Abstract
Type 2 diabetes has become a major health issue worldwide. Chronic hyperglycemia induces a low-grade inflammation that, on top of other mechanisms, leads to endothelial dysfunction. Mounting evidence suggests that DNA methylation, post-translational modifications of histones, and long non-coding RNAs play an important role in the initiation, maintenance, and progression of both macro- and micro-vascular complications of diabetes. Long-term exposure to hyperglycemia induces epigenetic changes that could become irreversible, a phenomenon known as the ‘metabolic memory.’ Whether epigenetic-based therapies could be used to slow or limit the progression of cardiovascular disease remains unclear. While non-coding RNAs are currently investigated as potential biomarkers that predict diabetic cardiovascular disease incidence and progression, their therapeutic role is only hypothetical. In this review, we highlight the latest findings in experimental and clinical studies relevant to epigenetics and cardiovascular disease in diabetes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes ranks a close second on burden of cardiovascular health after ischemic heart disease, costing an estimated $245 billion annually in the US alone [1•, 2]. Although cardiovascular events in patients with diabetes significantly decreased in the past decade, mortality is still driven by cardiovascular complications in those patients [3•].
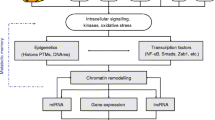
The pathogenesis of diabetic cardiovascular disease (CVD) has been widely studied during the past two decades. Vascular damage resulting from chronic exposure to hyperglycemia could be initially induced by the release of reactive oxygen species (ROS) [4]. Several other mechanisms participate in the assault, mainly the formation of advanced glycation end-products (AGEs), the activation of protein kinase C (PKC), as well as the polyol and hexosamine flux [5••]. Together, these activate several inflammatory pathways such as the nuclear factor-κB (NF-κB) signaling pathway that, once activated, controls pro-inflammatory genes regulation and expression, and induces secretion of cytokines and other inflammatory proteins [6] (Fig. 1).
Signaling and epigenetic networks mediating cardiovascular complications of diabetes. Cardiovascular damage resulting from chronic exposure to hyperglycemia is induced by the release of reactive oxygen species (ROS). Secondary to that, four pathways (PW) are activated: the polyol PW flux, advanced glycation end-products (AGEs) formation, activation of protein kinase C (PKC), and the hexosamine PW flux. These all enhance inflammatory reactions in the vascular system; in particular, they activate the nuclear factor-κB (NF-κB), trigger multiple signaling pathways, transcription factors (TFs), and crosstalk with epigenetic networks. Epigenetics modifications including histone modifications, DNA methylation, and non-coding RNA in both forms: miRNAs and LncRNAs. All these modifications control gene expression leading to apoptosis, hypertrophy, proliferation, and remodeling at different cells and tissues. Persistence of such epigenetic aberrations and changes in the transcriptional regulation of key pathologic genes contribute to diabetic macro- and micro-vascular disease
However, all of the known pathways do not explain the complex pathophysiology behind cardiovascular complications of diabetes. Even different variants of genes described in major genome-wide association studies (GWAS) have thus far had little impact on risk prediction of cardiovascular complications of diabetes [7]. Epigenetics, through its major components, DNA methylation, non-coding RNAs, and post-translational changes, play an important role in gene transcription [8]. Hence, the epigenome comprises all chemical modifications to DNA that influence the activity of genes within the genome [9]. Major efforts are currently underway to explore the human epigenome in an attempt to understand its role in the modulation of major diseases [9, 10].
Recent data support the role of epigenetics in the dysregulation of beta cells in patients with diabetes [11]. Additionally, epigenetics mechanisms participate in the process of inflammation [12], oxidative stress [13], and endothelial dysfunction [14], all representing the hallmark of cardiovascular complications of diabetes. Current therapies for diabetes are aimed to optimize glycemic control and reduce the associated cardiovascular risk factors. However, the success is moderate when it comes to diabetic CVD, as many complications are irreversible. It is therefore important to understand the epigenetic mechanisms of cardiovascular complications of diabetes as they can potentially be used to develop novel drugs. The current review aims to understand the emerging role of epigenetics in diabetic CVD.
Mechanisms of Epigenetics
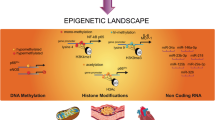
Epigenetics is the study of heritable changes in gene expression that are not due to changes in the DNA sequence [15]. Chromosomal DNA is tightly wrapped into chromatin by histone proteins and other chromatin assembly factors. These histone proteins were long considered passive supporting structures for chromatin [16, 17], but during the past decade, it has been clearly established that they contribute to gene expression by switching from an active (euchromatin) to inactive (heterochromatin) states, allowing regulation of the transcriptional state of the target gene [16, 18]. Epigenetic modifications allow both non-heritable changes that enable cells to rapidly adapt to any sudden variation of their environment and heritable modifications in response to extended stimuli that can be transmitted as memory to the next generation of cells [19]. The major epigenetic mechanisms include DNA cytosine methylation, covalent post-translational modifications of histones and non-coding RNAs [8].
