Abstract
Type 2 diabetes (T2DM) is a chronic disease with a rapidly increasing global burden. An early event in the disease is deregulation of glycaemic control resulting in periods of hyperglycaemia. Large-scale clinical studies have shown that complications resulting from this hyperglycaemia can be manifest long after glycaemic control has been restored (UKPDS, Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33) UK Prospective Diabetes Study (UKPDS) Group. Lancet 352:837–852, 1998; Chalmers J, Cooper ME, UKPDS and the legacy effect. N Engl J Med 359:1618–1620, 2008), a phenomenon known as the “legacy effect” (Holman RR et al., 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med 359:1577–1589, 2008). Such continued development of cardiovascular complications, which result from prior exposure to hyperglycaemia, has led to the proposal of a “metabolic memory” (Cooper ME, Metabolic memory: implications for diabetic vascular complications. Pediatr Diabetes 10:343–346, 2009). Such a hypothesis suggests that a transient exposure to hyperglycaemia results in persistent changes in gene expression that are not reversed merely by restoring glycaemic control. Support for early, persistent changes came from the Diabetes Control and Complications Trial (DCCT) which revealed that early glycaemic control in diabetic patients led to sustained benefits and better outcomes (Cooper ME, Metabolic memory: implications for diabetic vascular complications. Pediatr Diabetes 10:343–346, 2009), and it has recently been proposed that minimising early exposure to hyperglycaemia is paramount (Aizawa T, Funase Y, Intervention at the very early stage of type 2 diabetes. Diabetologia 54:703–704; author reply 707–708, 2011). Currently, the most attractive potential mechanism responsible for the “legacy effect” is epigenetic, manifested by changes in DNA methylation and/or posttranslational modifications on histones. Over the last decade, numerous studies have identified correlations of specific epigenetic marks with type 2 diabetes, and more recently the mechanisms by which these changes lead to persistent alterations in gene expression levels have been interrogated.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Crosstalk Disease
- Diabetes
- DNA methylation
- Histone acetylation
- Histone methylation
- Hyperglycaemia
- Metabolic memory
- Vascular
1 Introduction
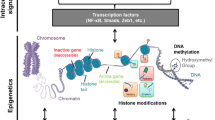
The proposal that epigenetic mechanisms contribute to the progression and complications associated with type 2 diabetes has been around for many years. In 1990, Roy et al. reported a persistent change in the levels of fibronectin mRNA in tissues from diabetic rats and cultured human endothelial cells after a short period of exposure to high levels of glucose (50 mmol/l) and suggested heritable changes in chromatin structure as a potential mechanism underlying their observations (Roy et al. 1990). More recently, such a suggestion has been gaining support with further studies that provide evidence supporting the notion that changes in chromatin are induced during diabetes and by exposure to hyperglycaemia (Maier and Olek 2002; Ling and Groop 2009; Reddy and Natarajan 2011; Villeneuve et al. 2011). Though there is still relatively limited understanding of the complete molecular details, changes in posttranslational modifications on histones have been associated with the gene expression changes seen in diabetes (Pirola et al. 2010). Alterations in several types of posttranslational modification including DNA methylation, histone acetylation and histone methylation have been observed in animal and cell models of diabetes and associated with changes in gene expression (Pirola et al. 2010).
