Abstract
A novel fully, automatic, adaptive, robust procedure for brain tissue classification from three-dimensional (3D) magnetic resonance head images (MRI) is described in this paper. We propose an automated scheme for magnetic resonance imaging (MRI) brain segmentation. An adaptive mean-shift methodology is utilized in order to categorize brain voxels into one of three main tissue types: gray matter, white matter, and cerebro spinal fluid. The MRI image space is characterized by a high dimensional feature space that includes multimodal intensity features in addition to spatial features. An adaptive mean-shift algorithm clusters the joint spatial-intensity feature space, thus extracting a representative set of high-density points within the feature space, otherwise known as modes. Tissue segmentation is obtained by a follow-up phase of intensity-based mode clustering into the three tissue categories. By its nonparametric nature, adaptive mean-shift can deal successfully with nonconvex clusters and produce convergence modes that are better applicant for intensity based categorization than the initial voxels. The performance of this brain tissue classification procedure is demonstrated through quantitative and qualitative validation experiments on both simulated MRI data (10 subjects) and real MRI data (43 subjects). The proposed method is validated on 3-D single and multimodal datasets, for both simulated and real MRI data. It is shown to perform well in comparison to other state-of-the-art methods without the use of a preregistered statistical brain atlas.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
Introduction
A magnetic resonance imaging instrument (MRI scanner) uses powerful magnets to polarize and excite hydrogen nuclei (single proton)) in water molecules in human tissue, producing a detectable signal which is spatially encoded resulting in images of the body. In brief, MRI involves the use of three kinds of electromagnetic field: a very strong (of the order of units of Tesla) static magnetic field to polarize the hydrogen nuclei, called the static field; a weaker time-varying (of the order of 1 kHz) for spatial encoding, called the gradient field(s); and a weak radio frequency (RF) field for manipulation of the hydrogen nuclei to produce measurable signals, collected through an RF antenna [1–3]. Like CT, MRI traditionally creates a two-dimensional image of a thin “slice” of the body and is therefore considered a tomographic imaging technique. Modern MRI instruments are capable of producing images in the form of three-dimensional (3D) blocks, which may be considered a generalization of the single-slice tomographic concept. Unlike CT, MRI does not involve the use of ionizing radiation and is therefore not associated with the same health hazards.
Fully automatic brain tissue classification from magnetic resonance images (MRI) is of great importance for research and clinical studies of the normal and diseased human brain. Operator assisted classification methods are non reproducible, and also are impractical for the large amounts of data Automated MRI segmentation systems classify brain voxels into one of three main tissue types: gray matter (Gm), white matter (Wm), and Cerebro-spinal fluid (Csf). Volumetric analysis of different parts of the brain is useful in assessing the progress or remission of various diseases, such as Alzheimer’s disease, epilepsy, multiple sclerosis, and schizophrenia. Both supervised and unsupervised approaches have been used for this task. In the supervised approach, intensity values of labeled voxel samples from each tissue (prototypes) must be provided during the learning phase. In a subsequent classification phase, unlabeled voxels are classified using a selected classifier. This method requires human interaction to select the prototypes and is therefore semi-automatic. To avoid re-training the classifier for each new scan, methods are required to normalize the intensity between MRI scans This allows selecting the prototypes and training the supervised classifier on a reference scan, following which voxels of any other scan, previously normalized with regard to the reference scan, can be classified using the same classifier without further human intervention. Unsupervised approaches often rely on a Gaussian approximation of the voxel intensity distribution for each tissue type. This is justified due to the Rician behavior of the noise present in the MRI intensity signal [4]. A Gaussian mixture model (GMM) is fitted to the voxels intensity using the expectation–maximization (EM) algorithm following which every voxel is assigned to the tissue class for which it gives the highest probability. The GMM–EM intensity-based framework has been refined to account for partial volume effects and blood vessel signals that may alter Csf segmentation. However, using intensity information alone has proven to be insufficient for a reliable automated segmentation of the brain tissues. Local signal perturbations caused by additive noise and multiplicative bias-fields are responsible for cluster overlaps in the intensity feature space, resulting in poor tissue-class separability. Due to the artifacts