DNA Methylation
DNA methylation refers to the biochemical reaction when a methyl group is added, by DNA methyl transferases, to the cytosine or adenine DNA nucleotides, and is one of the most stable epigenetic marks [20]. The principal targets of DNA methylation are specific dinucleotide sites in the genome called CpG islands (cytosine and guanine separated by one phosphate). All over the entire human genome, in normal cells, most of the CpG binucleotides are methylated [21], whereas CpG islands containing long stretches of CpG sites are usually unmethylated [22]. Bio-cellular states such as differentiation, degeneration, or imprinting, can induce methylation of promoter CpG islands resulting in gene repression [23]. Moreover, pathologic conditions can rise from this repression as in cancer cells where tumor suppressor genes are often silenced because of hypermethylation [24]. Protein–DNA interaction, gene expression, and chromatin stability can also be modified by methylation that occurs at sites different from CpG islands [25] such as in the brain, where CpH methylation-H could be adenine, guanine, or thymine-, was reported to cause transcriptional repression in neurons [26]. Abnormal DNA methylation or demethylation at the promoters of specific genes is one mechanism for the development of human diseases such as cancer, cardiovascular diseases (CVD), infections, or autoimmune diseases [27–29, 30•].
Post-Translational Modifications of Histones
Histones are the protein cores around which DNA wraps to form a tetrameric structure called nucleosome. Five histone families have been identified in humans: H1, H2A, H2B, H3, and H4 [31]. Histones play an important role in epigenetics as they regulate gene expression. The N-terminal part of histones is exposed to chemical reactions such as acetylation, methylation, ubiquitination, sumoylation, or phosphorylation, a phenomenon known as post-translational modifications (PTMs) of histones [18, 32•]. Such modifications change chromatin structure and, therefore, alter the binding of the transcription factors at different locations [33]. Additionally, PTMs of histones have a major role in the regulation of gene expression by producing anchoring sites for co-activators, co-suppressors, and other chromatin proteins regulators. Thereafter, histone changes can modulate the euchromatin (accessible) or heterochromatin (inaccessible) state of chromatin in combination with other epigenetics mechanisms [32•]. PTMs of histones control transcription, leading to either gene activation or silencing, which interferes with normal cellular cycles such as apoptosis, DNA damage repair; or even contribute to the pathogenesis of diseases [34, 35]. Moreover, histone modification levels are predictive for gene expression [36].
Noncoding RNAs
Non-coding RNAs (ncRNAs) are functional RNAs that are not translated into polypeptides or proteins [37]. Two forms represent the cornerstone of epigenetic signature: Micro-RNAs (miRNAs) and long non-coding RNAs (lncRNAs). miRNAs are approximately 20–22 nucleotides long [38] and lncRNAs are more than 200 nucleotides [39•]. They regulate gene expression post-transcriptionally by targeting the 3′ untranslated regions of mRNA; hence they can inhibit translation, degrade, or sequestrate mRNAs [40]. miRNAs are important regulators of up to 60 % of human genes. However, the function and impact of lncRNAs are still not completely understood. Recent data suggest that lncRNAs are able to regulate chromatin structure and organize proteins complexes across chromosomes [39•], as well as activate transcription at distal promoters [41].
Epigenetics of Cardiovascular Disease in Diabetes
Hyperglycemia, Inflammation, and Epigenetic Regulation of Cardiovascular Cell Types
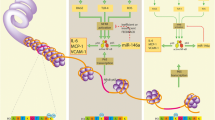
Factors that contribute to cardiovascular complications of diabetes such as chronic hyperglycemia, inflammation, and oxidative stress also affect DNA methylation, PTMs of histones, and ncRNAs in cardiovascular cells (see Picture 1). Experimental studies have shown that epigenetic changes secondary to hyperglycemia and inflammation are not only detected in various cardiovascular cell types but also in peripheral blood mononuclear cells (PBMCs) [13, 42, 43••].
Previous experimental studies have shown that the histone methyltransferase Set-7 induces monomethylation of lysine 4 of histone 3 on the promoter gene encoding for the transcription factor NF-κB p65 in endothelial cells exposed to hyperglycemia, which up-regulates the NF-κB p65 and increases the secretion of inflammatory molecules [12]. Paneni et al. recently replicated the same observations in monocytes of patients with T2D [13], and reported that Set-7 hyperglycemia-induced methylation was associated with increased plasmatic levels of intercellular cell adhesion molecule-1, monocyte chemo-attractant protein-1, and oxidative marker 8-isoprostaglandin F2α [13]. Miao et al. reported that epigenetic changes induced by hyperglycemia in PBMCs are cell-specific since lymphocytes and monocytes have distinct patterns of histone methylation in healthy individuals [42]. In monocytes of patients with T2D, hyperacetylation of inflammatory transcription factor promoters was predominant and correlated with inflammatory genes TNF-α and COX-2 activity, compared with normoglycemic individuals [44]. Miao et al. [43••] recently compared patients previously included in the conventional treatment arm of the Diabetes Control and Complications Trial (DCCT) who developed diabetic microangiopathy (cases) to patients who were allocated the intensive treatment and had no progression of microvascular complications (controls). They reported a significantly greater number of promoter regions with enrichment in H3K9Ac (hyperacetylation) in monocytes, but not in lymphocytes, in cases versus controls [43••]; this confirms the existence of an epigenetic component in the metabolic memory—the concept that early glycemic control is a major determinant of diabetic complications later in life. Experimental studies have demonstrated the implication of epigenetics in the metabolic memory of cultured cells exposed to diabetic conditions, as these cells continue to express pro-inflammatory and fibrotic genes long after restoration of normoglycemia [14, 45, 46].