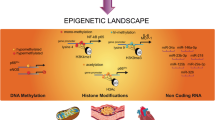
2 DNA Methylation
Methylation of DNA involves the addition of a methyl group to specific cytosine residues of DNA by DNA methyltransferase enzymes (DNMTs). DNA methylation was the first described epigenetic mark and methylation of the DNA for a specific gene generally leads to silencing of expression of that gene. DNA methylation has long been known to function as a cellular memory, and the mechanisms by which DNA methylation can be inherited over many cell generations are well understood (Jaenisch and Bird 2003). There is some good correlative data which, collectively, points an accusing finger at DNA methylation for a role in metabolic memory and diabetes, reviewed in Maier and Olek 2002. DNMTs use the donor S-adenosylmethionine (SAM) for the methyl group, and perturbations in the level of available SAM, which is influenced by diet, can have some profound effects on the extent of DNA methylation. Low levels of SAM are associated with global hypomethylation and may contribute to formation of cancers (Singh et al. 2003). Individuals with diabetes show reduced concentrations of SAM in their blood suggesting a potential hypomethylation of genes in these individuals and potential for perturbed gene expression levels (Poirier et al. 2001). In addition to global effects on methylation resulting from fluctuations in the levels of SAM, DNA methylation is also influenced by many cell signalling pathways. Methylation of cytosines within specific genes is controlled by the recruitment of DNMTs by regulatory cofactors in response to local environmental cues (Szyf and Detich 2001; Turek-Plewa and Jagodzinski 2005). DNA methylation was originally thought to be an essentially irreversible modification that could only be removed through the process of cell division and DNA replication. However, there is now compelling evidence supporting a mechanism of active demethylation, though a precise molecular explanation for this process is still the subject of some conjecture (Wu and Zhang 2010). Given that DNA methylation can be regulated by the local cellular environment and, once deposited, methyl marks have the potential to be stably maintained over many cell generations, it is an excellent candidate for a mechanism responsible for cellular memory in type 2 diabetes. Consistent with such notion, a role for DNA methylation has recently been identified as a contributory mechanism to learning and memory in the brain, and memories can be maintained many years after the event that initiated them (Day and Sweatt 2010). The discovery of active DNA demethylation does provide some optimism that any cellular memory encoded by DNA methylation could eventually be reset with the use of an appropriate intervention.
Though there is no definitive experimental data that has unequivocally demonstrated that DNA methylation is responsible for metabolic memory in type 2 diabetes, many studies have reported associations between altered methylation in models of diabetes and as a result of hyperglycaemia in culture. In the Zucker (type 2) diabetic fatty rat, increased levels of global DNA methylation were observed at 12 and 21 weeks in the liver although not in the kidney or heart (Williams and Schalinske 2011). The authors also observed an increase in expression of DNMT1 in the liver at 21 weeks, but not at 12 weeks. The significance of this increase is not entirely clear, as it follows the increase in DNA methylation; it cannot be the driver for it though it may be involved in maintaining high methylation levels. Cultures of the human hepatocellular carcinoma cell line, HepG2, also show an increase in DNMT activity and global DNA methylation in response to a 72-h exposure to high (16.7 mmol/l) glucose (Chiang et al. 2009).
The db/db mouse contains a natural mutation in the leptin receptor gene and is a well-established model of diabetes, providing an in vivo model to study the progression of and mechanisms underlying diabetes (Lee et al. 1996). In these mice, the gene encoding the insulin-like growth factor 1 receptor (Igfr1) shows enhanced levels of DNA methylation and reduced levels of mRNA expression in skeletal muscle compared to wild-type mice (Nikoshkov et al. 2011). Human patients with type 2 diabetes also show increased methylation within the gene for peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) within skeletal muscle, and the levels of methylation showed inverse correlation with mRNA levels (Barrès et al. 2009). In pancreatic islet cells from patients with type 2 diabetes, the insulin gene shows increased levels of DNA methylation and decreased levels of mRNA compared to nondiabetic individuals (Yang et al. 2011). Consistent with a role for hyperglycaemia in this response is the observation that rat pancreatic beta cells showed similar changes when cultured in high glucose (16.7 mmol/l) for 72 h (Yang et al. 2011). These recent data provide some suggestions that perturbation in DNA methylation resulting directly or indirectly from hyperglycaemia results in changes of gene expression relevant to type 2 diabetes in different tissues.
Technological advances such as microarray analysis and high-throughput sequencing techniques over recent years now allow researchers to interrogate the entire genome and uncover novel target genes whose involvement was previously unrecognised. One recent study used a high-throughput sequencing approach to examine the epigenetic landscape of cultured human aortic endothelial cells exposed to hyperglycaemia (30 mmol/l glucose). The analysis revealed an increase in DNA methylation across many sites within the genome (Pirola et al. 2011). Such methylation would be predicted to result in reduced gene expression, and although this was not directly tested, it was shown that the regions of increased DNA methylation did not overlap with regions of high levels of histone acetylation which were associated with gene expression (Pirola et al. 2011).
These data suggest an involvement of DNA methylation in defining the pathophysiological phenotype of cells from individuals with type 2 diabetes. Such changes in DNA methylation may be the direct result of a short period of hyperglycaemia, and the hypothesis that such changes are responsible for, or at least contribute to, the observed metabolic memory is attractive. To date much of the data collected on DNA methylation and type 2 diabetes is correlative, and it is not clearly understood if DNA methylation changes cause or are the result of complications associated with type 2 diabetes. Hopefully such questions will be addressed in the future.
