka J et al (2010) The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci USA 107:5563–5568
Balakrishnan S, Tastan O, Carbonell J, Klein-Seetharaman J (2009) Alternative paths in HIV-1 targeted human signal transduction pathways. BMC Genomics 10(Suppl 3):S30
Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Luger K, Kaye KM (2006) Kaposi’s sarcoma-associated herpesvirus LANA hitches a ride on the chromosome. Cell Cycle 5:1048–1052
Belshan M, Schweitzer CJ, Donnellan MR, Lu R, Engelman A (2009) In vivo biotinylation and capture of HIV-1 matrix and integrase proteins. J Virol Methods 159:178–184
Bera S, Pandey KK, Vora AC, Grandgenett DP (2009) Molecular interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol 389:183–198
Blaise S, de Parseval N, Benit L, Heidmann T (2003) Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA 100:13013–13018
Blazkova J, Trejbalova K, Gondois-Rey F et al (2009) CpG methylation controls reactivation of HIV from latency. PLoS Pathog 5:e1000554
Brady T, Lee YN, Ronen K et al (2009) Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev 23:633–642
Brady T, Roth SL, Malani N et al (2011) A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res 39:e72
Brister JR, Muzyczka N (1999) Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol 73:9325–9336
Bushman F, Lewinski M, Ciuffi A et al (2005) Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol 3:848–858
Busschots K, Voet A, De Maeyer M et al (2007) Identification of the LEDGF/p75 binding site in HIV-1 integrase. J Mol Biol 365:1480–1492
Callahan R, Smith GH (2000) MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene 19:992–1001
Callahan R, Smith GH (2008) Common integration sites for MMTV in viral induced mouse mammary tumors. J Mammary Gland Biol Neoplasia 13:309–321
Cavazzana-Calvo M, Hacein-Bey S, de Saint BG et al (2000) Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288:669–672
Chami M, Gozuacik D, Saigo K et al (2000) Hepatitis B virus-related insertional mutagenesis implicates SERCA1 gene in the control of apoptosis. Oncogene 19:2877–2886
Cherepanov P (2007) LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res 35:113–124
Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A (2005a) Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA 102:17308–17313
Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A (2005b) Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol 12:526–532
Cherepanov P, Maertens GN, Hare S (2011) Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol 21:249–256
Chomont N, El-Far M, Ancuta P et al (2009) HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900
Christ F, Thys W, De Rijck J et al (2008) Transportin-SR2 imports HIV into the nucleus. Curr Biol 18:1192–1202
Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF (1995) In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1:1284–1290
Chun TW, Carruth L, Finzi D et al (1997a) Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188
Chun TW, Stuyver L, Mizell SB et al (1997b) Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA 94:13193–13197
Ciuffi A, Bushman FD (2006) Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet 22:388–395
Ciuffi A, Llano M, Poeschla E et al (2005) A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11:1287–1289
Ciuffi A, Diamond TL, Hwang Y, Marshall HM, Bushman FD (2006) Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum Gene Ther 17:960–967
Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J (2009) Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7:798–812
Daniel R, Katz RA, Skalka AM (1999) A role for DNA-PK in retroviral DNA integration. Science 284:644–647
Delelis O, Parissi V, Leh H et al (2007) Efficient and specific internal cleavage of a retroviral palindromic DNA sequence by tetrameric HIV-1 integrase. PLoS One 2:e608
Delelis O, Zamborlini A, Thierry S, Saib A (2010) Chromosomal tethering and proviral integration. Biochim Biophys Acta 1799:207–216
Derse D, Crise B, Li Y et al (2007) Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J Virol 81:6731–6741
Desfarges S, Ciuffi A (2010) Retroviral integration site selection. Viruses 2:111–130
