Abstract
Density functional theory (DFT) provides some of the most important methods used in computational theory today. They allow one to determine the electronic structure of finite chemical systems, be they molecules or clusters, using a quantum-mechanical model, and exposes, thus, the great majority of the systems’ properties relevant to chemical applications. However, the numerical treatment of large chemical systems proves to be expensive, requiring elaborate parallelisation strategies.This paper presents two recent developments which aim at improving the parallel scalability of the quantum chemistry code ParaGauss. First, we introduce a new Fortran interface to parallel matrix algebra and its library implementation. This interface specifies a set of distributed data objects, combined with a set of linear algebra operators. Thus, complicated algebraic expressions can be expressed efficiently in pseudo-mathematical notation, while the numerical computations are carried out by back-end parallel routines. This technique is evaluated on relativistic transformations, as implemented in ParaGauss.The second development addresses the solution of the generalized matrix eigenvalue problem—an inherent step in electronic structure calculations. In the case the symmetry of a molecule is exploited, pertinent matrices expose a block-diagonal structure which makes the efficient use of existing parallel eigenvalue solvers difficult. We discuss a technique that uses a malleable parallel task scheduling (MPTS) algorithm for scheduling instances of parallel ScaLAPACK-routines on the available processor resources. This technique significantly improves the parallel performance of this numerical step, reducing the corresponding execution time to below 1 s in most applications considered.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- High performance computing
- Parallel numerical algebra
- Density functional theory
- Relativistic quantum chemistry
- Scheduling algorithms
1 Introduction
Many current implementations of first-principles electronic structure methods for molecules, clusters, and local models of surfaces and solids are limited in their parallel scalability due to the intrinsic diversity of structures and algorithms involved in various subtasks. We refer here to methods that aim at exploiting advantages of expansion techniques based on localized (Gaussian-type) functions. The inherent structure of the iterative self-consistent field (SCF) problem does not admit a homogeneous parallelisation strategy, a fact that severely limits current opportunities in modelling complex chemical systems like catalysts, nano-structured materials as well as large complexes in solution. This paper discusses two recent developments, which address this problem, and provide parallelisation strategies for numerical problems that appear specifically in electronic structure codes. The presented approaches are accompanied by library implementations that are invoked by the Gaussian-based density-functional code ParaGauss [4].
Scientific codes often contain linear algebra expressions, systems of linear equations, and eigenvalue problems. An example is the relativistic transformations and solving the block-diagonal eigenvalue problem for the resulting Hamiltonian as implemented in the quantum chemistry program package ParaGauss [4]. An abstraction for linear algebra operations facilitates a separation of concerns, where mathematics is separated from details of the technical implementation. This allows a quick implementation of matrix and vector transformations, thus contributing to the productivity of developing numerical routines. Furthermore, as these transformations may have the appearance of mathematical expressions, the application semantics is easily comprehensible, hence improving the readability of the code.
A number of languages or library extensions exist which provide such a functionality: Matlab and Octave are high-level scripting languages which operate only with mathematical objects, but are usually not suitable for high-performance codes and large software projects. The C++ libraries Armadillo [27], uBLAS (as part of BOOST [19]) and MTL [13], among others, offer template-based matrix classes with comprehensive functionality, and partially also advanced linear algebra operations, such as factorisations or eigenvalue problems. Fortran provides an intrinsic programming model, which allows to formulate basic matrix algebra in the source code, using variables and one- and two-dimensional arrays as representations for scalars, vectors and matrices, respectively. The Matran [29] library yields further matrix functionality for Fortran, together with advanced operations. However, except for a commercial version of the MTL (Supercomputing Edition), software or literature about abstractions supporting parallelism and data distribution, especially for Fortran, is scarce.
In data-intensive applications, it is often desired to distribute the underlying arrays over compute nodes, to avoid memory bottlenecks. Furthermore, expensive operations, especially BLAS Level 3 and eigenvalue computations, are often executed by parallel routines. However, common available parallelisation techniques, such as MPI or Co-Arrays in Fortran 2008, provide a rather low-level interface to express data distribution, and available parallel numerical routines, such as PBLAS and ScaLAPACK [5], work with specific distributed data types and do not allow to express parallel matrix algebra in mathematical notation, as described above. The PLAPACK package [31] addresses this issue and defines more object-style distributed data types along with automatic distribution routines, allowing thus a more modern programming style. However, even though the APIs of those linear algebra libraries are carefully designed, the routine signatures are still complicated, making elaborate algebraic code tedious to write. Hence, separation of concerns is difficult to comply with, which can easily lead to poor code quality.