3 Histone Methylation
Like DNA, histones are also a substrate for methylation. Both lysine and arginine residues on histones can be methylated and multiple methyl groups can be added to each. Lysine can be mono-, di- or trimethylated, while arginine can be mono-, symmetrically di- or asymmetrically dimethylated (Fig. 10.1). The consequence of the methylation is dependent on the exact nature of methylation. For example, trimethylation of histone H3 lysine 4 (H3K4me3) is associated with active gene promoters, while methylation of H3K9 is associated with gene repression. Histones are methylated by a series of enzymes known as histone methyltransferases (HMTs) and histone methylation was originally thought to be irreversible. However, over the last few years, several enzymes have been identified that that can actively remove methyl groups from histones, and it is now appreciated that histone methylation is very dynamic and contributes to many physiological responses (Shi et al. 2004; Tsukada et al. 2006). Unlike DNA methylation, the process by which posttranslational modifications such as histone methylation can be preserved across multiple cell divisions remains unclear. However, some data suggest that during DNA replication, the histones are partitioned between the old and newly synthesised DNA and that newly inserted histones are modified to match their neighbours (Xu et al. 2010). Histone methylation impacts on chromatin function by influencing the set of proteins that are recruited to a specific gene. Adding or removing methyl groups on specific histone residues creates, or destroys, binding sites for specific proteins via their chromodomains, PHD fingers or Tudor domains (Chen et al. 2011; Sanchez and Zhou 2011; Yap and Zhou 2011). Ultimately the proteins recruited induce changes in the expression of the underlying genes, either directly by influencing the activity of the RNA polymerase or its associated subunits or indirectly by initiating further chromatin modifications or changes to the overall structure.
Posttranslational modification of lysine and arginine residues in histones. (a) Illustration shows the results of posttranslational modifications on the structure of lysine. Acetyltransferases can add an acetyl moiety onto the lysine to give acetyl lysine (left) which not only provides a structural change but also removes the positive charge carried by the native lysine. Methyltransferases can add one, two or three methyl groups onto the lysine to produce three forms of methylated lysine (right). Unlike acetylation, the addition of methyl groups does not affect the positive charge carried by the lysine. (b) Illustration showing the results of posttranslational modifications on the structure of arginine
To uncover the consequences of metabolic disturbance on histone methylation, Miao et al. profiled the levels of dimethylated histone H3 lysine 4 (H3K4me, which is preferentially, but not exclusively, associated with active genes) and dimethylated histone H3 lysine 9 (H3K9me2, which is associated with repressed genes) of monocytes cultured in normal or high (25 mmol/l) glucose (Miao et al. 2007). This study identified 9 and 26 genes that showed decreased and increased H3K4me2 levels in high glucose conditions, respectively. One of the genes showing increased levels of H3K4me2 was the histone K9 demethylase JMJD2A. Increased expression of JMJD2A could reduce epigenetic silencing at other loci by removing H3K9 methylation, inhibiting the H3K9me dependant repression at these genes and thus providing a feed forward mechanism. It is not clear that JMJD2A expression is actually increased though as no measurements of this were reported and overall the changes in H3K4me2 levels at genes did not correlate well with changes in expression of the associated mRNA. Thus, although the H3K4me levels were increased, there is not really a strong argument that the levels of JMJD2A would necessarily go up as a result. The levels of H3K9me2, which is associated with gene repression, were increased in 39 genes and decreased in 11 genes. Unlike H3K4me2 and consistent with many other studies, the changes in the levels of H3K9me2 did correlate very well with changes in gene expression (Miao et al. 2007).