Deuring R, Doerfler W (1983) Proof of recombination between viral and cellular genomes in human KB cells productively infected by adenovirus type 12: structure of the junction site in a symmetric recombinant (SYREC). Gene 26:283–289
Deuring R, Klotz G, Doerfler W (1981) An unusual symmetric recombinant between adenovirus type 12 DNA and human cell DNA. Proc Natl Acad Sci USA 78:3142–3146
Dhepakson P, Mori Y, Jiang YB et al (2002) Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J Gen Virol 83:847–854
Diamond TL, Bushman FD (2005) Division of labor within human immunodeficiency virus integrase complexes: determinants of catalysis and target DNA capture. J Virol 79:15376–15387
Doerfler W (2009) Epigenetic mechanisms in human adenovirus type 12 oncogenesis. Semin Cancer Biol 19:136–143
Doerfler W (2011) Epigenetic consequences of foreign DNA insertions: de novo methylation and global alterations of methylation patterns in recipient genomes. Rev Med Virol 21:336–346
Doerfler W (2012) Impact of foreign DNA integration on tumor biology and on evolution via epigenetic alterations. Epigenomics 4:41–49
Duelli D, Lazebnik Y (2007) Cell-to-cell fusion as a link between viruses and cancer. Nat Rev Cancer 7:968–976
Emery DW (2011) The use of chromatin insulators to improve the expression and safety of integrating gene transfer vectors. Hum Gene Ther 22:761–774
Emonet SE, Urata S, de la Torre JC (2011) Arenavirus reverse genetics: new approaches for the investigation of arenavirus biology and development of antiviral strategies. Virology 411:416–425
Engelman A, Cherepanov P (2008) The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog 4:e1000046
Epstein MA, Achong BG, Barr YM (1964) Virus particles in cultured lymphoblasts from burkitt’s lymphoma. Lancet 1:702–703
Farnet CM, Haseltine WA (1991) Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol 65:1910–1915
Faschinger A, Rouault F, Sollner J et al (2008) Mouse mammary tumor virus integration site selection in human and mouse genomes. J Virol 82:1360–1367
Fassati A, Goff SP (2001) Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635
Faure A, Calmels C, Desjobert C et al (2005) HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res 33:977–986
Ferber MJ, Montoya DP, Yu C et al (2003) Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene 22:3813–3820
Ferris AL, Wu X, Hughes CM et al (2010) Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci USA 107:3135–3140
Finzi D, Hermankova M, Pierson T et al (1997) Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300
Finzi D, Blankson J, Siliciano JD et al (1999) Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517
Frank AC, Wolfe KH (2009) Evolutionary capture of viral and plasmid DNA by yeast nuclear chromosomes. Eukaryot Cell 8:1521–1531
Friedrich BM, Dziuba N, Li G, Endsley MA, Murray JL, Ferguson MR (2011) Host factors mediating HIV-1 replication. Virus Res 161:101–114
Gao J, Luo X, Tang K, Li X, Li G (2006) Epstein-Barr virus integrates frequently into chromosome 4q, 2q, 1q and 7q of burkitt’s lymphoma cell line (raji). J Virol Methods 136:193–199
Geuking MB, Weber J, Dewannieux M et al (2009) Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323:393–396
Gijsbers R, Ronen K, Vets S et al (2010) LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther 18:552–560
Gijsbers R, Vets S, De Rijck J et al (2011) Role of the PWWP domain of lens epithelium-derived growth factor (LEDGF)/p75 cofactor in lentiviral integration targeting. J Biol Chem 286:41812–41825
Gompels UA, Macaulay HA (1995) Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J Gen Virol 76(Pt 2):451–458
Guiot E, Carayon K, Delelis O et al (2006) Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J Biol Chem 281:22707–22719
Gutensohn N, Cole P (1980) Epidemiology of Hodgkin’s disease. Semin Oncol 7:92–102
Hacein-Bey-Abina S, Garrigue A, Wang GP et al (2008) Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 118:3132–3142
Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P (2010) Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236
Henckaerts E, Linden RM (2010) Adeno-associated virus: a key to the human genome? Future Virol 5:555–574
