Abstract
A wide variety of aromatic compounds are aerobically degraded by bacteria. Aromatic-ring hydroxylation is one of the most common initial degradation step and thus is an essential catalytic reaction for aromatic-ring degradation by bacteria in nature. Most of the cases, the hydroxylation is catalyzed by an oxygenase family known as Rieske nonheme iron (di)oxygenase or Rieske oxygenase (RO). ROs catalyze a broad range of aromatic-ring compounds including mono- and polycyclic aromatic and hetero-aromatic compounds. ROs are composed of terminal oxygenase and electron transfer components. The terminal oxygenase component has Rieske [2Fe-2S] cluster as a redox center for receive electrons from the electron transfer component(s) and mononuclear iron as a catalytic site for dioxygen activation. In addition to genetic and biochemical studies, their crystal structures have been extensively studied recently. To date (11/7/2012), the structures of 15 different terminal oxygenases and 11 electron transfer components have been determined and deposited to Protein Data Bank (76 structures including their variants in total).
This chapter will address structures and functions of aromatic-ring hydroxylating dioxygenases. The overviews of ROs (Sect. 9.1) and overall structure of ROs containing their electron transfer components (Sect. 9.2), inter- and intramolecular electron transfer in ROs (Sect. 9.3), catalytic mechanism (Sect. 9.4), and substrate specificity and enzyme engineering (Sect. 9.5) are reviewed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Aromatic ring
- Crystal structure
- Dioxygenation
- Electron transfer
- Hydroxylation
- Nonheme iron
- Protein–protein interaction
- Rieske oxygenase
- Substrate specificity
1 Aromatic-Ring Hydroxylating Dioxygenases
Aromatic compounds are widely distributed and abundant organic compounds in nature. They are derived from aromatic amino acids, lignin components, flavonoids, quinones, crude oils, etc. Some of the toxic xenobiotics, such as dioxins, polychlorinated biphenyls, and nitroaromatics, also contain aromatic ring(s). Aromatic compounds are utilized by microorganisms as the carbon source and/or electron donor for respiration. Bacterial aerobic metabolism of aromatic compounds has been extensively studied for their applications of bioremediation and chemical and pharmaceutical industries. Initial reaction of aromatic-ring degradation is generally dihydroxylation by Rieske nonheme iron dioxygenase (RO) to yield cis-dihydrodiols and derivatives with strict stereo- and regio-specificity (Boyd and Sheldrake 1998; Gibson and Parales 2000; Ullrich and Hofrichter 2007). ROs catalyze not only dioxygenation but also monooxygenation, sulfoxidation, denitrification, and demethylation (Fig. 9.1) (Rosche et al. 1995; Herman et al. 2005; Nojiri 2012). In contrast to bacterial ROs, less numbers of these family enzymes in higher organisms are found, but known ROs in plants and insects play essential roles in biological processes, such as chlorophyll metabolism, cell death, development, and cholesterol metabolism (Gray et al. 1997; Tanaka et al. 1998; Yoshiyama et al. 2006; Yoshiyama-Yanagawa et al. 2011). Among the ROs, relatively simple compounds, such as naphthalene and biphenyl, are the substrates for the best-studied RO members. In these ROs, enzymatic mechanisms based three-dimensional structures are understood in detail.
Oxidation of aromatic compounds catalyzed by Rieske oxygenases. Compounds in parenthesis are unstable and spontaneous reaction (dotted arrow) to form the diol compounds is shown, except for the dicamba monooxygenation reaction (in the monooxygenated compound of dicamba in parenthesis is proposed intermediate of the monooxygenation reaction and the 3,6-dichlorosalicylic acid as the final product is shown)
2 RO Structures
2.1 Classification
ROs are composed of terminal oxygenase and electron transfer components. Oxygenation reaction is catalyzed by the terminal oxygenase component and two electrons are required for the reaction (Wolfe et al. 2001). Electron transfer components transfer the electrons from NAD(P)H to terminal oxygenase component. Electron transfer components are only reductase or ferredoxin and ferredoxin reductase (Table 9.1). All the components contain redox center(s) for storage of electrons. According to Batie’s classification system, ROs are divided into five groups by their component numbers and prosthetic groups (Table 9.1) (Batie et al. 1991). In class I ROs, the electron transfer component is only reductase which contains FMN (class IA) or FAD (class IB) and plant-type [2Fe-2S] cluster as the cofactors. The reductase component possesses NAD(P)H binding site for transferring electrons from NAD(P)H to terminal oxygenase via FMN or FAD and the plant-type [2Fe-2S] cluster. ROs in class II have two electron transfer components, ferredoxin and ferredoxin reductase. The ferredoxin reductase in class II has only FAD as a cofactor. Ferredoxin components in class IIA and IIB have putidaredoxin-type [2Fe-2S] cluster and Rieske-type [2Fe-2S] cluster, respectively. The [2Fe-2S] cluster of putidaredoxin-type ferredoxin is coordinated by four cysteine residues, whereas that of Rieske-type ferredoxin is coordinated by two cysteine and two histidine residues. There are few examples of RO systems with [3Fe-4S]-type ferredoxin as the electron transfer component (Martin and Mohn 1999; Kim et al. 2006; Takagi et al. 2005). However, so far, they have not been classified into any class in Batie’s classification. Therefore, here we propose “class IIC” into which ROs with [3Fe-4S]-type ferredoxin as the examples are classified (Table 9.1). Class III ROs have both ferredoxin reductase and ferredoxin component as well as class II ROs, but the ferredoxin reductase component contains plant-type [2Fe-2S] cluster and the FAD cofactor similarly to the reductase component of class IB ROs.