Increased inflammatory gene expression has long been associated with type 2 diabetes and insulin resistance (Dandona et al. 2004), and many groups focus on the mechanisms responsible for such changes. A major driver of inflammatory gene expression is the transcription factor NF-κB (though it also has other roles too) which is regulated both by expression levels and through a cytoplasmic sequestration mechanism. In bovine endothelial cells, a transient (16 h) exposure to hyperglycaemia (30 mmol/l glucose) increased levels of NF-κB mRNA which were sustained over a 6-day period (Brasacchio et al. 2009). Increased NF-κB levels were associated with monomethylation of H3 lysine 4 (H3K4me1) most likely brought about as a result of recruitment of the histone methyltransferase SET7 to the NF-κβ promoter (Brasacchio et al. 2009). Indeed knockdown of SET7 abolished the increase in NF-κΒ in response to hyperglycaemia. As well as the addition of an activating histone modification, hyperglycaemia produced a reduction of the repressive H3K9 methylation mark by the histone demethylase, LSD1 (El-Osta et al. 2008; Brasacchio et al. 2009). Induction of NF-κΒ activity may also be important in modulating the VSMC phenotype during diabetes, and cultured porcine VSMCs show increased NF-κΒ function when cultured in high (25 mmol/l) glucose (Yerneni et al. 1999). NF-κB activity drives expression of many inflammatory genes such as tumour necrosis factor (TNF)-α and interleukin (IL)-1β and plays a contributory role to vascular complications such as atherosclerosis. Hyperglycaemia also increases expression of a second transcription factor, NFAT, associated with proliferation and migration of VSMC as well as neointima formation. Mouse cerebral arteries cultured in 20 mmol/l glucose and blood glucose levels of 17.6 mmol/l in mice resulted in an increase of nuclear NFAT activity and potentially contribute to vascular dysfunction in patients with type 2 diabetes (Nilsson et al. 2006; Nilsson-Berglund et al. 2010).
Consistent with increased function of transcription factors that regulate inflammatory associated genes, VSMC from type 2 diabetic mice (db/db) show persistent changes in inflammatory gene expression even after isolation and culturing in vitro (Li et al. 2006). Correlating with this increased gene expression are reduced levels of the repressive histone mark H3K9me3 at the promoters of inflammatory genes (Villeneuve et al. 2008). The reduced levels of H3K9me3 are associated with reduced expression and a reduced recruitment of the histone methyltransferase Suv39h1 (Fig. 10.2a), which is the key methyltransferase responsible for the deposition of this mark, to the promoters of inflammatory genes. Ectopic expression of Suv39h1 in VSMC from db/db mice could reverse the inflammatory phenotype and led to the decrease in inflammatory gene expression. When cultured for a 2-week period in high glucose (25 mmol/l), human VSMC also showed a decrease in the levels of H3K9me3 at inflammatory gene promoters, and consistent with the mouse model, SUV39H1 is implicated in this response. In this case, knockdown of SUV39H1 by shRNA potentiated inflammatory gene expression, while ectopic expression of SUV39H1 reduced the induction of inflammatory gene expression by TNF-α (Villeneuve et al. 2008).
Changes in chromatin modifications in response to hyperglycaemia. Exposure of endothelial cells to high concentrations of glucose leads to changes in the recruitment of different chromatin-modifying enzymes to specific gene promoters and a subsequent change in the expression levels of those genes. (a) In the presence of high glucose, the histone methyltransferase Set7/9 and the histone demethylase LSD1 both show increased recruitment to chromatin in endothelial cells, while recruitment of the histone methyltransferase SUV39H1 is reduced. Changes in the recruitment of these enzymes result in an increase of H3K4me and decrease in H3K9me and increased expression of genes including NF-κb, TNF-α, TGF-β and IL-β. (b) In the presence of high glucose, the histone acetyltransferase p300 shows enhanced recruitment to some genes and leads to increased levels of H3K9 and H3K14 acetylation levels and expression of genes such as NF-κb, CREB and AP-1. The precise molecular details connecting high glucose levels to the recruitment of these chromatin-modifying enzymes are yet to be uncovered
Mesangial cells are smooth muscle cells which regulate capillary blood flow in the kidney and show a loss of contractility in diabetes. When exposed to high glucose (30 mmol/l) for 48 h, cultured mesangial cells showed an increased expression of several extracellular matrix-encoding genes. This increased gene expression could be inhibited by an antibody to TGF-β1 (a secreted peptide known to play a role in type 1 diabetes) suggesting the increase involves TGF-β1 activity (Sun et al. 2010). The increases in gene expression correlated with a decrease in the repressive marks H3K9me2 and H3K9me3 and an increase in the active marks H3K4me1, 2 and 3 across those genes. The changes seen in H3K4 methylation appear to involve the histone methyltransferase Set7/9 as recruitment of Set7/9 is enhanced in response to high glucose and knockdown of Set7/9 inhibits the increase in expression (Sun et al. 2010). Clearly other chromatin-modifying factors must also be involved as Set7/9 is only able to monomethylate H3K4 and has no direct effect on the levels of H3K9 methylation.