Hoglund M, Siden T, Rohme D (1992) Different pathways for chromosomal integration of transfected circular pSVneo plasmids in normal and established rodent cells. Gene 116:215–222
Hurley EA, Klaman LD, Agger S, Lawrence JB, Thorley-Lawson DA (1991) The prototypical Epstein-Barr virus-transformed lymphoblastoid cell line IB4 is an unusual variant containing integrated but no episomal viral DNA. J Virol 65:3958–3963
Huser D, Gogol-Doring A, Lutter T, Weger S, Winter K, Hammer EM, Cathomen T, Reinert K, Heilbronn R (2010) Integration preferences of wildtype AAV-2 for consensus rep-binding sites at numerous loci in the human genome. PLoS Pathog, 6, e1000985
Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A (2009) Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J 276:2926–2946
Jern P, Coffin JM (2008) Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732
Joos B, Fischer M, Kuster H et al (2008) HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci USA 105:16725–16730
Jordan A, Bisgrove D, Verdin E (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22:1868–1877
Jourdan N, Jobart-Malfait A, Dos Reis G, Quignon F, Piolot T, Klein C, Tramier M, Coppey M, Marechal V (2012) Live-cell imaging reveals multiple interactions between Epstein-Barr Nuclear Antigen 1 (EBNA-1) and cellular chromatin during interphase and mitosis. J Virol 86(9):5314–5329
Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E (2009) Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 5:e1000495
Kaufer BB, Arndt S, Trapp S, Osterrieder N, Jarosinski KW (2011) Herpesvirus telomerase RNA (vTR) with a mutated template sequence abrogates herpesvirus-induced lymphomagenesis. PLoS Pathog 7:e1002333
Kim S, Kim N, Dong B et al (2008) Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J Virol 82:9964–9977
Kim HH, van den Heuvel AP, Schmidt JW, Ross SR (2011) Novel common integration sites targeted by mouse mammary tumor virus insertion in mammary tumors have oncogenic activity. PLoS One 6:e27425
Klenerman P, Hengartner H, Zinkernagel RM (1997) A non-retroviral RNA virus persists in DNA form. Nature 390:298–301
Kondo K, Kondo T, Okuno T, Takahashi M, Yamanishi K (1991) Latent human herpesvirus 6 infection of human monocytes/macrophages. J Gen Virol 72(Pt 6):1401–1408
Kondo K, Kondo T, Shimada K, Amo K, Miyagawa H, Yamanishi K (2002) Strong interaction between human herpesvirus 6 and peripheral blood monocytes/macrophages during acute infection. J Med Virol 67:364–369
Kotin RM, Menninger JC, Ward DC, Berns KI (1991) Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics 10:831–834
Lamartina S, Ciliberto G, Toniatti C (2000) Selective cleavage of AAVS1 substrates by the adeno-associated virus type 2 rep68 protein is dependent on topological and sequence constraints. J Virol 74:8831–8842
Lestou VS, De Braekeleer M, Strehl S, Ott G, Gadner H, Ambros PF (1993) Non-random integration of Epstein-Barr virus in lymphoblastoid cell lines. Genes Chromosomes Cancer 8:38–48
Levin A, Hayouka Z, Friedler A, Loyter A (2010) Transportin 3 and importin alpha are required for effective nuclear import of HIV-1 integrase in virus-infected cells. Nucleus 1:422–431
Lewinski MK, Bushman FD (2005) Retroviral DNA integration–mechanism and consequences. Adv Genet 55:147–181
Lewinski MK, Bisgrove D, Shinn P et al (2005) Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol 79:6610–6619
Lewinski MK, Yamashita M, Emerman M et al (2006) Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog 2:e60
Lewis PF, Emerman M (1994) Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol 68:510–516
Li M, Craigie R (2005) Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem 280:29334–29339
Li X, Krishnan L, Cherepanov P, Engelman A (2011) Structural biology of retroviral DNA integration. Virology 411:194–205
Lin CW, Engelman A (2003) The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J Virol 77:5030–5036
Liu H, Fu Y, Jiang D et al (2010) Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol 84:11876–11887
Llano M, Saenz DT, Meehan A et al (2006) An essential role for LEDGF/p75 in HIV integration. Science 314:461–464
Mahmoudi T, Parra M, Vries RG et al (2006) The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem 281:19960–19968