In Sect. 2 we review a Fortran interface to linear algebra routines and its implementation, as introduced in Ref. [25]. The interface specifies distributed data types, representing matrix operands, and overloads the existing operators \(\{+,-,{\ast},{\ast}{\ast}\}\) to accept them. We show how this combination makes it is possible to express matrix algebra in clear mathematical notation directly in the code, while operating with distributed data and parallel routines at its back-end. We demonstrate how the newly established interface can be used to parallelise an existing sequential implementation of relativistic transformations in a quantum chemistry code and include a few benchmarks.
Furthermore, in Sect. 3 we present an approach to solve the generalized matrix eigenvalue problem
with symmetric block-diagonal matrices H and S > 0. The matrix E is diagonal, and contains the eigenvalues, and the columns of C the corresponding eigenvectors. Equation (1) for dense symmetric matrices can be solved by existing parallel solvers, provided e.g. by the libraries ScaLAPACK [5], PLAPACK [31] or ELPA [2]. However, a more complicated situation arises when the spatial symmetry of a molecule or cluster is exploited to reduce the dense eigenvalue problem to a few subproblems of smaller dimensions [24].
While the majority of current quantum chemistry problems are solved without invoking spatial symmetry constraints, exploiting point group symmetry can offer significant advantages in the field of nanoparticles [16, 26]. Clusters of various materials in the size range of nanoparticles (1–10 nm diameter) typically contain about 50 to several thousand atoms. Molecules with more than 100 heavy atoms still represent a challenge to accurate quantum chemistry methods and symmetry thus yields a convenient way of applying these methods to nano-sized model species in the relevant size range. Such nanoparticles represent prototypical applications, where a generalized matrix eigenvalue problem, decomposed into several sub-matrices (typically 4–10) of different sizes, has to be solved. This simplification significantly reduces the computational effort; however the implied block-diagonal matrix structure cannot be handled efficiently by existing (parallel) eigenvalue solvers. An approach to solving this problem is discussed in Sect. 3.
2 A Fortran Interface to Parallel Matrix Algebra
This section presents the specification and implementation of a set of high-level Fortran data types, functions, subroutines, and operator interfaces, which facilitate the handling of distributed data objects as a representation of matrix operands, as well as parallel arithmetic operations on them. Basic linear algebra can be expressed directly in common mathematical notation, more advanced operations, such as eigenvalue computations and domain specific operations, by subroutine calls. We have targeted and achieved a programming model which allows a separation of concerns: mathematical expressions are stated as simple as possible in the source code, while the library takes care of the low-level parallelisation, reliability and performance. As Sect. 2.3 shows, the library has been tested, and integrated into the quantum chemistry software ParaGauss [4].
In addition, the following requirements were considered in the interface design: (i) easy data object management (creation, disposal of, etc.) with technical details, such as physical data distribution, being hidden from the user, and (ii) an interface implementable with Fortran, MPI, and performance-optimized external libraries.
The interface realizes a single instruction multiple data (SIMD) programming model, in contrast to the single program multiple data (SPMD) style, often applied in pure MPI codes. Here, each library call represents a collective operation, encouraging synchronous control flow. The resulting source code looks like a sequential program, parallelism is achieved by operations on distributed objects by library routines. The internals of the opaque distributed objects, including the actual numeric data, are not exposed. These design features entail the advantage of good readability. Furthermore, adoption of serial code basically requires a change of the affected data types, as well as invocation of the library with a USE-statement.
The user is given the opportunity to choose the processes, involved in the data distribution and computation, via a communication context. This allows combining different paradigms and parallelisation technologies—the user is not restricted to a single paradigm for the overall application.
We defined two data types representing distributed dense and diagonal matrices: rmatrix and rdmatrix. The generic constructor MATRIX converts a 1- or 2-dimensional array into the distributed matrix object [25]. The complementary generic function ARRAY converts a matrix object back into a plain array. Furthermore, we overload some of Fortran intrinsic unary and binary arithmetic operators to express parallel operations between the distributed data types. These operators include: binary addition, binary subtraction, unary negation, binary (matrix) multiplication, exponentiation. Matrix transposition is implemented as a unary function tr(). The generic constructors MATRIX and ARRAY are sufficient to implement arbitrary matrix functions. For example, a reference implementation for the function tr() is a composition of Fortran intrinsic function, array and matrix constructors: tr(A) = matrix(transpose(array(A))). Figure 1 shows a simple code example, which demonstrates the general usage of the API presented here.
Demonstration of the API usage. Line 1 declares arrays whereas lines 2–3 declare two types of distributed matrices. In lines 5–6, distributed data objects for the diagonal and the dense matrix, respectively, are created. Line 8 shows a simple arithmetic operation. Finally, in line 10 the computed dense matrix is stored in a plain array. In this particular case the current implementation requires communication only in line 10
2.1 Interface Implementation
The API has been implemented as a library with Fortran bindings. The implementation language is Fortran 95 with the enhanced data type facilities (EDF), documented in the technical report TR15581 [11]. For our purposes, allocatable components, documented in this report, provide automatic deallocation upon object destruction and the “move semantics” similar to that of “rvalues” formalized in the C++11 standard. Additionally, we rely on MPI for communication primitives, and external, parallel library routines for expensive operations: PDGEMM from the PBLAS library for the dense matrix multiplication, PDSYGVX and PDTRAN from the ScaLAPACK library for the dense generalized eigenvalue problem, and the matrix transposition, respectively [5].