These data identify some changes in histone methylation levels at specific genes associated with type 2 diabetes. Short periods of hyperglycaemia have been suggested as the priming event for long-term progression of complications in type 2 diabetes, and it is clear that increasing the glucose levels to which cells are exposed is sufficient to trigger changes in histone methylation in some cell types. The identification of specific histone methyltransferases and demethylases that respond to hyperglycaemia should facilitate future investigations into the nature of the signalling pathways involved.
4 Histone Acetylation
Lysine residues on histones are subject to the addition of an acetyl group (Fig. 10.1a) by enzymes known as histone acetyltransferases (HATs), and this acetylation can be reversed by a group of enzymes known as histone deacetylases (HDACs). Inappropriate regulation of acetylation is associated with a host of human disease states; mutations within the histone acetyltransferase and CREB-binding protein (CBP) result in Rubinstein-Taybi syndrome, while HDAC inhibitors are used as anticancer agents (Kalkhoven et al. 2003; Witt et al. 2009). The addition of the acetyl group to the lysine residue neutralises the positive charge of the amino group of the lysine residue and is thought to promote a more open chromatin structure (Hansen et al. 1998). Such a change allows increased accessibility to transcription factors and RNA polymerase and thus enhances levels of gene transcription. Acetylated lysine residues are also recognised by bromodomain-containing proteins, and thus, a second mechanism by which acetylation regulates gene expression is by the recruitment of proteins and/or complexes that regulate gene expression (Zeng and Zhou 2002).
As with histone methylation, there are a selection of studies that implicate a role for histone acetylation changes in regulating the phenotype of cells in individuals with type 2 diabetes. Rats fed a high-fat diet to induce hyperglycaemia showed increased levels of histone acetylation of the fibrillin 1 gene in both the heart and the kidney. The increased levels of acetylation were associated with increased levels of fibrillin mRNA (Gaikwad et al. 2010). Monocytes from patients with diabetes also show enhanced levels of histone acetylation (Miao et al. 2004) and increased histone acetylation of some genes, e.g. Mll3 correlates with increased mRNA levels (Miao et al. 2007). In a genome-wide study using human aortic endothelial cells, Pirola et al. identified a number of genes that were associated with increased levels of H3K9 and H3K14 acetylation subsequent to high glucose (30 mmol/l) exposure as well as a number of genes that were associated with reduced levels of acetylation (Pirola et al. 2011). Analysis of the cohort of genes that showed hyperacetylation identified regulation of apoptosis and the NF-κβ pathways as common pathways that are influenced by these gene sets. The authors however could not uncover any clear association with the hypoacetylated gene set and a specific cellular pathway. Incubation of the endothelial cells with the histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), mimicked at least some of the hyperacetylation seen with hyperglycaemia. How much overlap there is between hyperglycaemia-induced hyperacetylation and that induced as a result of histone deacetylase inhibition is not known; presumably the latter would be more widespread but it may be important to interrogate the response in the future given the clinical use of histone deacetylase inhibitors to treat diseases such as cancer. In general, histone acetylation of H3K9 and H3K14 is associated with gene expression and hyperacetylation of genes in endothelial cells correlated well with an increase in mRNA levels (Pirola et al. 2011). A mechanism leading to hyperacetylated histones is suggested by experiments with human umbilical vein endothelial cells (HUVECS). Incubation of cultured HUVECS in high glucose (25 mmol/l) resulted in the increase of mRNA and protein level of the histone acetyltransferase p300 and enhanced recruitment of p300 at the NF-κB and fibronectin genes (Chen et al. 2010). Several transcription factors, including NF-κB, CREB and AP-1, were all induced by high glucose, and this induction was inhibited by knockdown of p300 while ectopic expression of p300 resulted in the increased expression of fibronectin, endothelin 1 and VEGF mRNA levels under normal levels of glucose (Chen et al. 2010).
Hyperglycaemia induces the expression of some genes in VSMC though whether VSMC shows increases in histone acetylation in type 2 diabetes similar to other cell types is not clear. However, it is likely and changes in acetylation do play an important role in VSMC proliferation and neointima formation. Inhibition of HDAC enzymes (which should presumably result in hyperacetylation) with the inhibitor Scriptaid reduced the proliferation of cultured rat VSMC by causing them to arrest at the G1 phase of the cell cycle. Additionally, in vivo administration of Scriptaid reduced the level of neointima in a mouse blood vessel injury model (Findeisen et al. 2011). Such evidence is apparently contradictory, and taken at face value, it would suggest that hyperacetylation in VSMC would be beneficial. Of course actively promoting acetylation is not quite the same as inhibiting deacetylation and these two approaches may not target the exact same set of genes.