A- Endothelial Cells
Endothelial cells (ECs) are sparsely distributed all over the cardiovascular system; hence they are directed affected by systemic hyperglycemia [47]. Endothelial dysfunction could rapidly develop without any clinical signs at the beginning. Epigenetics of cardiovascular complications of diabetes start primarily in ECs [14, 46, 48]. El-Osta et al. [14] reported that transient exposure to hyperglycemia could induce epigenetic changes in the p65 subunit promoter of the NF-κB, leading to permanent epigenomic changes in cultured aortic cells. Similarly, Brasacchio et al. [46] showed that hyperglycemia resulted in persistent histone modifications such as enhanced H3K4 and reduced H3K9 in aortic ECs. Moreover, quantitative measurements of histone PTMs and gene expression show that changes are glucose-dependent [46]. Recently, Paneni et al. [49] found that in human ECs exposed to hyperglycemia, the mitochondrial adaptor p66Shc was epigenetically upregulated by promoter CpG demethylation and H3 acetylation. Moreover the overexpression continued even after returning to normoglycemia and could only be inhibited after pharmacologic intervention [49]. In retinal ECs of diabetic mice, histone acetylation was increased along with inflammatory markers in mice presenting with diabetic retinopathy (DR) [50]. However, this mark was reversed by minocycline, which is usually used to treat DR in experimental studies [50]. miRNAs play a crucial role in mediating the endothelial dysfunction along with DNA methylation and PTMs of histones (Tables 1 and 2). However, the contribution of lncRNAs still remains to be clarified. Recently, Yan et al. showed that the expression of lncRNA-MIAT (myocardial infarction-associated transcript) is significantly increased in retinal ECs exposed to hyperglycemia. Additionally, its inhibition limited the damage of neovascularization [51].
Vascular Smooth Muscle Cells
Studies extrapolated from mouse models have shown that hyperglycemia increases the transcription and expression of inflammatory genes and induces pro-atherogenic responses in vascular smooth muscle cells (VSMCs). Villeneuve et al. showed that in VSMCs exposed to hyperglycemia, the CVD complications-protective epigenetic mark H3K9me3 was decreased at the promoters of inflammatory genes in diabetic mice [45]. In primary human vascular cells, the acetylation of histone H3 on K9 and K14 were both also induced by hyperglycemia [52]. Additionally, these changes were conversely correlated to DNA methylation [52]. Further, the same group reported that high levels of miR-125b correlated with reduced H3K9me3 at the promoter regions of inflammatory genes, concomitant with an increased expression of cytokines [53].
Cardiac Cells
Although emerging data linked some aspects of hypertrophy, heart failure, and arrhythmias in cardiomyocytes (CMs) to DNA methylation and PTMs of histones [30•], less evidence has been reported in hearts of diabetic patients. Monkemann et al. reported that oxidative stress damages CMs via p53-dependent apoptosis in diabetic cardiomyopathy [54]. Interestingly, methylation of the p21(WAF1/CIP1) gene that encodes several protein kinases at p53 is an early step in the development of hyperglycemia-induced cardiomyopathy in diabetic rats [54]. Gaikwad et al. reported deacetylation and dephosphorylation of histone H3 in the heart and kidney of diabetic Sprague-Dawley rats leading to changes in gene expression in the extracellular matrix (ECM) and therefore hypertrophy [55]. In all, several miRNAs involved in hypertrophy, fibrosis, and apoptosis have been identified in the diabetic heart (Table 1).
Renal Cells
Several experimental studies in diabetic mice and rats have demonstrated the role of histone modifications in diabetic nephropathy (DN) [56–58]. In a cell culture model, Sun et al. showed that immunoprecipitation of soluble chromatin from high-glucose and TGF-β-stimulated rat mesangial cells revealed enrichment of H3 methylation at K4 and depletion of the repressive K9 methylation mark at several pro-fibrotic promoters [56]. Gaikwad et al. reported in db/db mice that kidney failure induced H3 acetylation, dimethylation, and phosphorylation in CMs of T2D mice, predisposing them to hypertrophy [58]. Several miRNAs play a key role in the injury of podocytes and mesangial cells in the diabetic kidney, hence regulating initial changes in the ECM and later fibrosis. While most studies focused on the identification of miRNAs that are differentially expressed in different kidney tissues at different stages of DN, none of these have been validated yet in miRNA knockout mouse models and a few only were transfected to check functionality (Table 2).