The matrix operands are represented by data types, containing a (distributed) allocatable array, and meta data, necessary for MPI, PBLAS, and ScaLAPACK usage, see Fig. 2. The arrays representing dense matrices are distributed in a block-cyclic manner, a native distribution scheme of PBLAS and ScaLAPACK (see the ScaLAPACK user’s guide [5]). With this data distribution scheme, the array constructor instance array_r of the generic ARRAY function, assembling a full 2D array on each process, is the only operation that requires communication [25].
For arithmetic operations between these data types, we overload the existing intrinsic operators with the interface construct. Figure 3 demonstrates this for the multiplication operator *. Here, the compiler does a static type check and resolves an operation to a specific function call, see Fig. 3. The storage for the function result is explicitly allocated in the function body. For the automatic deallocation of intermediate results, we rely on the allocatable semantics of the EDF, as explained at the beginning of this subsection. This important feature facilitates arithmetic expressions without introducing memory leaks and, together with the move semantics, allows an optimizing Fortran compiler to avoid unnecessary copying of data from the right-hand side of a matrix-valued expression in an assignment statement.
The diagonal matrix, represented by the data type rdmatrix, is stored in the 1D array d(:), see Fig. 2. As the storage requirement for a diagonal matrix is far less demanding than that for a dense matrix, we replicate the data on every process. This significantly simplifies implementation of operations involving diagonal matrices, and saves communication overhead at the cost of slightly higher memory consumption.
2.2 The Test Case: Background
We evaluated the approach to matrix algebra just described with a test case in the context of the quantum-chemistry code ParaGauss [4], which inspired us to develop this library abstraction. The test case covers dense matrix algebra, multiplications of dense and diagonal matrices, generalized eigenvalue problem and some domain specific operations on matrices. This section describes the background of the test case in abstract mathematical notation. In the next section we will show how this formalism translates into source code, and present some benchmarks.
Relativistic quantum chemistry covers a variety of approximations that account for the fact that the velocities of (core) electrons in heavy atoms is sufficiently close to the speed of light. A popular relativistic approximation is derived from the Douglas–Kroll approach [10] enabled by a practical scheme of building matrix representations of integro-differential operators [8]. The transformation starts with the four dense matrices, T, S, V, and O, comprising matrix representations of kinetic energy T, overlap S of basis functions, and matrix representations of two potential terms, V and O [8, 10]. The output consists of the relativistic counterparts of the kinetic energy T rel and potential matrix V rel. The first subproblem to address is a generalized eigenvalue problem for the matrices T and S which amounts to finding a matrix U and a diagonal matrix t such that U T TU = t and U T SU = 1 [8]. The kinetic energy eigenvalues, t, are further used to compute relativistic factors, formally diagonal matrices of the same dimension: \(t_{\text{rel}},e,a,b,r = \text{factors}(2t)\).
The next step of the second-order DKH transformation is to transform the input matrices V and O: \(\tilde{V } = {U}^{T}\mathit{VU}\), and \(\tilde{O} = {U}^{T}\mathit{OU}\). The central piece of the transformation is the matrix equation for the relativistic potential:
where we introduce the intermediate matrix \(R = \text{rpt}(e,{r}^{2}a\tilde{V }a - b\tilde{O}b)\) using the matrix valued function, rpt(e,X), defined as rpt: (e,X)↦Y, \(Y _{\mathit{mn}} = X_{\mathit{mn}}/(e_{m} + e_{n})\). Finally, a back-transformation follows: \(T_{\text{rel}} = {U}^{-T}t_{\text{rel}}{U}^{-1}\), \(V _{\text{rel}} = {U}^{-T}\tilde{V }_{\text{rel}}{U}^{-1}\).
2.3 Evaluation
Translated into Fortran, the central part of the DKH transformation, Eq. (2), is shown in Fig. 4. The code initiates two dense matrix multiplications, five dense matrix additions and a few multiplications of dense and diagonal matrices. Note that declarations of distributed matrices do not reserve space for actual data. It is the responsibility of the primitive operations that return an rmatrix or an rdmatrix to reserve space, initialize and finally fill those structures with data. Upon return from the subroutine, all intermediate data structures, declared in the scope of the subroutine, will be automatically freed by a standard TR15581 conforming compiler. Apart from the module name in the USE statement, no other modifications were necessary to parallelise the serial version of this code. Imperative coding style with destructive matrix updates (not shown here) is supported as well [25].