present, voxel-wise intensity-based classification methods, such as K-means modeling and GMMs may give unrealistic results, with tissue class regions appearing granular, fragmented, or violating anatomical constraints. Incorporating spatial information via a statistical atlas provides a means for improving the segmentation results. The statistical atlas provides the prior probability for each pixel to originate from a particular tissue class [5–7]. Each tissue’s intensity is modeled by a Parzen density fitted to voxels selected from an affine-registered atlas. Co-registration of the input image and the atlas is critical in this scenario. It is important to stress that an appropriate atlas does not always exist for the data at hand. Two such cases are brain data with pathologies or brain data obtained from young infants. An additional conventional method to improve segmentation smoothness is to model neighboring voxels interactions using a Markov random field (MRF) statistical spatial model. MRF-based algorithms are computationally intensive, requiring critical parameter settings. It is possible to use MRF with predefined settings, which is faster but possibly less accurate. Statistical modeling frameworks can include the spatial information by augmenting the input feature space. By appending the spatial position coordinates [x, y, z] to the intensity features, a higher dimensional feature space is obtained where clusters represent both voxels’ intensity and spatial position distribution. Clusters in the augmented feature space are more closely related to the brain anatomy. This observation may prove problematic for parametric models such as GMMs that implicitly assume cluster convexity, as the brain anatomy cannot be decomposed into a small number of convex regions in the joint spatial-intensity feature space. A recently published solution includes using a large number of Gaussians per brain tissue, in order to capture the complicated spatial layout of the individual tissues. Supervised classification by the K-nearest-neighbors algorithm in an augmented position-intensity feature space has also been proposed In this case a manually segmented prototype atlas or an anatomical template is aligned with the considered brain in order to obtain the segmentation. An alternative to statistical parametric approaches is the use of unsupervised nonparametric schemes. One such approach is the mean-shift algorithm. Here, adaptive gradient ascent is used to detect local maxima of data density in feature space. Data points are associated with local maxima, or modes, thereby defining the clusters. Key characteristics of the mean-shift algorithm include the fact that no initial cluster positions are required, as well as the fact that the final number of extracted clusters is a result of the algorithm. A description of the mean shift algorithm is provided. In recent years, mean-shift has been used for image segmentation, object tracking, and medical image analysis applications in the current work; the objective is to utilize the mean-shift formalism to provide a robust segmentation framework for brain tissues in MRI that integrates spatial information in a simple way without requiring the use of a statistical brain atlas or HMRF modeling [8]. The developed framework, initially proposed is based on a variation of the mean-shift algorithm, termed the adaptive mean-shift algorithm by assigning a distinct bandwidth to every data point, the adaptive mean-shift allows for increased sensitivity to local data structure even in a higher dimensional feature space corresponding to multimodal MRI.
The issues in the existing work are (1) A GMM is fitted to the voxels intensity using the EM algorithm following which every voxel is assigned to the tissue class for which it gives the highest probability. The GMM-EM intensity based framework has been refined to account for partial volume effects and blood vessel signals that may alter Csf segmentation. (2) A Markov random field (MRF) based algorithms are computationally intensive, requiring critical parameter settings. It is possible to use MRF with predefined settings, which is faster but possibly less accurate. Statistical modeling frameworks can include the spatial information by augmenting the input feature space [9, 10].
Our proposed work is that the developed framework, initially proposed is based on a variation of the mean-shift algorithm, termed the adaptive mean-shift algorithm By assigning a distinct bandwidth to every data point, the adaptive mean-shift allows for increased sensitivity to local data structure even in a higher dimensional feature space corresponding to multimodal MRI. We present an automated segmentation framework for brain MRI volumes based on adaptive mean-shift clustering in the joint spatial and intensity feature space. The method was validated both on simulated and real brain datasets, and the results were compared with state-of-the-art algorithms. The advantage over intensity-based GMM EM schemes as well as additional state-of-the-art methods was demonstrated. We will show that using the AMS framework, segmentation of the normal tissues is not degraded by the presence of abnormal tissues.