Epigenetic Involvement in Clinical Studies of Cardiovascular Complications of Diabetes
Follow-up of the DCCT trial showed that T1D patients originally assigned to an intensive treatment developed less microvascular complications than patients who were assigned to the conventional treatment, despite narrowing of HBA1C difference between both groups at the end of the 4-year follow-up [59]. This epidemiologic study was the first to prove that an early and intensive glycemic control is crucial to preventing long-term cardiovascular complications of diabetes, and that the benefit remains even after loss of initial good glycemic control, thereafter defining the “legacy effect” in diabetes. The United Kingdom Prospective Diabetes Study (UKPDS) [60] reported almost two decades ago that patients with T2D assigned to an intensive diabetic treatment had less microvascular complications and no differences in macrovascular complications. However, the 10-year follow-up of patients who were initially allocated to the intensive treatment had fewer macro- and micro- vascular complications, even if there were no more differences in glycemic control [61]. Interestingly, this was also the case of the 10-year follow-up of the Veterans Affairs Diabetes Trial (VADT) study where participants did not initially show any improvement in the cardiovascular outcome [62] but 10 years later demonstrated a reduction in major cardiovascular events [63••].
However, Action in Diabetes and Vascular Disease: Preterax and Diamicron MR Controlled Evaluation (ADVANCE) [64] and Action to Control CardiOvascular Risk in Diabetes (ACCORD) [65] studies did not show any benefit of glucose-treatment intensification in patients with established complications or with high cardiovascular risk profiles in terms of preventing or limiting cardiovascular complications of diabetes; this might be due to the relatively short duration of follow-up of patients.
Epigenetic findings relevant to the metabolic memory in endothelial and vascular cells are consistent with the epidemiologic and clinical observations of the legacy effect [14, 46, 48]. Moreover, hyperglycemia induces epigenetic changes that are persistent even after returning to normoglycemia, a phenomenon many believe to be responsible for the irreversibility of cardiovascular complications of diabetes in individuals with poor long-standing glycemic control.
Clinical studies related to epigenetics of cardiovascular complications of diabetes, although rare, confirm some of the earlier findings of experimental studies (see related sections). In terms of DNA methylation and PTMs of histones, most studies were performed on PBMCs that could only be considered as surrogate endpoints since accessibility to cardiovascular cells could be technically and ethically difficult in patients with T2D [66]. Even within PBMC sub-populations, previous data demonstrated that epigenomics of lymphocytes and monocytes are different in normoglycemia and hyperglycemia conditions. The few available translational studies examined the epigenomes of smooth muscle cells and adipocytes in healthy, obese, and diabetic individuals [67, 68]. However, efforts are currently concentrated on the identification of plasma and urinary miRNAs for their potential use as biomarkers in the diagnosis and prognosis of cardiovascular complications of diabetic patients [69•].
We report here the main clinical studies related to epigenetics of macro- and micro-vascular complications of diabetes.
Epigenetics of Macrovascular Complications of Diabetes
Changes in DNA methylation and PTMs of histones have been related to coronary artery disease (CAD), heart failure, hypertrophic cardiomyopathy, dilated cardiomyopathy, arrhythmias, and other cardiovascular pathologies in the general population [30•]. miRNAs have been proposed as potential prognostic biomarkers for CAD in large cohorts of patients that also includes patients with diabetes [70, 71•, 72], implying a role for miRNAs in diabetic CAD. For instance, miR-4513 rs2168518 polymorphism has been associated with higher prevalence of traditional CV risk factors such as fasting glucose, incidence of T2D, and poor survival of CAD patients [70]. Motawae et al. recently showed that miR-9 and miR-370 were significantly elevated in patients with CAD and T2D versus patients with diabetes or CAD separately [73]. Zampetaki et al. reported that miR-126 was a strong predictor of myocardial infarction in a 10-year follow-up of 832 patients [71•]. Interestingly, miR-126 levels were low in circulating endothelial microparticles of diabetic patients with CAD [74].
Data on DNA methylation or histone acetylation in diabetic peripheral artery disease (PAD) is lacking [75]. Nevertheless, the potential of miRNAs for the diagnosis of PAD has recently been demonstrated. Initially, miR-503 has been highlighted as a regulator of diabetic PAD in experimental studies as it is increased in ischemic limb of diabetic mice [76]. Recently, 12 different circulating miRNAs were identified and characterized in peripheral blood of patients with T2D and PAD [77]. Although these findings were replicated in two separate samples of patients, the study was not designed to test the prognostic utility of the identified miRNAs in a longitudinal follow-up of diabetic patients with atherosclerotic PAD [77].
Epigenetics of Microvascular Complication of Diabetes
Clinical studies and/or studies done on human tissues consolidate the implication of epigenetics in microangiopathy pathophysiology and their progression with earlier experimental and animal findings. Sapienza et al. [78] reported 187 genes differentially methylated in DNA extracted from saliva of diabetic patients with renal failure compared with patients without DN. In another study by Stenvinkel et al. [79], DNA hypermethylation was found to correlate with C-reactive protein and interleukin-6 in patients with renal failure regardless of diabetes status. Genomic profiling of renal tubuli collected from patients with end-stage renal failure (ESRD), including diabetic patients, revealed differentially methylated enhancer regions of pro-fibrotic genes that are characteristics of nephrosclerosis [80]. In a small cohort of T1D patients, Bell et al. [81] identified 19 CpG sites differentially methylated in nephropathy compared with patients with T1D and normal kidney function. Moreover, the duration of diabetes prior to the onset of DN correlated with the changes in methylation of CpG sites [81]. We previously investigated adult individuals born from T1D mothers as a model of fetal exposure to maternal hyperglycemia, taking those born from T1D fathers as controls [82•]. We demonstrated that adult offspring of T1D mothers have a reduced renal functional reserve, which predisposes these individuals to hypertension, renal, and vascular diseases later in life [82•]. Further, we showed that global DNA was less methylated in offspring of T1D mothers versus T1D fathers [83]. Moreover, the gene encoding for DNA methyltransferase 1 (DNMT1)—a key enzyme involved in gene expression during early organs development—was under-methylated in offspring of T1D mothers [83].