2.2 Terminal Oxygenase Component
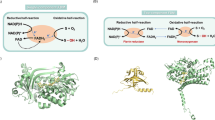
Terminal oxygenase in RO has Rieske-type [2Fe-2S] cluster and mononuclear nonheme iron. Crystal structures of naphthalene dioxygenase, biphenyl dioxygenase, carbazole dioxygenase, nitrobenzene dioxygenase, dicamba monooxygenase, 2-oxoquinoline 8-monooxygenase, toluene dioxygenase, cumene dioxygenase, and polycyclic hydrocarbon dioxygenase have been determined (Table 9.2). Terminal oxygenase component is usually an α3 or α3β3 subunit configuration (Fig. 9.2). The terminal oxygenase component of naphthalene dioxygenase (NDO) is the first example of terminal oxygenase structure of ROs (Kauppi et al. 1998). The terminal oxygenase of NDO is α3β3 configuration and resembles shape of mushroom. The height of the mushroom is 75 Å and the diameters of the cap and stem are 102 Å and 50 Å, respectively. The terminal oxygenase component of carbazole dioxygenase (CARDO) and 2-oxoquinoline 8-monooxygenase are the first examples of RO which have α3 configuration (Nojiri et al. 2005; Martins et al. 2005). The terminal oxygenase of CARDO is a doughnut shape with 100 Å in diameter, a central hole in 30 Å, and 45 Å thickness. Both of the redox centers (Rieske [2Fe-2S] cluster and mononuclear nonheme iron) and substrate-binding site are in the α-subunit. The β-subunit is thought to have a structural role in the holoenzyme but, in biphenyl dioxygenase (BDO) from Pandoraea pnomenusa (formerly Comamonas testosteroni) strain B-356, the β-subunit influences substrate specificity (Hurtubise et al. 1998). The α-subunit is consist of two domains, Rieske and catalytic domains. One iron (Fe1) of the Rieske cluster is coordinated by two cysteine residues and the other iron (Fe2) is coordinated by two histidine residues. Two inorganic sulfide ions bridge the two iron ions, forming a flat rhombic arrangement. This domain of ROs is similar to the Rieske iron-sulfur protein of the cytochrome bc 1 complex in the electron transport chain for ATP generation (Iwata et al. 1996) and chloroplast b 6 f complex (Carrell et al. 1997). The Rieske domain of terminal oxygenase component is likely to be derived from a common ancestral Rieske protein with these Rieske proteins because of their similarities. The distance between the Rieske center and the mononuclear iron within a single α subunit is further from the distance between the Rieske center and the iron in a neighboring α subunit. Therefore, electrons are thought to transfer from a Rieske center to a mononuclear iron in the neighboring α subunit (Fig. 9.2). The catalytic domain of RO is dominated by a stranded antiparallel β-sheet which extends into the middle of the domain. One side of the large β-sheet is covered with α-helices that contain ligands, an aspartate, and two histidines, to the mononuclear iron. A channel from molecular surface, which enables substrates to access to the active site, is formed by the sheet and the helices. The region close to the active site iron is primarily composed of hydrophobic residues so that the terminal oxygenase of ROs can accommodate hydrophobic substrates.
Overall structures of Rieske oxygenase components. The iron atom, sulfur atom, FAD, and carbazole are shown in brown sphere, yellow sphere, yellow sticks, and yellow and blue sticks (imino nitrogen), respectively. (a) The terminal oxygenase component with α3 configuration (blue; PDB entry code: 2DE7), ferredoxin (red; PDB entry code: 1VCK), and ferredoxin reductase (orange; unpublished) of carbazole 1,9a-dioxygenase. Schematic electron transfer is also presented. Magnified view of the substrate-binding pocket is shown in the in set. (b) The terminal oxygenase of naphthalene dioxygenase with α3β3 configuration (PDB entry code: 1NDO). The α and β subunits of the terminal oxygenase component are colored in blue and green, respectively
2.3 Electron Transfer Components
The crystal structures of the electron transfer components of ROs have not been determined as many as the terminal oxygenase components (Table 9.2), but the structures have offered important insights into structure-function relationships of electron carrier proteins.
2.3.1 Reductase
The first three-dimensional structure of class I reductase component is phthalate dioxygenase from Burkholderia cepacia strain DB01 (Table 9.2) (Correll et al. 1992). This protein has three individual domains: FMN, NAD, and [2Fe-2S] domains (N-terminal to C-terminal). The FMN-binding, [2Fe-2S] cluster, and NADH-binding domains are brought together near a central cleft in the molecule with only 4.9 Å separating the 8-methyl group of FMN and a cysteine sulfur ligated to iron. The phthalate dioxygenase belongs to class IA but another example of reductase structure of class I, benzoate dioxygenase, belongs to class IB. The reductase component of benzoate dioxygenase from Acinetobacter baylyi strain ADP1 also has three domains, [2Fe-2S] cluster, FAD-binding, and NADH-binding domains (N-terminal to C-terminal), with different order from that of phthalate dioxygenase (Karlsson et al. 2002).