Methodology
Proposed Algorithm
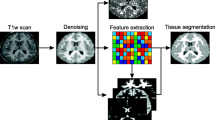
The fast adaptive mean-shift algorithm (FAMS) is utilized to analyze 3-D multimodal MRI data and provide segmentation maps of the three main tissue types (Gm, Wm, Csf). One to four MRI modalities are available per segmentation task. Standard preprocessing steps include: (1) brain parenchyma extraction using the brain extraction tool (BET). The obtained brain masks were visually inspected and corrected for outliers when needed. When a binary mask was available from the dataset, it was used instead of applying BET. (2) Intensity in-homogeneities (bias) correction by homomorphic low pass filtering and (3) Intensity values normalization across input channels (input modalities) via linear histogram stretching based on the darkest and brightest percentage points. The normalization sets the darkest percent of voxels to zero and rescales the brightest percent to 4095. The purpose is to obtain similar dynamic ranges for all the considered modalities. Following the initial data processing, feature-vectors are extracted per input voxel. The set of feature-vectors is input to the adaptive mean-shift clustering stage of the framework. The output of the clustering step is a set of modes which provides a compact representation of the data. A follow-up merging stage is proposed to further prune the initial set of modes. Finally the categorization of the resultant modes into three categories, as defined in the brain segmentation task, is achieved via an intensity-based clustering stage. Figure 1 shows a summarizing block diagram for the proposed algorithm.
-
A.
Pre processing module.
The initial data processing, feature-vectors are extracted per input voxel. Intensity as well as spatial features (x, y, z voxel coordinates) is used for an overall dimensionality of 3 + n, where n is the number of input intensity channels (modalities). The set of feature vectors is input to the adaptive mean-shift clustering stage of the framework.
-
B.
Fast adaptive mean-shift clustering.
The set of feature vectors is input to the adaptive mean-shift clustering stage of the framework. The process starts by clustering the input feature vectors, which represent the multimodal MRI brain data using the FAMS implementation of the AMS algorithm. In that AMS, we can include Locality-Sensitive Hashing. That can produce optimal approximate neighborhood with radius. This stage, each feature vector bears the label of its convergence mode, or cluster. Each mode obtained by the clustering process expresses the local structure of the data in a given region of the feature space. It should be emphasized that modes define clusters of arbitrary shape, without any convexity constraints. The number of obtained modes is an output of the fast adaptive mean-shift algorithm.
-
C.
Iterative mode pruning module.
The number of modes is a large compression of the initial data but it is still much larger than the targeted number of classes. A mode pruning step is therefore required. In fact, we have used the nonparametric adaptive mean-shift for clustering in the joint spatial-intensity feature space as the clusters are inherently no convex. For the pruning of the modes, however, we switch to an intensity-only feature space for which clusters can be conveniently approximated as convex, enabling the use of parametric models (i.e., multivariate Gaussians). For this purpose, a pruning mechanism is added as follows. A fixed-radius window is shifted across the intensity feature space (ignoring spatial features), centered on each mode. Modes that co-exist within the window are merged. Mahalanobis distance is utilized for the distance computation. For the computation of the Mahalanobis distance, a covariance matrix is computed per mode from the intensity values of its corresponding voxels. Therefore, the pruning of the initial modes list is performed until the largest variance among all pruned modes exceeds a preset threshold. That can produce Pruned mode list.
-
D.
Voxel-weighted clustering module.
That results from the preceding clustering and pruning steps, is indicated by an arrow in the color of the corresponding segmentation map. It can be observed that the intensity value for each sample mode is closer to a peak of the whole brain intensity distribution than for most of the voxels it represents. Therefore, the modes intensity provides a higher probability classification into one of the three tissue types than the intensities of the voxels they represent. The effect observed at a single mode level is the sharpening of the intensity distribution peaks (similar to the effect of a bias correction algorithm) which results in a stronger intensity separation between the different tissue types. It can produce the Final segmentation results.
Results and Discussion
In this section, we present the performance of the proposed segmentation framework on 3-D simulated and real datasets. Simulated data was downloaded from the Brain web Simulated Brain Database (SBD) repository. Real data was downloaded from the center for Morph metric Analysis. Mass-achusetts General Hospital Repository, which is a standard repository for algorithm comparison (hereon termed IBSR). Both qualitative and quantitative validation is conducted, with a comparison to additional state of-the-art segmentation algorithms and the ground truth data when available. The AMS framework performance on real data sets is demonstrated. A set of 20 normal T1-weighted real brain data was downloaded from the IBSR repository. Each volume consists of around 60 coronal T1 slices. Segmentation overlap index values obtained with several segmentation algorithms are available for comparison in the IBSR site. Three Sample slices are shown in Fig. 2. The original data; the ground truth and the AMS segmentation are presented in Fig. 2 (a)–(c), respectively.