Very few studies have addressed the usefulness of plasmatic and urinary miRNAs in clinical DN. Data regarding patients with diabetes are usually extrapolated from even fewer studies with limited number of participants. Neal et al. reported that total miRNAs were significantly reduced in patients with ESRD compared with patients with mild kidney impairment [84]. Moreover, miR-210 and miR-16 correlated with the progression of nephropathy [84]. In a cross-section study of patients with nephrotic syndrome and various underlying diseases—DN, minimal change and focal membranous—miR-200, miR-29a, and miR-192 were significantly different according to the groups versus healthy controls [85]. Argyropoulos et al. [86] identified 27 urinary miRNAs whose expressions and quantity are correlated to the severity of urinary albumin excretion and creatinine clearance. In individuals with T1D, urinary miRNA profiling revealed that miR-145 and miR-130a were elevated whereas miR-424 and miR-155 were decreased in patients with microalbuminuria versus patients with normoalbuminuria [87].
As we previously mentioned, in vitro and in vivo models showed that increased histone acetylation is an important epigenetic reaction to hyperglycemia in retinal ECs. In a case control study that included patients with various stages of diabetic retinal damage, Maghbooli et al. reported that DNA methylation levels were globally higher in patients with DR compared with controls. Moreover, higher levels were observed in advanced stages, which was not explained by conventional factors such as hypertension and diabetes duration [88]. Over 300 miRNAs have been reported to be dysregulated in retinal cells in experimental studies [89] but only a few have been validated (Table 2) and a tiny number have been tested in clinical settings. In patients with T2D, miR-21 is known to protect ECs from glucotoxicity [90]. Qing et al. recently reported that a combination of 3 miRNAs: miR-21, miR-181c and miR-1179; had an 82 % accuracy of predicting the development of proliferative DR in patients with non-proliferative retinopathy [91].
Implications
With the recent advances in sequencing technologies and bioinformatics analysis platforms, epigenetic discoveries are moving towards an exponential increase. This would offer the opportunity to discover novel drugs and potential diagnostic biomarkers.
Experimental models in diabetes and CVD show that inhibition or activation of enzymes such as histone acetylases, deacetylases, or DNA demethylases may reverse the damage caused by hyperglycemia in specific cells. TGF-β, hyperglycemia, and diabetes-induced changes lead to sustained expression of pro-fibrotic and pro-inflammatory genes [57]; thus, agents that reverse them may be useful for chronic renal fibrotic disorders such as DN. Yuan et al. showed that TGF-β antibody can reverse high-glucose induced enrichment of the active H3K4me2 mark at ECM gene promoters in mesangial cells [92], whereas Reddy et al. showed that losartan, an angiotensin receptor antagonist, reverses epigenetic modifications in renal cells of diabetic mice [93]. However, it is wise to remember that hyperglycemia targets different cells that do not share the same epigenome as the cardiovascular system. It is therefore unlikely that a systematic pharmacologic therapy targeting DNA methylation or histone PTMs would treat or even reverse cardiovascular complications of diabetes. Other alternatives may offer the potential to use epigenetics in the treatment of diabetes and related CVD. Induced pluripotent stem cells (IPSCs) offer a regenerative possibility for the cardiovascular system and the heart itself; it has been suggested that miRNAs could be easily used to produce induced IPSCs after modification of the programming of adult cells [94]. Macrophages are key cells in the development of occlusive atherosclerotic artery disease as they form harmful foam cells that are embedded with cholesterol. It has been suggested that in-vitro inhibition of histone deacetylase-3 in macrophages reduced atherogenicity [95]. Although this kind of translational approach is only starting to emerge, it may offer the potential to use epigenetics not only as a science to understand the pathophysiology of cardiovascular complications of diabetes but also as a therapeutic model in personalized medicine on the long-term.
As discussed earlier, several circulating miRNAs have been identified in patients with diabetes and CVD; however, replication in studies that include a large number of patients is lacking. Most importantly, there is a clear need to test the prognostic utility of miRNAs in order to obtain various micro-RNAs “signatures,” each predicting accurately the likelihood of developing a specific cardiovascular complication in patients with T2D.
Conclusions
It is widely accepted now that epigenetics participate in the initiation and progression of cardiovascular complications of diabetes as reported in experimental studies. However, the impact of epigenetics in clinical diabetes is still moderate. More anticipated studies should confirm methylation and histone PTMs of various human cells affected by hyperglycemia, other than peripheral blood cell mononuclear cells. On the other hand, many cohorts and trials with a larger number of participants are required to replicate initial diagnostic and prognostic potential of extra-cellular miRNAs. Recent follow-up of earlier neutral trials on glycemic intensification, such as UKPDS and VADT, indicate that epigenetic mechanisms that could be behind cardiovascular complications of diabetes could be reversible, even after some time. This may offer a therapeutic perspective for diabetic patients with already established cardiovascular disease using tissue-specific epigenetic modifiers drugs.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. This paper shows the evolution of communicable and non-communicable diseases all over the globe during the last 2 decades.