Fortran code for the popular form of relativistic transformation, Eq. (2), illustrating the use of the matrix algebra API. A function returning more than one matrix, here the diagonal matrix t rel and the dense matrix V rel, is commonly implemented as a subroutine with multiple output arguments. Declarations of distributed matrices do not lead to memory reservation for actual data. This is carried out by functions implementing primitive matrix operations. For local variables, such as e or aVa, these resources are freed on return from the function
Run time performance has been evaluated at the facilities provided by Leibniz Rechenzentrum, Munich, on the migration system SuperMIG, built by IBM [23]. The machine comprises 205 nodes, each hosting four 10-core Intel Xeon Westmere-EX processors. The nodes are equipped with 256 GB of memory and interconnected by an Infiniband QDR network. With this topology calculations involving, for example, 36, 64 and 81 processor cores were scheduled on 1, 2, or 3 nodes, respectively. For the benchmarks, we employed the default vendor-supplied MPI library implementation, IBM MPI v5.2, and BLACS/ScaLAPACK v1.8, linking to the highly optimized BLAS library distributed with Intel’s MKL v10.3.
Log-log plot of the wall-clock time required for a matrix multiplication using PBLAS procedure PDGEMM for matrix dimensions 4,000 (left pane) and 2,000 (right pane) as a function of the number of cores involved (black circles) in comparison to the time required to convert a distributed matrix to a plain array (gray squares). Linear regression suggests a power law, T ∼ P −α, 0.8 < α < 0.9 for the dependence of matrix multiplication time, T, on core number, P. Ideal scaling, T ∼ P −1, is shown for comparison (dashed line). The times required for matrix multiplication and building a replicated array scale with the matrix dimension N as N 3 and N 2, respectively
In Fig. 5 we show the scaling behaviour of a dense matrix multiplication as one of the primitive linear algebra operations (black circles). The parallel efficiency is limited by the underlying PBLAS implementation [5]. For a typical matrix dimension of 2,000–4,000, the implementation provided by the local computing centre shows an efficiency above 50 % for a core number below 100. In this example, a linear regression of the data displayed in Fig. 5 suggests a scaling behaviour of the time, T, required by the operation with the number of cores, P, as a power law, T ∼ P −α with α = 0.8…0.9. Note that for these matrix dimensions, the cost of a conversion of a distributed (resulting) matrix to a plain two-dimensional array, replicated on all processes requires an overhead that becomes comparable with the costs of a single matrix multiplication at about 40 cores (gray squares, Fig. 5). For higher matrix dimensions, the crossover happens at higher core numbers. Of course, this overhead is amortized when processing more involved linear algebra expressions before, if at all, the results will have to be converted to native arrays.
This is the case for the core of the second-order DKH transformation, see the source code in Fig. 4 implementing Eq. (2), for which both the input and output are distributed matrices. Figure 6 shows how the wall-clock time required for a DKH transformation for two matrix dimensions, N = 4,000 and N = 2,000, depends on the number of cores used. A linear regression of the data on the log-log scale suggests a power law for the timespan, T ∼ P −0.9. The parallelisation efficiency is limited primarily by the PBLAS library implementation, cf. Fig. 5. This efficiency approaches 50 % as the number of cores is increased along the horizontal axis. Note that in practical applications, end users very often consider the total timespan as a more relevant metric of the parallelisation efficiency.
Log-log plot of the wall-clock time required for the central piece of the second-order DKH transformation for matrix dimensions 4,000 (black circles) and 2,000 (gray squares) as a function of number of cores. Linear regression suggests that the wall-clock time, T, scales with the number of cores, P, as a power law, T ∼ P −α, with 0.8 < α < 0.9. Ideal scaling, T ∼ P −1, is shown for comparison (dashed lines)
3 MPTS-Scheduling of Parallel Eigenvalue Computations
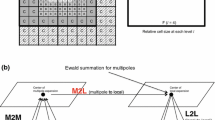
This section discusses a solution to the symmetric, block-diagonal, generalized eigenvalue problem HC = SCE, as introduced in Sect. 1. The specific block-diagonal structure of the matrices H and S have the important property that the distinct blocks can be treated as independent subproblems. This property already implies a possibility to introduce parallelism to the diagonalisation step [3]: the sub-matrices are sorted by their size in descending order; whenever a processor becomes available, it is employed to carry out a sequential diagonalisation of the first sub-matrix in the list. However, this approach, also known as LPT algorithm [15] as a special form of list scheduling [14], shows very limited scalability: the sequential execution time for the largest matrix is a lower bound on the overall execution time, and the number of matrices is an upper bound on the number of processors which can be used. See Fig. 7a.