Since it was first uncovered, regulation of histone acetylation has proven to be an important mechanism by which expression levels of genes are regulated in all tissues and in response to many pathophysiological stimuli. Clinically approved HDAC inhibitors are already available to treat some cancers so a more detailed understanding of acetylation in the progression of type 2 diabetes may uncover some potential opportunities for alternative uses of them.
5 Combinatorial Interactions
Histone proteins can be modified by acetylation, methylation, ubiquitination, phosphorylation and sumoylation. Many enzymes exist within the cell that can add or remove specific marks, usually only at a subset of the potential amino acids within a single histone. The availability of such an array of modifications led to the proposal of a histone code (Turner 1993; Strahl and Allis 2000) – histone modifications are read, either individually or in combination, by other proteins to bring about distinct functional outcomes. Since then, researchers have come a long way in understanding how single modifications are deposited and removed; however, it is clear that knowledge of a single epigenetic mark is not sufficient to predict functional changes in gene activity. Such observations suggest that combinations of histone modifications must interact, though our understanding of such interactions, or “crosstalk,” is still in its infancy. It is known that some modifications on histones are dependent on or enhanced by the presence of others. An initial study identified a requirement for the ubiquitination of histone H2B for the COMPASS complex to methylate histone H3 in yeast (Fig. 10.3) (Dover et al. 2002). Enhanced acetylation of histones can promote the methylation of H3K4 in cardiac myocytes (Fig. 10.3) (Bingham et al. 2007), while histone deacetylation is required for demethylation of H3K4 by LSD1 (Lee et al. 2005; Shi et al. 2005).
Combinatorial interactions between chromatin modifications. Shown are two examples of the presence of one chromatin modification influencing the deposition of another. Ubiquitination of H2BK123 is required for methylation of H3K4 by the COMPASS complex, while acetylation of histone H4 promotes the methylation of H3K4. Other interactions are known, and for a more in-depth discussion, see Wood (2011)
In type 2 diabetes, no specific interaction between histone modifications or association with combinatorial marks has yet been uncovered though it is clear that knowledge of a single mark is not sufficiently predictive of functional outcomes. For example, the mRNA levels of Mll3 are increased in monocytes from patients with type 2 diabetes, yet the gene is associated with an increase in the repressive mark H3K9me2. Similarly, histone acetylation levels of the SBF1 gene are also increased in monocytes which are exposed to high glucose, but the mRNA levels are decreased (Miao et al. 2007). Clearly our understanding of the precise relationship with specific epigenetic marks and gene activity is still incomplete, and we need to discover how individual marks interact with each other to produce a specific outcome. Advances in technology are enabling researchers to study a greater number of histone marks across entire genomes (Barski et al. 2007) as well as identifying epigenetic marks that coexist on the same nucleosome (Eberl et al. 2011), while interrogation of the vast amounts of data being generated is uncovering functional associations between marks and uncovering sets of chromatin marks or chromatin signatures (Hon et al. 2008, 2009).
6 Conclusions
Periods of transient hyperglycaemia in patients with type 2 diabetes appear to induce persistent changes that impact on an individual’s health after many years. There is compelling evidence for an epigenetic mechanism involving changes in the posttranslational modifications of DNA and histones around specific sets of genes which alter their long-term expression levels. Understanding which genes are affected and which specific epigenetic marks are involved should open up new avenues for treatment and/or management of complications associated with diabetes. Gaining an appreciation of the timeline of chromatin changes (and in which cells these are important) and how these relate to changes in gene expression levels and disease progression may also provide new opportunities for more accurate diagnoses that do not rely on quantifying the current metabolic state of an individual. Such data may provide a better predictive model regarding future potential complications for an individual who has suffered a period of metabolic disturbance.