American Diabetes A. Economic costs of diabetes in the U.S. in 2012. Diabetes Care. 2013;36:1033–46.
Abi Khalil C, Roussel R, Mohammedi K, Danchin N, Marre M. Cause-specific mortality in diabetes: recent changes in trend mortality. Eur J Prev Cardiol. 2012;19(3):374–81. This review paper focuses on recent changes in cardiovascular mortality in patients with diabetes.
Munzel T, Gori T, Bruno RM, Taddei S. Is oxidative stress a therapeutic target in cardiovascular disease? Eur Heart J. 2010;31(22):2741–8.
Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414(6865):813–20. This paper summarizes the biochemical process underlying cardiovascular complications of diabetes.
Baker RG, Hayden MS, Ghosh S. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 2011;13(1):11–22.
Prudente S, Dallapiccola B, Pellegrini F, Doria A, Trischitta V. Genetic prediction of common diseases. Still no help for the clinical diabetologist! Nutr Metab Cardiovasc Dis. 2012;22(11):929–36.
Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–63.
Maunakea AK, Chepelev I, Zhao K. Epigenome mapping in normal and disease States. Circ Res. 2010;107(3):327–39.
Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30.
Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31(6):1405–26.
Okabe J, Orlowski C, Balcerczyk A, Tikellis C, Thomas MC, Cooper ME, et al. Distinguishing hyperglycemic changes by Set-7 in vascular endothelial cells. Circ Res. 2012;110(8):1067–76.
Paneni F, Costantino S, Battista R, Castello L, Capretti G, Chiandotto S, et al. Adverse epigenetic signatures by histone methyltransferase Set-7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circ Cardiovasc Genet. 2015;8(1):150–8.
El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205(10):2409–17.
Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–8.
Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–19.
Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–79.
Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705.
Nightingale KP, O'Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16(2):125–36.
Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213(2):384–90.
Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8(7):1499–504.
Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22.
Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92.
Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2 Suppl 1:S4–11.
Fouse SD, Nagarajan RO, Costello JF. Genome-scale DNA methylation analysis. Epigenomics. 2010;2(1):105–17.
Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17(2):215–22.
Dang MN, Buzzetti R, Pozzilli P. Epigenetics in autoimmune diseases with focus on type 1 diabetes. Diabetes Metab Res Rev. 2013;29(1):8–18.
Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harb Perspect Med. 2012;2(12):a010272.
Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–59.
Abi Khalil C. The emerging role of epigenetics in cardiovascular disease. Ther Adv Chronic Dis. 2014;5(4):178–87. This recent review paper summarized the implication of epigenetics in atherosclerosis and major cardiac pathologies.
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60.
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95. This review paper explains accuratley PTMs of histones.
Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12.
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80.
Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–68.
Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107(7):2926–31.
Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5(4):e1000459.
Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–6.
Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. This is a recent review paper explaining the role of lncRNAs.
Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol Genomics. 2008;33(2):139–47.
Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154(6):1190–3.
Miao F, Wu X, Zhang L, Riggs AD, Natarajan R. Histone methylation patterns are cell-type specific in human monocytes and lymphocytes and well maintained at core genes. J Immunol. 2008;180(4):2264–9.
Miao F, Chen Z, Genuth S, Paterson A, Zhang L, Wu X, et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes. 2014;63(5):1748–62. It is the first study in humans to demonstrate the theory of the metabolic memory in patients with type 1 diabetes and nephropathy.
Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279(17):18091–7.
Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A. 2008;105(26):9047–52.
Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58(5):1229–36.
Paneni F, Beckman JA, Creager MA, Cosentino F. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. Eur Heart J. 2013;34(31):2436–43.
Feng B, Ruiz MA, Chakrabarti S. Oxidative-stress-induced epigenetic changes in chronic diabetic complications. Can J Physiol Pharmacol. 2013;91(3):213–20.
Paneni F, Mocharla P, Akhmedov A, Costantino S, Osto E, Volpe M, et al. Gene silencing of the mitochondrial adaptor p66(Shc) suppresses vascular hyperglycemic memory in diabetes. Circ Res. 2012;111(3):278–89.
Kadiyala CS, Zheng L, Du Y, Yohannes E, Kao HY, Miyagi M, et al. Acetylation of retinal histones in diabetes increases inflammatory proteins: effects of minocycline and manipulation of histone acetyltransferase (HAT) and histone deacetylase (HDAC). J Biol Chem. 2012;287(31):25869–80.
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143–56.
Pirola L, Balcerczyk A, Tothill RW, Haviv I, Kaspi A, Lunke S, et al. Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res. 2011;21(10):1601–15.
Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59(11):2904–15.
Monkemann H, De Vriese AS, Blom HJ, Kluijtmans LA, Heil SG, Schild HH, et al. Early molecular events in the development of the diabetic cardiomyopathy. Amino Acids. 2002;23(1-3):331–6.