2.3.2 Ferredoxin
ROs in class II have a ferredoxin component which transfers electrons from ferredoxin reductase component to terminal oxygenase. Crystal structure of ferredoxin component of BDO (BphF) from Burkholderia xenovorans strain LB400 was determined as the first example of RO ferredoxin (Colbert et al. 2000). This small protein composed of 109 amino acid residues has a Rieske [2Fe-2S] cluster which is located at the apex of this molecule with its two histidine ligands exposed to solvent. Redox potential of BphF was approximately −150 mV, whereas Rieske proteins from the bc 1 or b 6 f complexes have redox potentials near +300 mV. Some other ferredoxin structures of ROs; two different CARDOs, NDO, toluene dioxygenase; and other two different BDOs have been determined (Table 9.2). They are primarily similar to BphF protein at structural level except for ferredoxin of CARDO from Novosphingobium sp. strain KA1, which is not Rieske-type ferredoxin but putidaredoxin type. This protein structure is round shape rather than wedge shape as the Rieske-type ferredoxins (Umeda et al. unpublished). This difference probably influences interaction with terminal oxygenase and ferredoxin reductase component.
2.3.3 Ferredoxin Reductase
Crystal structure of ferredoxin reductase (BphA4) in class IIB from Acidovorax sp. strain KKS102 with and without NADH has been determined as the first example of ferredoxin reductase in this class (Senda et al. 2000). This protein has significant similarity to not only ferredoxin reductases of ROs in other bacteria but also putidaredoxin reductase of P450cam system of Pseudomonas putida, and it has essentially the same fold as the enzymes of the glutathione reductase family. This protein has three domains, FAD-binding, NADH-binding, and C-terminal domain. The FAD-binding and C-terminal domains have interacting surface areas for its counterpart, ferredoxin component (BphA3) (Senda et al. 2007). Crystal structures of ferredoxin reductase components of CARDOs from Janthinobacterium sp. strain J3 and Novosphingobium sp. strain KA1 and toluene dioxygenase from Pseudomonas putida strain F1 have been also determined (Table 9.2). They are essentially similar to BphA4 structure.
3 Electron Transfer in Rieske Oxygenase
Both intra- and intermolecular electron transfers in ROs are essential for catalytic activity. Redox potential of each redox center is an important factor for electron transfer but limited numbers of reports which demonstrated experimentally measured redox potentials (Table 9.3). The Rieske proteins have reduction potentials ranging from −150 to +400 mV which is proposed to be due to electrostatic environment of the cluster and/or structural differences, whereas the redox centers in RO are relatively lower (Colbert et al. 2000; Brown et al. 2008). The redox potential of Rieske proteins is known to be dependent on pH probably because of the two iron-coordinating histidines. The wide range redox potential of Rieske proteins is first proposed to be due to solvent accessibility or protein structure. However, determined structures of Rieske proteins are significantly similar and do not have large variations in solvent accessibility. Thus, the redox potential is likely to be determined by structural difference rather than solvent accessibility. This is supported by biochemical studies using mutated enzymes (Klingen and Ullmann 2004).
One redox center must be in close proximity to the other so that electron transfer occurs. In class II or III ROs, the ferredoxin component shuttles between the ferredoxin reductase and terminal oxygenase components and thus the ferredoxin noncovalently binds to the other components. To date, two electron transfer complexes have been reported: ferredoxin reductase (BphA4) and ferredoxin (BphA3) of BDO from Acidovorax sp. strain KKS102 and ferredoxin and terminal oxygenase of CARDO (from Pseudomonas resinovorans strain CA10 and Janthinobacterium sp. strain J3, respectively). These crystal structures have revealed interacting site, binding force, and conformational changes by complex formation. The crystal structure of BphA4 and BphA3 complex demonstrated BphA3 binds to middle of BphA4 by hydrophobic interaction (four tryptophan residues in BphA4 and two proline residues in BphA3), electrostatic-like interaction (negatively charged surface of BphA4 interacts with positively charged BphA3 surface), and hydrogen bonds (Fig. 9.3a) (Senda et al. 2007). Electrons are likely to be transferred from FAD in BphA4 to the [2Fe-2S] cluster in BphA3 via Trp320 of BphA4 and His66 of BphA3. Structural comparison between the free and complex forms of BphA3 and BphA4 revealed conformational changes, clockwise rotation of NADH and C-terminal domain of BphA4, side chain rotation of His66 (a ligand in [2Fe-2S] cluster) of BphA3, and the peptide bond between Gly46 and Glu47 of BphA3. This report also presents the butterfly-like motion of the isoalloxazine ring of FAD along with redox status. The other electron transfer complex of ferredoxin and terminal oxygenase of CARDO revealed interacting amino acid residues between the components (Fig. 9.3b, c) (Ashikawa et al. 2006). Ferredoxin binds to boundary of the terminal oxygenase subunits by electrostatic and hydrophobic interactions. Free forms of ferredoxin and terminal oxygenase component of CARDO have also been determined. Detailed comparison revealed two significant conformational changes of main chain, Lys12-Trp15 and Asp347-Asn352 of the terminal oxygenase component. These amino acid residues are located at the component boundary to form hydrophobic interactions and the hydrophobic residues: Trp15 and Val351 of Oxy interact with Phe67 and Pro83 of Fd, respectively. On the interacting molecular surfaces, electrostatic interactions (Lys13, Arg118, Arg210, and Glu353 of the terminal oxygenase interact with Glu64, Glu43, Glu55, and His68 of the ferredoxin component, respectively) were also observed. According to the complex structure, electron transfer pathway from the terminal oxygenase and the ferredoxin was proposed (Fig. 9.3c). The distance between the Rieske cluster of ferredoxin and mononuclear iron of the terminal oxygenase is approximately 22 Å. This is too far for the direct electron transfer without Rieske cluster of the terminal oxygenase. However, the average distance between the ligand atoms of the Rieske clusters of terminal oxygenase and ferredoxin was approximately 12–13 Å which is shorter than 14 Å threshold, the limit of electron tunneling in a protein medium (Page et al. 1999).