For a typical application, the dimension N of the basis set, which corresponds to the size of H, ranges from 500 up to several thousand. The corresponding generalized eigenvalue problem represents a fraction of the cost of the electronic structure calculation, but it is a fraction growing with \(\mathcal{O}({N}^{3})\) and is intrinsically difficult to parallelise efficiently. Thus, it becomes more important with increasing problem size and CPU count. Moreover, the solution of Eq. (1) is usually part of an iterative scheme with \(1{0}^{3} - 1{0}^{5}\) diagonalisations necessary in a single typical application. Thus, an efficient parallel algorithm is required to avoid bottlenecks in large-scale applications.
There exist previous approaches to employ ScaLAPACK [5] as parallel eigensolver in electronic structure calculations, see Refs. [17, 32]. However, in these calculations, the molecular symmetry was not exploited, so H was a single dense, symmetric matrix, which can be treated by standard parallel eigensolvers, such as PDSYEVX or PDSYGVX from ScaLAPACK [5]. In our case, the diagonalisation of \(H \in {\mathrm{I\!R}}^{N\times N}\) can be divided into several smaller sub-problems. Thus, to benefit from these computational advantages, a different parallelisation strategy is required.
Here we describe an approach to tackle this problem, originally reported in Ref. [24]: the sequential and parallel routines DSYGV and PDSYGV from LAPACK [1] and ScaLAPACK [5], respectively, are employed to diagonalise independently the set of sub-matrices. A malleable task scheduling algorithm allots to each matrix a processor count and schedules the instances of the eigensolvers to achieve good load balancing. Thus, the existing parallel eigensolvers can be used efficiently, to reduce the overall execution time.
3.1 Problem Discussion
The objective is to find all eigenvalues and eigenvectors of a set of symmetric matrices of different size. The term “task” will be used as synonym for a single matrix diagonalisation which is processed by an eigensolver.
Hence, the problem can be formulated as follows: given is a set of n tasks \(\mathcal{T} =\{ T_{1},\ldots,T_{n}\}\) and a set of m identical processors. Each task is assigned a matrix of defined size, so there is a set of sizes \(\mathcal{S} =\{ S_{1},\ldots S_{n}\}\) with the relation \(T_{i}\mapsto S_{i}\). The matrix sizes in \(\mathcal{S}\) can vary arbitrarily. The tasks are independent and nonpreemptable. Furthermore they are malleable, i.e., a task may be executed by an arbitrary number of processors p ≤ m, resulting in different execution times. As the employed routine—the eigensolver—is identical for each task, there is only one cost function denoted as
which predicts the execution time of task T i with size S i when executed on p processors.
Furthermore, there is a set of processor counts \(\mathcal{P}\), where its elements P represent a possible number of processors on which a task can be executed. The makespan is the overall time required to process all tasks from \(\mathcal{T}\). The goal is to find a scheduling with a minimal makespan. As the problem arises in many practical applications, it is well studied and known as malleable parallel task scheduling (MPTS). An example is depicted in Fig. 7b.
Hence, our approach to a solution is as follows: as a first step, we generate an offline scheduling from the task set \(\mathcal{T}\) and the process count m, using an MPTS algorithm, see Sect. 3.2. This algorithm requires a cost function, which predicts the time required by a task—here the execution time of the employed eigensolver. Its practical implementation is discussed in Sect. 3.3. This established scheduling is then used in ParaGauss for the efficient parallel computation of the eigenvalue problems.
3.2 Malleable Parallel Task Scheduling
3.2.1 Related Work
The MPTS problem introduced in Sect. 3.1 is a common scheduling problem and was frequently discussed over the last decades, see Refs. [6, 7, 21]. MPTS is a generalisation of a sequential scheduling problem which is NP-complete in the strong sense [12]. Therefore, a variety of approximation algorithms with polynomial runtime exist. A common approach is based on a two-phase strategy, first introduced by Turek et al. [30]. The idea is to find a processor allotment for each task in a first step and to solve the resulting nonmalleable parallel task scheduling (NPTS) problem in a second step. Ludwig and Tiwari [20] suggested such an algorithm with approximation factor 2, which shall be the basis of the strategy proposed here. Mounié et al. [22] followed a different approach by formulating a Knapsack problem as core component. They provide the currently best practical algorithm with approximation factor \(\frac{3} {2} + \epsilon \) for any fixed ε > 0. When the tasks have to be executed on processors with successive indices, Steinberg [28] proposed an adapted strip-packing algorithm with approximation factor 2. Furthermore, Jansen [18] gave an approximation scheme with makespan at most 1 + ε for any fixed ε > 0. For special cases of the MPTS, algorithms exist with approximation factors close to one (e.g. Ref. [9] for identical malleable tasks), but those do not apply in our case.
The problem described above is a standard MPTS problem, so the algorithms mentioned could in principle be applied. However, as we will see, the number of sub-matrices to be processed is small due to fundamental symmetry rules. This allows us to modify the algorithm from Ref. [20] by introducing a combinatorial approach in order to find the optimal solution of the NPTS problem.