Gaikwad AB, Gupta J, Tikoo K. Epigenetic changes and alteration of Fbn1 and Col3A1 gene expression under hyperglycaemic and hyperinsulinaemic conditions. Biochem J. 2010;432(2):333–41.
Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010;21(12):2069–80.
Sayyed SG, Gaikwad AB, Lichtnekert J, Kulkarni O, Eulberg D, Klussmann S, et al. Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transplant. 2010;25(6):1811–7.
Gaikwad AB, Sayyed SG, Lichtnekert J, Tikoo K, Anders HJ. Renal failure increases cardiac histone h3 acetylation, dimethylation, and phosphorylation and the induction of cardiomyopathy-related genes in type 2 diabetes. Am J Pathol. 2010;176(3):1079–83.
Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. N Engl J Med. 2000;342(6):381–9.
Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet. 1998;352(9131):837–53.
Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HAW. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 2008;359(15):1577–89.
Duckworth W, Abraira C, Moritz T, Reda D, Emanuele N, Reaven PD, et al. Glucose control and vascular complications in veterans with type 2 diabetes. N Engl J Med. 2009;360(2):129–39.
Hayward RA, Reaven PD, Wiitala WL, Bahn GD, Rda DJ, Ge L, et al. Follow-up of glycemic control and cardiovascular outcomes in type 2 diabetes. N Engl J Med. 2015;372(23):2197–206. This recently published follow-up of the VADT trial showed that early intensification of diabetes might be benefitial, but only on the long term.
Group AC, Patel A, MacMahon S, Chalmers J, Neal B, Billot L, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358(24):2560–72.
Action to Control Cardiovascular Risk in Diabetes Study G, Gerstein HC, Miller ME, Byington RP, Goff Jr DC, Bigger JT, et al. Effects of intensive glucose lowering in type 2 diabetes. N Engl J Med. 2008;358(24):2545–59.
Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6(7):828–37.
Drogan D, Boeing H, Janke J, Schmitt B, Zhou Y, Walter J, et al. Regional distribution of body fat in relation to DNA methylation within the LPL, ADIPOQ and PPARgamma promoters in subcutaneous adipose tissue. Nutr Diabetes. 2015;5:e168.
Liu F, Sun Q, Wang L, Nie S, Li J. Bioinformatics analysis of abnormal DNA methylation in muscle samples from monozygotic twins discordant for type 2 diabetes. Mol Med Rep. 2015;12(1):351–6.
Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol. 2013;9(9):513–21. A comprehensive review paper relating to circulating microRNAs in patients with diabetes.
Li Q, Chen L, Chen D, Wu X, Chen M. Influence of microRNA-related polymorphisms on clinical outcomes in coronary artery disease. Am J Transl Res. 2015;7(2):393–400.
Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60(4):290–9. This study is the most comprehensive on the prognostic role of miRNAs in coronary artery disease.
Goretti E, Wagner DR, Devaux Y. miRNAs as biomarkers of myocardial infarction: a step forward towards personalized medicine? Trends Mol Med. 2014;20(12):716–25.
Motawae TM, Ismail MF, Shabayek MI, Seleem MM. MicroRNAs 9 and 370 Association with Biochemical Markers in T2D and CAD Complication of T2D. PLoS ONE. 2015;10(5):e0126957.
Jansen F, Yang X, Hoelscher M, Cattelan A, Schmitz T, Proebsting S, et al. Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 2013;128(18):2026–38.
Kullo IJ, Leeper NJ. The genetic basis of peripheral arterial disease: current knowledge, challenges, and future directions. Circ Res. 2015;116(9):1551–60.
Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, et al. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123(3):282–91.
Stather PW, Sylvius N, Wild JB, Choke E, Sayers RD, Bown MJ. Differential microRNA expression profiles in peripheral arterial disease. Circ Cardiovasc Genet. 2013;6(5):490–7.
Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2014;6(1):20–8.
Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, et al. Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease? J Intern Med. 2007;261(5):488–99.
Ko YA, Mohtat D, Suzuki M, Park AS, Izquierdo MC, Han SY, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14(10):R108.
Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3(5):33–38
Abi Khalil C, Travert F, Fetita S, Rouzet F, Porcher R, Riveline J-P, et al. Fetal exposure to maternal type 1 diabetes is associated with renal dysfunction at adult age. Diabetes. 2010;59(10):2631–6. This study is aligned with the recent concept of programmation of adult disease during fetal life.
Gautier JF, Porcher R, Abi Khalil C, Bellili-Munoz N, Fetita LS, Travert F, et al. Kidney dysfunction in adult offspring exposed in utero to type 1 diabetes is associated with alterations in genome-wide DNA Mmthylation. PLoS ONE. 2015;10(8):e0134654.
Neal CS, Michael MZ, Pimlott LK, Yong TY, Li JY, Gleadle JM. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant. 2011;26(11):3794–802.
Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Urinary sediment miRNA levels in adult nephrotic syndrome. Clin Chim Acta. 2013;418:5–11.
Argyropoulos C, Wang K, McClarty S, Huang D, Bernardo J, Ellis D, et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS ONE. 2013;8(1):e54662.