The crystal structures of the electron transfer complexes. (a) The ferredoxin and ferredoxin reductase complex of biphenyl dioxygenase from Acidovorax sp. strain KKS102 (PDB entry code: 2YVJ). The ferredoxin and ferredoxin reductase are colored in red and orange, respectively. The [2Fe-2S] cluster, FAD, and NAD+ molecules are shown as the brown and yellow balls, yellow sticks, and green sticks, respectively. The Rieske [2Fe-2S] cluster of the ferredoxin binds closely to the FAD and NAD+ molecules in the middle of ferredoxin reductase component. (b) The terminal oxygenase and ferredoxin complex of carbazole dioxygenase from Janthinobacterium sp. strain J3 and Pseudomonas resinovorans strain CA10 (PDB entry code: 2DE7). Chains A, B, C (terminal oxygenase) and chains D, E, F (ferredoxin) are colored in light blue, marine, deep blue, light pink, red, and dark pink, respectively. The iron atom, sulfur atom, and carbazole are shown in brown sphere, yellow sphere, and yellow and blue (imino nitrogen) sticks, respectively. The ferredoxin components bind to the terminal oxygenase one for one in close proximity between each Rieske [2Fe-2S] cluster. (c) Stereoview of proposed electron transfer pathways between the ferredoxin and terminal oxygenase of carbazole dioxygenase. Two possible routes are presented as orange and red arrows, according to a report by Ashikawa et al. (2006)
The distance from Rieske [2Fe-2S] cluster of one terminal oxygenase α subunit and mononuclear iron of the neighboring α subunit is shorter than 14 Å as well. A conserved aspartate residue (D180 in CARDO; Fig. 9.3c) binds to a histidine ligand to the Rieske cluster and a histidine ligand to the mononuclear iron. A mutational study of the corresponding residue to the aspartate residue in toluene dioxygenase from Pseudomonas putida strain F1 (Asp219) demonstrated this residue is essential for enzyme activity (Jiang et al. 1996; Friemann et al. 2009), indicating the aspartate residue construct a main path for electron transfer from the Rieske cluster to mononuclear iron.
4 Catalytic Mechanism
In ROs, dioxygen is reductively activated as well as other oxidase and oxygenase enzymes. Biochemical studies of the ROs, especially for single turnover studies by regulating the enzymes to be oxidized and reduced, have provided important insights into reaction mechanisms. A scheme of proposed catalytic mechanism of ROs based on the previous studies (Ashikawa et al. 2012; Karlsson et al. 2003; Neibergall et al. 2007; Wolfe and Lipscomb 2003; Wolfe et al. 2001, 2002; Kovaleva and Lipscomb 2008) is presented in Fig. 9.4. Single turnover studies of NDO have revealed two electrons (reduction of one Fe of Rieske [2Fe-2S] cluster and the mononuclear iron), and substrate binding is required for oxygen reactivity (Wolfe et al. 2001). After substrate binding and oxygen reactivation, an Fe(III)-OOH intermediate is likely to undergo O–O bond homolytic fission to yield an Fe(V)-oxo-hydroxo intermediate. The cis-hydroxylated product is released when one or two electron(s) come from the electron transfer component. ROs with the Rieske cluster oxidized and the mononuclear iron reduced are under resting state. Addition of H2O2 to NDO under resting state allows reaction with the substrate to yield the product (Fig. 9.4) (Wolfe and Lipscomb 2003). This is so-called “peroxide shunt” reaction that does not require a reduced Rieske cluster. In benzoate 1,2-dioxygenase, even fully oxidized form with the Rieske cluster oxidized and mononuclear iron in the Fe(III) state can utilized H2O2 as a source of reduced oxygen atom to form the appropriate product from benzoate (Neibergall et al. 2007). Several crystal structures with substrate and oxygen and reaction intermediates suggested further detailed reaction mechanisms. Karlsson and coworkers revealed that a molecular oxygen species bound to the mononuclear iron in a side-on fashion by determining crystal structures of NDO with substrate and dioxygen (Karlsson et al. 2003). The side-on binding of dioxygen enables each oxygen to attach neighboring carbons from the same face of the aromatic ring to produce the cis-dihydrodiols. However, when NO is introduced instead of O2, NO is bound end-on to the mononuclear iron, whereas NO is commonly used as an analogue for dioxygen (Karlsson et al. 2005). Crystal structures of CARDO with substrate, dioxygen, and both carbazole and dioxygen revealed that a room for oxygen binding in the substrate-binding pocket is created by substrate binding, suggesting the substrate binds to the active site before dioxygen binding (Ashikawa et al. 2012). On the other hand, CARDO with the substrate revealed conformational changes with closure of the entranceway of the substrate by substrate binding (Fig. 9.5b, c) (Ashikawa et al. 2006). This conformational change seems to trap the substrate at the substrate-binding site and to exclude water from the active site during turnover in order to minimize the risk of leakage of partially processed substrates.