Marshall HM, Ronen K, Berry C et al (2007) Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340
Matsumura S, Persson LM, Wong L, Wilson AC (2010) The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains. J Virol 84:2318–2330
McCarty DM, Pereira DJ, Zolotukhin I, Zhou X, Ryan JH, Muzyczka N (1994a) Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol 68:4988–4997
McCarty DM, Ryan JH, Zolotukhin S, Zhou X, Muzyczka N (1994b) Interaction of the adeno-associated virus Rep protein with a sequence within the a palindrome of the viral terminal repeat. J Virol 68:4998–5006
Meehan AM, Poeschla EM (2010) Chromatin tethering and retroviral integration: recent discoveries and parallels with DNA viruses. Biochim Biophys Acta 1799:182–191
Meehan AM, Saenz DT, Morrison JH et al (2009) LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog 5:e1000522
Mi S, Lee X, Li X et al (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789
Miller MD, Farnet CM, Bushman FD (1997) Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol 71:5382–5390
Mitchell RS, Beitzel BF, Schroder AR et al (2004) Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2:E234
Murakami Y, Saigo K, Takashima H et al (2005) Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 54:1162–1168
Nacheva EP, Ward KN, Brazma D et al (2008) Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J Med Virol 80:1952–1958
Neuveut C, Wei Y, Buendia MA (2010) Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol 52:594–604
Nevins JR (2007) Cell transformation by viruses. In: Knipe (PM) DMH (ed) Fields virology. Lippincott Williams & Wilkins, Philadelphia, pp 211–250
Newrzela S, Cornils K, Heinrich T (2011) Retroviral insertional mutagenesis can contribute to immortalization of mature T lymphocytes. Mol Med 17(11-12):1223–1232
O’Brien V (1998) Viruses and apoptosis. J Gen Virol 79(Pt 8):1833–1845
Ocwieja KE, Brady TL, Ronen K et al (2011) HIV integration targeting: a pathway involving transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog 7:e1001313
Pellett PE, Ablashi DV, Ambros PF et al (2011) Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol.
Poeschla EM (2008) Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci 65:1403–1424
Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56:1–14
Raghavendra NK, Shkriabai N, Graham R, Hess S, Kvaratskhelia M, Wu L (2010) Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology 7:66
Rawn SM, Cross JC (2008) The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol 24:159–181
Roe T, Reynolds TC, Yu G, Brown PO (1993) Integration of murine leukemia virus DNA depends on mitosis. EMBO J 12:2099–2108
Roth SL, Malani N, Bushman FD (2011) Gammaretroviral integration into nucleosomal target DNA in vivo. J Virol 85:7393–7401
Roulston A, Marcellus RC, Branton PE (1999) Viruses and apoptosis. Annu Rev Microbiol 53:577–628
Schaller T, Ocwieja KE, Rasaiyaah J et al (2011) HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439
Schrijvers R, De Rijck J, Demeulemeester J et al (2012) LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog 8:e1002558
Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521–529
Shan L, Yang HC, Rabi SA et al (2011) Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J Virol 85:5384–5393
Shun MC, Raghavendra NK, Vandegraaff N et al (2007) LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21:1767–1778
Siliciano RF (2010) What do we need to do to cure HIV infection. Top HIV Med 18:104–108
Silvers RM, Smith JA, Schowalter M et al (2010) Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum Gene Ther 21:337–349
Sinha S, Grandgenett DP (2005) Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J Virol 79:8208–8216
Sinha S, Pursley MH, Grandgenett DP (2002) Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J Virol 76:3105–3113
Soto-Giron MJ, Garcia-Vallejo F (2012) Changes in the topology of gene expression networks by human immunodeficiency virus type 1 (HIV-1) integration in macrophages. Virus Res, 163:91–7
Stabel S, Doerfler W (1982) Nucleotide sequence at the site of junction between adenovirus type 12 DNA and repetitive hamster cell DNA in transformed cell line CLAC1. Nucleic Acids Res 10:8007–8023