3.2.2 The Employed Algorithm
As in the approximate algorithm by Ludwig and Tiwari [20], we also follow a two-phase approach. The first phase determines a number of allotted processors for each task using an approximate algorithm where the processor allotment is generated in \(\mathcal{O}(mn)\) operations. Thus, the original MPTS problem is transformed into an NPTS problem. In the second phase, a scheduling for the resulting nonmalleable tasks is generated. This can be achieved by applying any (optimal or approximate) NPTS algorithm. An important characteristic of this approach is that the approximation factor of the MPTS problem is equal to the approximation factor of the NPTS algorithm applied in the second phase. Ludwig and Tiwari proposed an algorithm that provides an approximation factor of 2.
In our specific case, the number of tasks is bounded above by 10. This allows an alternative strategy for solving the NPTS problem exactly by employing a combinatorial algorithm, different from the algorithm by Ludwig and Tiwari [20]. In our combinatorial approach, the maximum number of possible permutations is \(10! \approx 3.6 \cdot 1{0}^{6}\). However, we also introduced optimisations, which make this worst case very rare. For the detailed algorithm, please refer to Ref. [24].
3.3 Cost Function
The scheduling algorithm described requires a cost function which estimates the execution time of the employed eigensolver routines—in our case DSYGV and PDSYGV from the (Sca)LAPACK library. It is difficult to determine upfront how accurate the estimates have to be. However, the validation of the algorithm will show whether the error bounds are tight enough for the algorithm to work in practice.
There exist analytic models for the performance prediction of parallel routines, see for example the ScaLAPACK User’s Guide [5]. However, validation of the models showed that the prediction error usually lies between 10 and 30 %. Furthermore, it does not provide a solution for the practical use in a scheduling algorithm: basically, each routine needs its own model. Therefore, if the routine changes (e.g. due to a revision or the use of a different library), the model has to be adapted as well, which requires in-depth knowledge of the employed routine.
Here we follow a different approach: the routine is handled as a “black box”. Predictions of its execution time are based on empirical data which are generated ahead of time on a given computing resource by test runs with a set of randomly generated matrices on a set of possible processor allotments \(\mathcal{P}\). Then, with a one-dimensional curve-fitting algorithm, a continuous cost function t is generated for each element of \(\mathcal{P}\), Fig. 8. Thus, each \(P \in \mathcal{P}\) has a related cost function \(t_{P}: S\mapsto t_{P,S}\), not to be confused with Eq. (3).
Finally, we combine the emerging set of P-related cost functions to form the general cost function, Eq. (3). However, in practice, when a certain number of allotted processors is exceeded, parallel routines no longer feature a speedup or even slow down [32]. This behaviour does not comply with the assumption of a general monotonic cost function. To satisfy this constraint, we define the cost function, Eq. (3), as follows:
All possible processor counts \(P \in \mathcal{P}\) are considered which are smaller than or equal to p. The P which results in the smallest execution time for the given S also determines the P-related cost function and thus the result t of the general cost function, Eq. (4).
3.4 Evaluation
We evaluated the scheduler just described for two molecular systems as example applications: the palladium clusters Pd489 and Pd670 in symmetry O h . Both test systems entail 10 matrices, resulting from the applied point group symmetry, with dimensions ranging from 305 to 2,105. On average, the matrix dimensions of the test system Pd670 are about 40 % larger than those of the system Pd489.
For the test runs, the migration system SuperMIG, built by IBM and installed at Leibniz Rechenzentrum, was used; see Sect. 2.3 for more details.
We measured the execution times of the complete eigensolver step in a single SCF iteration, see Fig. 9. Recall that typical quantum chemical applications require between 103 and 105 eigenvalue computations. This should be taken into consideration when examining the real-time savings achieved by this parallelisation technique.
Log-log time diagrams of the complete eigensolver step of the test systems Pd489 and Pd670. Considered is the wall-clock time of the diagonalisation module during one SCF iteration. The curves labelled “predicted” show the makespan of the scheduling algorithm, predicted by the cost function. The curves labelled “computed” provide the real execution time of the scheduled eigensolvers. The lines labelled “LPT” indicate the execution time of the sequential LAPACK routine that computes the largest matrix and yield, thus, the best possible performance of the previously used LPT scheduler. (a) Pd489. (b) Pd670
Figure 9 shows that the cost function works accurate in most cases, with an error well below 10 %. Interestingly, this does not apply to small processor numbers (m ≤ 2), where the error is quite significant (up to \(\approx \)40 % in the Pd670 test runs). Fortunately, these cases still provide good scheduling results, even with poorly predicted execution times. For higher processor numbers m, the cost function works sufficiently accurate to facilitate the practical use of the scheduling algorithm.