Scheme of proposed catalytic cycles of Rieske oxygenases. Oxidized Rieske [2Fe-2S] cluster, reduced Rieske [2Fe-2S] cluster, oxidized nonheme iron, and reduced nonheme iron are shown as [2Fe-2S]ox, [2Fe-2S]red, Fe3+, and Fe2+, respectively. Oxidized, reduced, and resting state of the redox centers in the terminal oxygenase are shown in red, blue, and purple background. Solid arrows indicate the proposed main pathway taking account into the biochemically and structurally well-characterized ROs. Broken arrow shows the possible pathway based on biochemical studies and the complex structures determined
Substrate-binding site of RO with a substrate (crystal structure of carbazole 1,9 a-dioxygenase and carbazole complex by Ashikawa et al. 2006). (a) Stereoview of carbazole bound to the substrate-binding pocket of CARDO. The amino acid residues which contact the substrate are shown in light pink sticks and carbazole is shown in a white and blue (imino nitrogen) sticks. (b) Superposition of the entrance of the substrate-binding pocket of CARDO. The carbazole-free and carbazole-binding structures are shown in blue green and green and orange and pink, respectively. (c and d) The surface plots of the carbazole-free (c) and carbazole-binding structures (d). White arrows indicate the entrance of the substrate-binding pocket. The insets show the molecular surfaces viewed from the orientation directed by the white arrows (Reprinted from Ashikawa et al. (2006), with permission of Elsevier)
5 Substrate Specificity
ROs are known for their broad range of substrate (not only aromatic compounds but cholesterols and chlorophylls as described above). ROs also catalyze monooxygenation, sulfoxidation, O- and N-dealkylation. Substrate specificity analyses and recent structural studies revealed what determines the substrate specificity and the relationship between structure and substrate specificity, which provides critical clues to the modification of the substrate-binding pocket to obtain an enhanced catalyst for bioremediation (degradation) of toxic contaminants and enzymatic synthesis of chiral precursors for the production of specialty compounds. Several examples of studies on mutant enzymes demonstrated that regio- and enantio-selectivity can be altered by the mutation as follows (Boyd and Sheldrake 1998).
BDO is one of the best-characterized RO by its mutant and chimeric enzymes (Furukawa et al. 2004). Randomly recombined terminal oxygenase components of BDO from Pseudomonas pseudoalcaligenes strain KF707 and Burkholderia xenovorans strain LB400 by DNA shuffling showed expanded substrate specificities not only for biphenyl derivatives and polychlorinated biphenyl (PCB) but for single aromatic compounds (Kumamaru et al. 1998). Subsequent random priming and site-directed mutants of BDO from strain KF707 showed expanded substrate range and different regio-specificities for various polychlorinated biphenyls from the wild-type enzyme (Suenaga et al. 2001, 2002). Many of the mutants which showed different substrate specificity from the wild-type enzyme had mutation at hydrophobic amino acid residues, such as Phe227, Ile335, and Phe377. According to structural studies of ROs, the substrate-binding pocket is commonly composed of hydrophobic residues consistent with RO’s preference for hydrophobic substrates (Fig. 9.5a). These mutational studies indicate that hydrophobic residues also play an important role in determining substrate-binding orientation. BDO from Burkholderia xenovorans strain LB400 is also well-studied enzyme by mutations. Mutated terminal oxygenase BphAEp4 (Thr335Ala/Phe336Met) and its derivative BphAERR41 metabolize dibenzofuran two and three times faster than wild-type enzyme, respectively (Mohammadi et al. 2011). The crystal structures of BphAEp4 and BphAERR41 revealed that replacement altered the constraints (hydrogen bonds) or plasticity of the substrate-binding pocket and increased available space for bulkier compounds than biphenyl, such as dibenzofuran and the doubly ortho-chlorinated congener 2,6-dichlorobiphenyl (Kumar et al. 2011; Mohammadi et al. 2011).
NDO is also one of the extensively studied ROs by mutational studies. Mutant enzymes of NDO from Pseudomonas putida strain NCIB9816-4 have demonstrated alteration of a series of amino acid residues composing the substrate-binding pocket can affect the regio-selectivity of product formation (Parales et al. 2000a, b; Yu et al. 2001). Detailed comparison between wild-type NDO and its amino acid replacement variants in which Phe352 is substituted by valine demonstrated region- and stereo-selectivity was principally dependent on the orientation of the substrate binding at the active site (Ferraro et al. 2006).
Another example of mutational study based on the crystal structure is CARDO from Janthinobacterium sp. strain J3. A series of site-directed mutagenesis variants in which the mutated amino acid residues are located at substrate-binding pocket wall showed different oxygenation site to hetero-aromatic compounds (Uchimura et al. 2008). Among the mutant enzymes, Ile262Val and Gln282Tyr converted carbazole to 1-hydroxycarbazole which is probably a dehydrated compound of lateral dioxygenation product (cis-1,2-dihydroxy-1,2-dihydrocarbazole) with higher yield than the wild-type enzyme. Another mutated variant Phe275Trp converted fluorene to 4-hydroxyfluorene (probable dehydration product of cis-3,4-dihydroxy-3,4-dihydrofluorene), whereas wild-type enzyme cannot catalyze this substrate. This variant also could oxidize fluoranthene to yield unidentified cis-dihydrodiol. Subsequent structural studies of mutated CARDO variants revealed difference in substrate-binding orientation between the variants and wild-type CARDO (Inoue et al. unpublished). In Ile262Val and Gln282Tyr variant structures with carbazole, the substrate bound to the active site rotated approximately 15° and 25°, respectively, from the substrate position of the wild type. In CARDO, imino nitrogen of carbazole forms hydrogen bonds with carbonyl oxygen of Gly178 at the active site (Fig. 9.5a). This hydrogen bond is likely to stabilize the substrate binding [in 2-oxoquinoline-8-monooxygenase from Pseudomonas putida, strain 86 also has similar hydrogen bond between NH group of the substrate, 2-oxoquinoline, and carbonyl oxygen of Gly216 (Martins et al. 2005)]. In the mutant enzymes, there is no “direct” hydrogen bonds between carbazole and the carbonyl oxygen of Gly178 but, instead, two water molecules were introduced into the space which created by the substrate rotation, and they formed different hydrogen bond networks with substrate and carbonyl oxygen of Gly178. The structural analysis of the mutant enzymes of CARDO suggested that different substrate orientation stabilized by the hydrogen bond networks increased formation of the lateral dioxygenation product.