Conclusion
We presented an automated segmentation framework for brain MRI volumes based on adaptive mean-shift clustering in the joint spatial and intensity feature space. The algorithm gave good results on noisy and biased data thanks to the adaptive mean-shift ability to work with non-convex clusters in the joint spatial intensity feature space as well as the mean-shift noise smoothing behavior. The advantages of our work are although only a rudimental bias field correction step is implemented and no spatial prior is extracted from an atlas, Moreover, by using the adaptive mean-shift instead of the constant bandwidth algorithm, we ensure an appropriate bandwidth value for each feature point without requiring per-dataset manual tuning In the current implementation (Matlab and C), the typical algorithm execution time is about 30 min for a four-modal 256 × 256 × 46 brain volume. As mean-shift runs a large loop on the whole feature vectors set, we believe that a full C/C\(++\) multithreaded implementation on a multicore PC can reduce by more than half the running time. In future research, we will examine ways to improve the current algorithm’s limitations. In particular, the current bandwidth selection algorithm based on the k-nearest neighbor makes no use of application specific information. Edge information, for instance, could help define the region of influence of a kernel by a given point since edges generally delimit regions corresponding to different tissue types. Another important issue regards the final mode merging step. Currently, it is based on the intensity clustering with k-means. This provides a robust and straightforward way of getting the desired number of classes tissue types but at the cost of losing some local spatial information contained in the modes found with mean-shift. The proposed framework will be extended to incorporate the detection of abnormal tissues such as sclerotic lesions and tumors.
References
Pham, D.L., Xu, C.Y., Prince, J.L.: A survey of current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2, 315–337 (2000)
Rouaïnia, M., Medjram, M.S., Doghmane, N.: Brain MRI segmentation and lesions detection by EM algorithm. World Academy of Science, Engineering and Technology 24 2006
Cocosco, C., Zijdenbos, A., Evans, A.: A fully automatic and robust brain MRI tissue classification method. Med. Image Anal. 7(4), 513–527 (2003)
Georgescu, B., Shimshoni, I., Meer, P.: Mean-shift based clustering in high dimensions: a texture classification example. In Proceedings of IEEE Conference on Computer Vision (ICCV), France, 2003, pp. 456–463
Derpanis, K.G.: Mean shift clustering. Shape Modeling and Applications, 2005 International Conference, Germany. August 15, 2005
Van Leemput, K., Maes, F., Vandeurmeulen, D., Suetens, P.: Auto-mated model-based tissue classification of MR images of the brain. IEEE Trans. Med. Imag. 18(10), 897–908 (1999)
Dugas-Phocion, G., González Ballester, M.Á., Malandain, G., Le-brun, C., Ayache, N.: Improved EM-based tissue segmentation and partial volume effect quantification in multi-sequence brain MRI,” in partial volume effect quantification in multi-sequence brain MRI. Paper presented at International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), France, pp. 26–33, 2004
Gudbjartsson, H., Patz, S.: The Rician distribution of noisy MRIdata. Magn. Reson. Med. 34, 910–914 (1995)
Dempster, A., Laird, N., Rubin, D.: Maximum likelihood from in-complete data via the EM algorithm. J. Roy. Stat. Soc. B 39, 1–38 (1977)
Van Leemput, K., Maes, F., Vandermeulen, D., Suetens, P.: A uni-fying framework for partial volume segmentation of brain MR images. IEEE Trans. Med. Image. 22(1), 105–119 (2003)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer India
About this paper
Cite this paper
Bethanney Janney, J., Aarthi, A., Rajesh Kumar Reddy, S. (2014). An Automatic MRI Brain Segmentation by Using Adaptive Mean-Shift Clustering Framework. In: Sathiakumar, S., Awasthi, L., Masillamani, M., Sridhar, S. (eds) Proceedings of International Conference on Internet Computing and Information Communications. Advances in Intelligent Systems and Computing, vol 216. Springer, New Delhi. https://doi.org/10.1007/978-81-322-1299-7_11
Download citation
DOI: https://doi.org/10.1007/978-81-322-1299-7_11
Published:
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-1298-0
Online ISBN: 978-81-322-1299-7
eBook Packages: EngineeringEngineering (R0)