Stephen SL, Sivanandam VG, Kochanek S (2008) Homologous and heterologous recombination between adenovirus vector DNA and chromosomal DNA. J Gene Med 10:1176–1189
Stephen SL, Montini E, Sivanandam VG et al (2010) Chromosomal integration of adenoviral vector DNA in vivo. J Virol 84:9987–9994
Surosky RT, Urabe M, Godwin SG et al (1997) Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol 71:7951–7959
Suzuki Y, Craigie R (2007) The road to chromatin – nuclear entry of retroviruses. Nat Rev Microbiol 5:187–196
Takakuwa T, Luo WJ, Ham MF, Sakane-Ishikawa F, Wada N, Aozasa K (2004) Integration of Epstein-Barr virus into chromosome 6q15 of burkitt lymphoma cell line (raji) induces loss of BACH2 expression. Am J Pathol 164:967–974
Tamori A, Nishiguchi S, Shiomi S et al (2005) Hepatitis B virus DNA integration in hepatocellular carcinoma after interferon-induced disappearance of hepatitis C virus. Am J Gastroenterol 100:1748–1753
Tanne E, Sela I (2005) Occurrence of a DNA sequence of a non-retro RNA virus in a host plant genome and its expression: evidence for recombination between viral and host RNAs. Virology 332:614–622
Tobaly-Tapiero J, Bittoun P, Lehmann-Che J et al (2008) Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 9:1717–1727
Tokino T, Matsubara K (1991) Chromosomal sites for hepatitis B virus integration in human hepatocellular carcinoma. J Virol 65:6761–6764
Treand C, du Chene I, Bres V et al (2006) Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 25:1690–1699
Trobridge GD, Miller DG, Jacobs MA et al (2006) Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA 103:1498–1503
Urcelay E, Ward P, Wiener SM, Safer B, Kotin RM (1995) Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol 69:2038–2046
Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z (2006) Cellular co-factors of HIV-1 integration. Trends Biochem Sci 31:98–105
Verma SC, Robertson ES (2003) ORF73 Of herpesvirus saimiri strain C488 tethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J Virol 77:12494–12506
Vu T, Carrum G, Hutton G, Heslop HE, Brenner MK, Kamble R (2007) Human herpesvirus-6 encephalitis following allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant 39:705–709
Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD (2007) HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17:1186–1194
Wang GP, Levine BL, Binder GK et al (2009) Analysis of lentiviral vector integration in HIV + study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther 17:844–850
Weitzman MD, Kyostio SR, Kotin RM, Owens RA (1994) Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA 91:5808–5812
Wronka G, Fechteler K, Schmitz B, Doerfler W (2002) Integrative recombination between adenovirus type 12 DNA and mammalian DNA in a cell-free system: joining by short sequence homologies. Virus Res 90:225–242
Wu X, Li Y, Crise B, Burgess SM (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749–1751
Yaginuma K, Kobayashi H, Kobayashi M, Morishima T, Matsuyama K, Koike K (1987) Multiple integration site of hepatitis B virus DNA in hepatocellular carcinoma and chronic active hepatitis tissues from children. J Virol 61:1808–1813
Yamada K, Tsukahara T, Yoshino K et al (2009) Identification of a high incidence region for retroviral vector integration near exon 1 of the LMO2 locus. Retrovirology 6:79
Zhang L, Chung C, Hu BS et al (2000) Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J Clin Invest 106:839–845
Zhao X, Liu Q, Cai Q (2012) Dr.VIS: a database of human disease-related viral integration sites. Nucleic Acids Res 40:D1041–D1046
Zur Hausen H, Schulte-Holthausen H (1970) Presence of EB virus nucleic acid homology in a “virus-free” line of burkitt tumour cells. Nature 227:245–248
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Desfarges, S., Ciuffi, A. (2012). Viral Integration and Consequences on Host Gene Expression. In: Witzany, G. (eds) Viruses: Essential Agents of Life. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-4899-6_7
Download citation
DOI: https://doi.org/10.1007/978-94-007-4899-6_7
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-4898-9
Online ISBN: 978-94-007-4899-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)