Protein engineering technology combined with three-dimensional structure information has enabled alteration of substrate specificity of ROs. However, limited numbers of modification of the substrate specificity of ROs were reported so far. Further structural and biochemical studies of ROs will develop novel catalytic reactions and applicable enzymes.
6 Outlook
In this chapter structural and functional properties of aromatic-ring hydroxylating dioxygenases which is known as Rieske oxygenases were presented. In addition to the genetic and biochemical studies, recent structural studies of ROs have demonstrated interactions between the components, the substrate recognition, and catalytic mechanisms at the atomic level. Considering that ROs have been found to be extensively diverse and widespread in nature (Capyk and Eltis 2012; Iwai et al. 2011), basic structural and functional studies on each enzyme are becoming more and more important. The structural information and mutational studies with genetic and biochemical approach help each other to generate enzyme which is attractive to application for bioremediation and industrial biocatalysis.
References
Armengaud J, Timmis KN (1997) Molecular characterization of Fdx1, a putidaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase of Sphingomonas sp. RW1. Eur J Biochem 247:833–842
Armengaud J, Gaillard J, Timmis KN (2000) A second [2Fe-2S] ferredoxin from Sphingomonas sp. Strain RW1 can function as an electron donor for the dioxin dioxygenase. J Bacteriol 182:2238–2244
Ashikawa Y, Fujimoto Z, Noguchi H, Habe H, Omori T, Yamane H, Nojiri H (2006) Electron transfer complex formation between oxygenase and ferredoxin components in Rieske nonheme iron oxygenase system. Structure 14:1779–1789
Ashikawa Y, Fujimoto Z, Usami Y, Inoue K, Noguchi H, Yamane H, Nojiri H (2012) Structural insight into the substrate- and dioxygen-binding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9a-dioxygenase. BMC Struct Biol 12:15
Batie CJ, Ballou DP, Correll CC (1991) In: Muller F (ed) Chemistry and biochemistry of flavoenzymes, vol 3. CRC Press, Boca Raton, pp 543–556
Beharry ZM, Eby DM, Coulter ED, Viswanathan R, Neidle EL, Phillips RS, Kurtz DM Jr (2003) Histidine ligand protonation and redox potential in the Rieske dioxygenases: role of a conserved aspartate in anthranilate 1,2-dioxygenase. Biochemistry 42:13625–13636
Boyd DR, Sheldrake GN (1998) The Dioxygenase-catalysed formation of vicinal cis-diols. Nat Prod Rep 15:309–324
Brown EN, Friemann R, Karlsson A, Parales JV, Couture MM, Eltis LD, Ramaswamy S (2008) Determining Rieske cluster reduction potentials. J Biol Inorg Chem 13:1301–1313
Capyk JK, Eltis LD (2012) Phylogenetic analysis reveals the surprising diversity of an oxygenase class. J Biol Inorg Chem 17:425–436
Carredano E, Karlsson A, Kauppi B, Choudhury D, Parales RE, Parales JV, Lee K, Gibson DT, Eklund H, Ramaswamy S (2000) Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J Mol Biol 296:701–712
Carrell CJ, Zhang H, Cramer WA, Smith JL (1997) Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein. Structure 5:1613–1625
Chakraborty S, Behrens M, Herman PL, Arendsen AF, Hagen WR, Carlson DL, Wang XZ, Weeks DP (2005) A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6: purification and characterization. Arch Biochem Biophys 437:20–28
Colbert CL, Couture MM, Eltis LD, Bolin JT (2000) A cluster exposed: structure of the Rieske ferredoxin from biphenyl dioxygenase and the redox properties of Rieske Fe-S proteins. Structure 8:1267–1278
Correll CC, Batie CJ, Ballou DP, Ludwig ML (1992) Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 258:1604–1610
Couture MM, Colbert CL, Babini E, Rosell FI, Mauk AG, Bolin JT, Eltis LD (2001) Characterization of BphF, a Rieske-type ferredoxin with a low reduction potential. Biochemistry 40:84–92
Dong X, Fushinobu S, Fukuda E, Terada T, Nakamura S, Shimizu K, Nojiri H, Omori T, Shoun H, Wakagi T (2005) Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J Bacteriol 187:2483–2490
D'Ordine RL, Rydel TJ, Storek MJ, Sturman EJ, Moshiri F, Bartlett RK, Brown GR, Eilers RJ, Dart C, Qi Y, Flasinski S, Franklin SJ (2009) Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation. J Mol Biol 392:481–497
Dumitru R, Jiang WZ, Weeks DP, Wilson MA (2009) Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation. J Mol Biol 392:498–510
Ferraro DJ, Okerlund AL, Mowers JC, Ramaswamy S (2006) Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase. J Bacteriol 188:6986–6994
Ferraro DJ, Brown EN, Yu CL, Parales RE, Gibson DT, Ramaswamy S (2007) Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1. BMC Struct Biol 7:10
Friemann R, Ivkovic-Jensen MM, Lessner DJ, Yu C-L, Gibson DT, Parales RE, Eklund H, Ramaswamy S (2005) Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase. J Mol Biol 348:1139–1151