Abbreviations
- CBP:
-
CREB-binding protein
- COMPASS:
-
Complex proteins associated with Set1
- CREB:
-
cAMP response element binding protein
- DCCT:
-
Diabetes Control and Complications Trial
- DNMT:
-
DNA methyltransferase
- H3K4me1:
-
Monomethylated Histone H3 lysine 4
- H3K4me2:
-
Dimethylated histone H3 lysine 4
- H3K4me3:
-
Trimethyl histone H3 lysine 4
- H3K9me2:
-
Dimethyl histone H3 lysine 9
- HDAC:
-
Histone deacetylase
- HAT:
-
Histone acetyltransferase
- HMT:
-
Histone methyltransferase
- HUVECS:
-
Human umbilical vein endothelial cells
- IL:
-
Interleukin
- JMJD2:
-
Jumonji domain 2
- LSD1:
-
Lysine-specific demethylase 1
- NFAT:
-
Nuclear factor of activated T cell
- PGC-1α:
-
Peroxisome proliferator-activated receptor gamma coactivator-1 alpha
- SAM:
-
S-adenosylmethionine
- shRNA:
-
Short hairpin RNA
- T2DM:
-
Type 2 diabetes mellitus
- TGF:
-
Transforming growth factor
- TNF:
-
Tumour necrosis factor
- VEGF:
-
Vascular endothelial growth factor
- VSMC:
-
Vascular smooth muscle cell
References
Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR (2009) Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab 10:189–198
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Bingham AJ, Ooi L, Kozera L, White E, Wood IC (2007) The repressor element 1-silencing transcription factor regulates heart-specific gene expression using multiple chromatin-modifying complexes. Mol Cell Biol 27:4082–4092
Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, El-Osta A (2009) Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 58:1229–1236
Chen S, Feng B, George B, Chakrabarti R, Chen M, Chakrabarti S (2010) Transcriptional coactivator p300 regulates glucose-induced gene expression in endothelial cells. Am J Physiol Endocrinol Metab 298:E127–E137
Chen C, Nott TJ, Jin J, Pawson T (2011) Deciphering arginine methylation: tudor tells the tale. Nat Rev Mol Cell Biol 12:629–642
Chiang EP, Wang YC, Chen WW, Tang FY (2009) Effects of insulin and glucose on cellular metabolic fluxes in homocysteine transsulfuration, remethylation, S-adenosylmethionine synthesis, and global deoxyribonucleic acid methylation. J Clin Endocrinol Metab 94:1017–1025
Dandona P, Aljada A, Bandyopadhyay A (2004) Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol 25:4–7
Day JJ, Sweatt JD (2010) DNA methylation and memory formation. Nat Neurosci 13:1319–1323
Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277:28368–28371
Eberl HC, Mann M, Vermeulen M (2011) Quantitative proteomics for epigenetics. Chembiochem 12:224–234
El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M (2008) Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205:2409–2417
Findeisen HM, Gizard F, Zhao Y, Qing H, Heywood EB, Jones KL, Cohn D, Bruemmer D (2011) Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition. Arterioscler Thromb Vasc Biol 31:851–860
Gaikwad AB, Gupta J, Tikoo K (2010) Epigenetic changes and alteration of Fbn1 and Col3A1 gene expression under hyperglycaemic and hyperinsulinaemic conditions. Biochem J 432:333–341
Hansen JC, Tse C, Wolffe AP (1998) Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37:17637–17641
Hon G, Ren B, Wang W (2008) ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol 4:e1000201
Hon G, Wang W, Ren B (2009) Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol 5:e1000566
Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33(Suppl):245–254
Kalkhoven E, Roelfsema JH, Teunissen H, den Boer A, Ariyurek Y, Zantema A, Breuning MH, Hennekam RC, Peters DJ (2003) Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum Mol Genet 12:441–450
Lee GH, Proenca R, Montez JM, Carroll KM, Darvishzadeh JG, Lee JI, Friedman JM (1996) Abnormal splicing of the leptin receptor in diabetic mice. Nature 379:632–635
Lee MG, Wynder C, Cooch N, Shiekhattar R (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437(7057):432–435
Li SL, Reddy MA, Cai Q, Meng L, Yuan H, Lanting L, Natarajan R (2006) Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes 55:2611–2619
Ling C, Groop L (2009) Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 58:2718–2725
Maier S, Olek A (2002) Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr 132(8 Suppl):2440S–2443S
Miao F, Gonzalo IG, Lanting L, Natarajan R (2004) In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem 279:18091–18097
Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R (2007) Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J Biol Chem 282:13854–13863