Barutta F, Tricarico M, Corbelli A, Annaratone L, Pinach S, Grimaldi S, et al. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS ONE. 2013;8(11):e73798.
Maghbooli Z, Hossein-nezhad A, Larijani B, Amini M, Keshtkar A. Global DNA methylation as a possible biomarker for diabetic retinopathy. Diabetes Metab Res Rev. 2015;31(2):183–9.
Kovacs B, Lumayag S, Cowan C, Xu S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011;52(7):4402–9.
Zeng J, Xiong Y, Li G, Liu M, He T, Tang Y, et al. MiR-21 is overexpressed in response to high glucose and protects endothelial cells from apoptosis. Exp Clin Endocrinol Diabetes. 2013;121(7):425–30.
Qing S, Yuan S, Yun C, Hui H, Mao P, Wen F, et al. Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell Physiol Biochem. 2014;34(5):1733–40.
Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol. 2013;304(5):F601–13.
Reddy MA, Sumanth P, Lanting L, Yuan H, Wang M, Mar D, Alpers CE, Bomsztyk K, Natarajan R. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int. 2014;85(2):362–73
Mallanna SK, Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344(1):16–25.
Van den Bossche J, Neele AE, Hoeksema MA, de Heij F, Boshuizen MC, van der Velden S, et al. Inhibiting epigenetic enzymes to improve atherogenic macrophage functions. Biochem Biophys Res Commun. 2014;455(3-4):396–402.
Zheng D, Ma J, Yu Y, Li M, Ni R, Wang G, Chen R, Li J, Fan GC, Lacefield JC, et al. Silencing of miR-195 reduces diabetic cardiomyopathy in C57BL/6 mice. Diabetologia. 2015.
Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36(2):181–8.
Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, et al. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010;584(16):3592–600.
Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res. 2010;86(3):410–20.
Greco S, Fasanaro P, Castelvecchio S, D'Alessandra Y, Arcelli D, Di Donato M, et al. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61(6):1633–41.
Baseler WA, Thapa D, Jagannathan R, Dabkowski ER, Croston TL, Hollander JM. miR-141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type 1 diabetic heart. Am J Physiol Cell Physiol. 2012;303(12):C1244–51.
Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23(2):252–65.
Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–7.
Long J, Wang Y, Wang W, Chang BH, Danesh FR: Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem 2010;285(30):23457–65
Dey N, Das F, Mariappan MM, Mandal CC, Ghosh-Choudhury N, Kasinath BS, et al. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J Biol Chem. 2011;286(29):25586–603.
Lai JY, Luo J, O'Connor C, Jing X, Nair V, Ju W, et al. MicroRNA-21 in glomerular injury. J Am Soc Nephrol. 2015;26(4):805–16.
Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008;22(12):4126–35.
Zhang Z, Luo X, Ding S, Chen J, Chen T, Chen X, et al. MicroRNA-451 regulates p38 MAPK signaling by targeting of Ywhaz and suppresses the mesangial hypertrophy in early diabetic nephropathy. FEBS Lett. 2012;586(1):20–6.
Alvarez ML, Khosroheidari M, Eddy E, Kiefer J. Role of MicroRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS ONE. 2013;8(10).
Bai Y, Bai X, Wang Z, Zhang X, Ruan C, Miao J. MicroRNA-126 inhibits ischemia-induced retinal neovascularization via regulating angiogenic growth factors. Exp Mol Pathol. 2011;91(1):471–7.
Feng B, Chen S, McArthur K, Wu Y, Sen S, Ding Q, et al. miR-146a-Mediated extracellular matrix protein production in chronic diabetes complications. Diabetes. 2011;60(11):2975–84.
Mortuza R, Feng B, Chakrabarti S. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia. 2014;57(5):1037–46.
Murray AR, Chen Q, Takahashi Y, Zhou KK, Park K, Ma JX. MicroRNA-200b downregulates oxidation resistance 1 (Oxr1) expression in the retina of type 1 diabetes model. Invest Ophthalmol Vis Sci. 2013;54(3):1689–97.
Silva VA, Polesskaya A, Sousa TA, Correa VM, Andre ND, Reis RI, et al. Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Mol Vis. 2011;17:2228–40.
Fulzele S, El-Sherbini A, Ahmad S, Sangani R, Matragoon S, El-Remessy A, et al. MicroRNA-146b-3p regulates retinal inflammation by suppressing adenosine deaminase-2 in diabetes. Biomed Res Int. 2015;2015:846501.
Acknowledgments
The authors gratefully acknowledge funding from the Qatar National Research Fund under its National Priorities Research Program award number NPRP 7-701-3 – 192 and the Biomedical Research Program at Weill Cornell Medical College in Qatar.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Jennifer Pasquier, Jessica Hoarau-Véchot, Khalid Fakhro, Arash Rafii, and Charbel Abi Khalil declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Macrovascular Complications in Diabetes
Rights and permissions
About this article
Cite this article
Pasquier, J., Hoarau-Véchot, J., Fakhro, K. et al. Epigenetics and Cardiovascular Disease in Diabetes. Curr Diab Rep 15, 108 (2015). https://doi.org/10.1007/s11892-015-0677-3
Published:
DOI: https://doi.org/10.1007/s11892-015-0677-3