Friemann R, Lee K, Brown EN, Gibson DT, Eklunda H, Ramaswamy S (2009) Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system. Acta Crystallogr Sect D 65:24–33
Furukawa K, Suenaga H, Goto M (2004) Biphenyl dioxygenases: functional versatilities and directed evolution. J Bacteriol 186:5189–5196
Furusawa Y, Nagarajan V, Tanokura M, Masai E, Fukuda M, Senda T (2004) Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J Mol Biol 342:1041–1052
Gakhar L, Malik ZA, Allen CC, Lipscomb DA, Larkin MJ, Ramaswamy S (2005) Structure and increased thermostability of Rhodococcus sp. naphthalene 1,2-dioxygenase. J Bacteriol 187:7222–7231
Gassner GT, Ludwig ML, Gatti DL, Correll CC, Ballou DP (1995) Structure and mechanism of the iron-sulfur flavoprotein phthalate dioxygenase reductase. FASEB J 9:1411–1418
Geary PJ, Saboowalla F, Patil D, Cammack R (1984) An investigation of the iron-sulphur proteins of benzene dioxygenase from Pseudomonas putida by electron-spin-resonance spectroscopy. Biochem J 217:667–673
Gibson DT, Parales RE (2000) Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol 11:236–243
Gray J, Close PS, Briggs SP, Johal GS (1997) A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89:25–31
Herman PL, Behrens M, Chakraborty S, Chrastil BM, Barycki J, Weeks DP (2005) A Three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6. J Biol Chem 280:24759–24767
Hurtubise Y, Barriault D, Sylvestre M (1998) Involvement of the terminal oxygenase beta subunit in the biphenyl dioxygenase reactivity pattern toward chlorobiphenyls. J Bacteriol 180:5828–5835
Inoue K, Ashikawa Y, Umeda T, Abo M, Katsuki J, Usami Y, Noguchi H, Fujimoto Z, Terada T, Yamane H, Nojiri H (2009) Specific interactions between the ferredoxin and terminal oxygenase components of a class IIB Rieske nonheme iron oxygenase, carbazole 1,9a-dioxygenase. J Mol Biol 392:436–451
Iwai S, Johnson TA, Chai B, Hashsham SA, Tiedje JM (2011) Comparison of the specificities and efficacies of primers for aromatic dioxygenase gene analysis of environmental samples. Appl Environ Microbiol 77:3551–3557
Iwata S, Saynovits M, Link TA, Michel H (1996) Structure of a water soluble fragment of the 'Rieske' iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 Å resolution. Structure 4:567–579
Jakoncic J, Jouanneau Y, Meyer C, Stojanoff V (2007) The crystal structure of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. FEBS J 274:2470–2481
Jiang H, Parales RE, Lynch NA, Gibson DT (1996) Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol 178:3133–3139
Karlsson A, Beharry ZM, Matthew Eby D, Coulter ED, Neidle EL, Kurtz DM Jr, Eklund H, Ramaswamy S (2002) X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1. J Mol Biol 318:261–272
Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S (2003) Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039–1042
Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S (2005) NO binding to naphthalene dioxygenase. J Biol Inorg Chem 10:483–489
Kauppi B, Lee K, Carredano E, Parales RE, Gibson DT, Eklund H, Ramaswamy S (1998) Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571–586
Kim SJ, Kweon O, Freeman JP, Jones RC, Adjei MD, Jhoo JW, Edmondson RD, Cerniglia CE (2006) Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 72:1045–1054
Klingen AR, Ullmann GM (2004) Negatively charged residues and hydrogen bonds tune the ligand histidine pKa values of Rieske iron-sulfur proteins. Biochemistry 43:12383–12389
Kovaleva EG, Lipscomb JD (2008) Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat Chem Biol 4:186–193
Kumamaru T, Suenaga H, Mitsuoka M, Watanabe T, Furukawa K (1998) Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat Biotechnol 16:663–666
Kumar P, Mohammadi M, Viger JF, Barriault D, Gomez-Gil L, Eltis LD, Bolin JT, Sylvestre M (2011) Structural insight into the expanded PCB-degrading abilities of a biphenyl dioxygenase obtained by directed evolution. J Mol Biol 405:531–547
Kumar P, Mohammadi M, Dhindwal S, Pham TT, Bolin JT, Sylvestre M (2012) Structural insights into the metabolism of 2-chlorodibenzofuran by an evolved biphenyl dioxygenase. Biochem Biophys Res Commun 421:757–762
Martin VJ, Mohn WW (1999) A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J Bacteriol 181:2675–2682
Martins BM, Svetlitchnaia T, Dobbek H (2005) 2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure 13:817–824
Mohammadi M, Viger JF, Kumar P, Barriault D, Bolin JT, Sylvestre M (2011) Retuning Rieske-type oxygenases to expand substrate range. J Biol Chem 286:27612–27621
Nam J-W, Noguchi H, Fujimoto Z, Mizuno H, Ashikawa Y, Abo M, Fushinobu S, Kobashi N, Wakagi T, Iwata K, Yoshida T, Habe H, Yamane H, Omori T, Nojiri H (2005) Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system. Proteins 58:779–789