The figure also shows a lower bound on the execution time of the previously used sequential scheduler (“LPT”-line). As one can see, this performance bound is now broken and the execution time is improved below this limit by our new algorithm. The diagonalisation step of the test system Pd489 was sped up by a factor of up to 11.1, compared to LPT, and by a factor of 31.6 compared to a sequential run. For the system Pd670 we achieved maximum speed-up factors of 10.5 and 40, compared to LPT and a sequential runs, respectively. For both test systems, the overall execution time of the eigensolver step in one SCF iteration now lies well below 1 s. Thus, we have shown that the execution time of this important step can be reduced down to a minor fraction of the overall electronic structure calculation.
4 Conclusions
We presented a parallel programming interface, which facilitates easy data management and the expression of matrix operations in almost mathematical notation. The evaluation shows that this technique is suitable for real-world problems—in our case relativistic transformations in quantum chemistry software. The resulting code has indeed the appearance of the original abstract mathematical formulation, while tedious, distracting low-level tasks, such as data organisation, are reduced to a few function calls before and after the matrix operations. We furthermore showed that with our implementation, which partly relies on the performance-optimized parallel libraries PBLAS and ScaLAPACK, the relativistic transformations scale up to 81 cores for input matrices of dimension 4,000, requiring less than 1 s for the overall relativistic transformation. This states a major improvement compared to the previous, sequential, implementation. Consequently, this work brings together programming productivity, code quality and parallel performance, which we consider a useful contribution to software engineering in high-performance computing.
Furthermore, a parallel bottleneck of the eigensolver step in ParaGauss, imposed by the previous LPT-based parallelisation approach, could be eliminated. This was achieved by employing a more sophisticated MPTS scheduler, together with parallel eigensolver routines from ScaLAPACK. The overall scalability of the eigensolver step was significantly improved, being now able to use processor core numbers up to 120 efficiently in large-scale chemical applications. The time spent in this step was reduced to a minor fraction of what was necessary for the previous solution, requiring less than 1 s in one SCF iteration for the test cases considered. This approach also goes beyond the use of parallel eigensolvers in other Gaussian-based DFT codes [2, 17]: to our knowledge, it is the first technique that allows an efficient parallel treatment of Hamilton matrices with a block-diagonal structure. Thus, DFT-based methods which achieve computational benefits by exploiting molecular symmetries have now been augmented with a specific, efficient parallelisation technique.
References
Anderson, E., Bai, Z., Bischof, C., Demmel, J., Dongarra, J., Du Croz, J., Greenbaum, A., Hammarling, S., McKenney, A., Ostrouchov, S., Sorensen, D.: LAPACK’s User’s Guide. SIAM, Philadelphia (1992)
Auckenthaler, T., Blum, V., Bungartz, H.J., Huckle, T., Johanni, R., Krämer, L., Lang, B., Lederer, H., Willems, P.R.: Parallel solution of partial symmetric eigenvalue problems from electronic structure calculations. Parallel Comput. 37, 783–794 (2011)
Belling, T., Grauschopf, T., Krüger, S., Mayer, M., Nörtemann, F., Staufer, M., Zenger, C., Rösch, N.: In: Bungartz, H.J., Durst, F., Zenger, C. (eds.) High Performance Scientific and Engineering Computing. Lecture Notes in Computational Science and Engineering, vol. 8, p. 439. Springer, Heidelberg (1999)
Belling, T., Grauschopf, T., Krüger, S., Nörtemann, F., Staufer, M., Mayer, M., Nasluzov, V.A., Birkenheuer, U., Hu, A., Matveev, A.V., Shor, A.V., Fuchs-Rohr, M.S.K., Neyman, K.M., Ganyushin, D.I., Kerdcharoen, T., Woiterski, A., Gordienko, A.B., Majumder, S., Rösch, N.: PARAGAUSS, version 3.1. Technische Universität München (2006)
Blackford, L.S., Choi, J., Cleary, A., D’Azevedo, E., Demmel, J., Dhillon, I., Dongarra, J., Hammarling, S., Henry, G., Petitet, A., Stanley, K., Walker, D., Whaley, R.C.: ScaLAPACK Users’ Guide. Society for Industrial and Applied Mathematics, Philadelphia (1997)
Blazewicz, J., Kovalyov, M.Y., Machowiak, M., Trystram, D., Weglarz, J.: Scheduling malleable tasks on parallel processors to minimize the makespan. Ann. Oper. Res. 129, 65–80 (2004)