Nikoshkov A, Sunkari V, Savu O, Forsberg E, Catrina SB, Brismar K (2011) Epigenetic DNA methylation in the promoters of the Igf1 receptor and insulin receptor genes in db/db mice. Epigenetics 6:405–409
Nilsson J, Nilsson LM, Chen YW, Molkentin JD, Erlinge D, Gomez MF (2006) High glucose activates nuclear factor of activated T cells in native vascular smooth muscle. Arterioscler Thromb Vasc Biol 26:794–800
Nilsson-Berglund LM, Zetterqvist AV, Nilsson-Ohman J, Sigvardsson M, González Bosc LV, Smith ML, Salehi A, Agardh E, Fredrikson GN, Agardh CD, Nilsson J, Wamhoff BR, Hultgårdh-Nilsson A, Gomez MF (2010) Nuclear factor of activated T cells regulates osteopontin expression in arterial smooth muscle in response to diabetes-induced hyperglycemia. Arterioscler Thromb Vasc Biol 30:218–224
Pirola L, Balcerczyk A, Okabe J, El-Osta A (2010) Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol 6:665–675
Pirola L, Balcerczyk A, Tothill RW, Haviv I, Kaspi A, Lunke S, Ziemann M, Karagiannis T, Tonna S, Kowalczyk A, Beresford-Smith B, Macintyre G, Kelong M, Hongyu Z, Zhu J, El-Osta A (2011) Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res 21:1601–1615
Poirier LA, Brown AT, Fink LM, Wise CK, Randolph CJ, Delongchamp RR, Fonseca VA (2001) Blood S-adenosylmethionine concentrations and lymphocyte methylenetetrahydrofolate reductase activity in diabetes mellitus and diabetic nephropathy. Metabolism 50:1014–1018
Reddy MA, Natarajan R (2011) Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res 90:421–429
Roy S, Sala R, Cagliero E, Lorenzi M (1990) Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci U S A 87:404–408
Sanchez R, Zhou MM (2011) The PHD finger: a versatile epigenome reader. Trends Biochem Sci 36:364–372
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953
Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19:857–864
Singh SM, Murphy B, O’Reilly RL (2003) Involvement of gene-diet/drug interaction in DNA methylation and its contribution to complex diseases: from cancer to schizophrenia. Clin Genet 64:451–460
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R (2010) Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 21:2069–2080
Szyf M, Detich N (2001) Regulation of the DNA methylation machinery and its role in cellular transformation. Prog Nucleic Acid Res Mol Biol 69:47–79
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811–816
Turek-Plewa J, Jagodzinski PP (2005) The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett 10:631–647
Turner BM (1993) Decoding the nucleosome. Cell 75:5–8
Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R (2008) Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A 105:9047–9052
Villeneuve LM, Reddy MA, Natarajan R (2011) Epigenetics: deciphering its role in diabetes and its chronic complications. Clin Exp Pharmacol Physiol 38:401–409
Williams KT, Schalinske KL (2011) Tissue-specific alterations of methyl group metabolism and DNA hypermethylation in the Zucker (type 2) diabetic fatty rat. Diabetes Metab Res Rev. 28:123–131
Witt O, Deubzer HE, Lodrini M, Milde T, Oehme I (2009) Targeting histone deacetylases in neuroblastoma. Curr Pharm Des 15:436–447
Wood IC. (2011) Uncovering combinatorial interactions in chromatin. Epigenomics. 3:371–9.
Wu SC, Zhang Y (2010) Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11:607–620
Xu M, Long C, Chen X, Huang C, Chen S, Zhu B (2010) Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328:94–98
Yang BT, Dayeh TA, Kirkpatrick CL, Taneera J, Kumar R, Groop L, Wollheim CB, Nitert MD, Ling C (2011) Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia 54:360–367
Yap KL, Zhou MM (2011) Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry 50:1966–1980
Yerneni KK, Bai W, Khan BV, Medford RM, Natarajan R (1999) Hyperglycemia-induced activation of nuclear transcription factor kappaB in vascular smooth muscle cells. Diabetes 48:855–864
Zeng L, Zhou MM (2002) Bromodomain: an acetyl-lysine binding domain. FEBS Lett 513:124–128
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Wood, I.C. (2013). Chromatin Switching and Gene Dynamics Associated with Type 2 Diabetes. In: Jirtle, R., Tyson, F. (eds) Environmental Epigenomics in Health and Disease. Epigenetics and Human Health. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-36827-1_10
Download citation
DOI: https://doi.org/10.1007/978-3-642-36827-1_10
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-36826-4
Online ISBN: 978-3-642-36827-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)