Neibergall MB, Stubna A, Mekmouche Y, Munck E, Lipscomb JD (2007) Hydrogen peroxide dependent cis-dihydroxylation of benzoate by fully oxidized benzoate 1,2-dioxygenase. Biochemistry 46:8004–8016
Nojiri H (2012) Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci Biotechnol Biochem 76:1–18
Nojiri H, Ashikawa Y, Noguchi H, Nam JW, Urata M, Fujimoto Z, Uchimura H, Terada T, Nakamura S, Shimizu K, Yoshida T, Habe H, Omori T (2005) Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase. J Mol Biol 351:355–370
Page CC, Moser CC, Chen X, Dutton PL (1999) Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402:47–52
Parales RE, Lee K, Resnick SM, Jiang H, Lessner DJ, Gibson DT (2000a) Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J Bacteriol 182:1641–1649
Parales RE, Resnick SM, Yu CL, Boyd DR, Sharma ND, Gibson DT (2000b) Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the alpha subunit. J Bacteriol 182:5495–5504
Riedel A, Fetzner S, Rampp M, Lingens F, Liebl U, Zimmermann JL, Nitschke W (1995) EPR, electron spin echo envelope modulation, and electron nuclear double resonance studies of the 2Fe2S centers of the 2-halobenzoate 1,2-dioxygenase from Burkholderia (Pseudomonas) cepacia 2CBS. J Biol Chem 270:30869–30873
Rosche B, Fetzner S, Lingens F, Nitschke W, Riedel A (1995) The 2Fe2S centres of the 2-oxo-1,2-dihydroquinoline 8-monooxygenase from Pseudomonas putida 86 studied by EPR spectroscopy. Biochim Biophys Acta 1252:177–179
Senda T, Yamada T, Sakurai N, Kubota M, Nishizaki T, Masai E, Fukuda M, Mitsuidagger Y (2000) Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase. J Mol Biol 304:397–410
Senda M, Kishigami S, Kimura S, Fukuda M, Ishida T, Senda T (2007) Molecular mechanism of the redox-dependent interaction between NADH-dependent ferredoxin reductase and Rieske-type [2Fe-2S] ferredoxin. J Mol Biol 373:382–400
Subramanian V, Liu TN, Yeh WK, Serdar CM, Wackett LP, Gibson DT (1985) Purification and properties of ferredoxin TOL. A component of toluene dioxygenase from Pseudomonas putida F1. J Biol Chem 260:2355–2363
Suenaga H, Goto M, Furukawa K (2001) Emergence of multifunctional oxygenase activities by random priming recombination. J Biol Chem 276:22500–22506
Suenaga H, Watanabe T, Sato M, Ngadiman FK (2002) Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J Bacteriol 184:3682–3688
Takagi T, Habe H, Yoshida T, Yamane H, Omori T, Nojiri H (2005) Characterization of [3Fe-4S] ferredoxin DbfA3, which functions in the angular dioxygenase system of Terrabacter sp. strain DBF63. Appl Microbiol Biotechnol 68:336–345
Tanaka A, Ito H, Tanaka R, Tanaka NK, Yoshida K, Okada K (1998) Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc Natl Acad Sci USA 95:12719–12723
Uchimura H, Horisaki T, Umeda T, Noguchi H, Usami Y, Li L, Terada T, Nakamura S, Shimizu K, Takemura T, Habe H, Furihata K, Omori T, Yamane H, Nojiri H (2008) Alteration of the substrate specificity of the angular dioxygenase carbazole 1,9a-dioxygenase. Biosci Biotechnol Biochem 72:3237–3248
Ullrich R, Hofrichter M (2007) Enzymatic hydroxylation of aromatic compounds. Cell Mol Life Sci 64:271–293
Wolfe MD, Lipscomb JD (2003) Hydrogen peroxide-coupled cis-diol formation catalyzed by naphthalene 1,2-dioxygenase. J Biol Chem 278:829–835
Wolfe MD, Parales JV, Gibson DT, Lipscomb JD (2001) Single turnover chemistry and regulation of O2 activation by the oxygenase component of naphthalene 1,2-dioxygenase. J Biol Chem 276:1945–1953
Wolfe MD, Altier DJ, Stubna A, Popescu CV, Münck E, Lipscomb JD (2002) Benzoate 1,2-dioxygenase from Pseudomonas putida: single turnover kinetics and regulation of a two-component Rieske dioxygenase. Biochemistry 41:9611–9626
Yoshiyama T, Namiki T, Mita K, Kataoka H, Niwa R (2006) Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133:2565–2574
Yoshiyama-Yanagawa T, Enya S, Shimada-Niwa Y, Yaguchi S, Haramoto Y, Matsuya T, Shiomi K, Sasakura Y, Takahashi S, Asashima M, Kataoka H, Niwa R (2011) The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J Biol Chem 286:25756–25762
Yu CL, Parales RE, Gibson DT (2001) Multiple mutations at the active site of naphthalene dioxygenase affect regioselectivity and enantioselectivity. J Ind Microbiol Biotechnol 27:94–103
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Japan
About this chapter
Cite this chapter
Inoue, K., Nojiri, H. (2014). Structure and Function of Aromatic-Ring Hydroxylating Dioxygenase System. In: Nojiri, H., Tsuda, M., Fukuda, M., Kamagata, Y. (eds) Biodegradative Bacteria. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54520-0_9
Download citation
DOI: https://doi.org/10.1007/978-4-431-54520-0_9
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54519-4
Online ISBN: 978-4-431-54520-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)