Blazewicz, J., Ecker, K., Pesch, E., Schmidt, G., Weglarz, J.: Handbook on Scheduling: From Theory to Applications. Springer, Heidelberg (2007)
Buenker, R.J., Chandra, P., Hess, B.A.: Matrix representation of the relativistic kinetic energy operator: two-component variational procedure for the treatment of many-electron atoms and molecules. Chem. Phys. 84, 1–9 (1984)
Decker, T., Lücking, T., Monien, B.: A 5/4-approximation algorithm for scheduling identical malleable tasks. Theor. Comput. Sci. 361(2), 226–240 (2006)
Douglas, M., Kroll, N.M.: Quantum electrodynamical corrections to the fine structure of helium. Ann. Phys. (NY) 82, 89 (1974)
Enhanced data type facilities. ISO/IEC TR 15581, 2nd edn. (1999). ftp://ftp.nag.co.uk/sc22wg5/N1351-N1400/N1379.pdf
Garey, M., Johnson, D.: Computers and Intractability: A Guide to the Theory of NP-Completeness. W.H. Freeman and Company, New York (1979)
Gottschling, P., Wise, D.S., Adams, M.D.: Representation-transparent matrix algorithms with scalable performance. In: Proceedings of the 21st Annual International Conference on Supercomputing, ICS ’07, Seattle, pp. 116–125. ACM, New York (2007)
Graham, R.L.: Bounds for certain multiprocessing anomalies. Bell Syst. Tech. J. 45, 1563–1581 (1966)
Graham, R.L.: Bounds on multiprocessing timing anomalities. SIAM J. Appl. Math. 17, 263–269 (1969)
Häberlen, O.D., Chung, S.C., Stener, M., Rösch, N.: From clusters to the bulk. A relativistic electronic structure investigation on a series of gold clusters Au n , \(n = 6,\ldots 147\). J. Chem. Phys. 106, 5189–5201 (1997)
Hein, J.: Improved parallel performance of SIESTA for the HPCx Phase2 system. Technical report, The University of Edinburgh (2004)
Jansen, K.: Scheduling malleable parallel tasks: an asymptotic fully polynomial time approximation scheme. Algorithmica 39, 59–81 (2004)
Ling, B.S.: The Boost C++ Libraries. XML Press (2011). http://books.google.de/books?id=xMH0XwAACAAJ
Ludwig, W., Tiwari, P.: Scheduling malleable and nonmalleable parallel tasks. In: SODA ’94, Arlington, pp. 167–176 (1994)
Mounié, G., Rapine, C., Trystram, D.: Efficient approximation algorithms for scheduling malleable tasks. In: SPAA ’99, Saint Malo, pp. 23–32 (1999)
Mounié, G., Rapine, C., Trystram, D.: A 3/2-approximation algorithm for scheduling independent monotonic malleable tasks. SIAM J. Comput. 37(2), 401–412 (2007)
National supercomputer HLRB-II. http://www.lrz-muenchen.de/. Retrieved on 10 Aug 2012
Roderus, M., Berariu, A., Bungartz, H.J., Krüger, S., Matveev, A.V., Rösch, N.: Scheduling parallel eigenvalue computations in a quantum chemistry code. In: Euro-Par (2)’10, Ischia, pp. 113–124 (2010)
Roderus, M., Matveev, A.V., Bungartz, H.J.: A high-level Fortran interface to parallel matrix algebra. In: CCSEIT-2012, International Conference Proceeding Series (ICPS), Coimbatore. ACM (2012, Accepted)
Rösch, N., Matveev, A., Nasluzov, V.A., Neyman, K.M., Moskaleva, L., Krüger, S.: Quantum chemistry with the Douglas–Kroll–Hess approach to relativistic density functional theory: efficient methods for molecules and materials. In: Schwerdtfeger, P. (ed.) Relativistic Electronic Structure Theory – Applications. Theoretical and Computational Chemistry Series, vol. 14, pp. 656–722. Elsevier, Amsterdam (2004)
Sanderson, C.: Armadillo: an open source C++ linear algebra library for fast prototyping and computationally intensive experiments. Technical report, NICTA (2010)
Steinberg, A.: A strip-packing algorithm with absolute performance bound 2. SIAM J. Comput. 26(2), 401–409 (1997)
Stewart, G.W.: Matran: a Fortran 95 matrix wrapper. Technical report, UMIACS (2003)
Turek, J., Wolf, J., Yu, P.: Approximate algorithms for scheduling parallelizable tasks. In: SPAA’92, San Diego, pp. 323–332 (1992)
van de Geijn, R.A.: Using PLAPACK: Parallel Linear Algebra Package. MIT, Cambridge (1997)
Ward, R.C., Bai, Y., Pratt, J.: Performance of parallel eigensolvers on electronic structure calculations II. Technical report, The University of Tennessee (2006)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Roderus, M., Matveev, A., Bungartz, HJ., Rösch, N. (2013). Advances in the Parallelisation of Software for Quantum Chemistry Applications. In: Bader, M., Bungartz, HJ., Weinzierl, T. (eds) Advanced Computing. Lecture Notes in Computational Science and Engineering, vol 93. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-38762-3_6
Download citation
DOI: https://doi.org/10.1007/978-3-642-38762-3_6
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-38761-6
Online ISBN: 978-3-642-38762-3
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)