Abstract
Cultivation of edible mushrooms is the premier agricultural application of mycology. The use of natural biodiversity and molecular genetics for breeding might contribute significantly to the successful outcome of several challenges concerning the management of fruiting induction, fungal diseases, or food quality which growers and mushroom industry participants are faced with. This chapter deals mainly with the saprophytic edible mushrooms belonging to the genus Agaricus, and highlights the button mushroom Agaricus bisporus. The importance of wild germplasm in Agaricus breeding is discussed by stressing the interest of phylogeny for identifying new interesting species or varieties, and the lack of diversity in the cultivated strains, whereas a genetic and phenotypical diversity is now available in Agaricus collections. Breeding strategies adapted to their life cycles and using molecular markers and quantitative genetics are proposed for genetic improvement of Agaricus strains in the era of genomics.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Mitochondrial Genome
- Single Nucleotide Polymorphism Marker
- Edible Mushroom
- Restriction Fragment Length Polymorphism
- Nuclear Migration
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
I. Introduction
Cultivation of edible mushrooms is the premier agricultural application of mycology. Humanity’s use of mushrooms extends back to Palaeolithic times. According to Boa (2004), the archaeological record reveals edible species associated with people living 13,000 years ago in Chile, but it is in China where the eating of wild fungi is first reliably noted, several hundred years before the birth of Christ. Edible fungi were collected from forests in ancient Greek and Roman times and highly valued, though more by high-ranking people than by peasants. At the beginning of the twenty-first century, gourmet mushrooms may contribute to the development of a new agriculture by addressing the consumer demand for healthy and sustainable products and some of the non-nutritional use of agricultural productions in developed countries, as well as making substantial contributions to the diets of poor people in developing countries. They may either be commercially collected in forests or cultivated, and present an interesting biodiversity.
By compiling more than 200 different sources from 110 countries, but excluding detailed review of species from developed countries, over 2,300 wild useful species of mushrooms were identified by Boa (2004). Because of a decline in forest-based industries in some countries, wild mushrooms are now considered as new sources of income even in northern countries (Román and Boa 2006). A mushroom is defined as a macrofungus with a distinctive fruiting body that is large enough to be seen with the naked eye and to be picked up by hand, and can be either a Basidiomycete or an Ascomycete, aerial or underground (Chang and Miles 1992). Mushrooms can be roughly divided into various categories depending on their ecology. Saprophytic mushrooms play an important role in the cycling of carbon and other elements through the breakdown of lignocellulosic plant residues and animal dung, whilst ectomycorrhizal mushrooms are involved in symbiotic associations with plant roots. Despite the important ecological and economical role of mushrooms, forest planning and management has paid little or no attention to the harvesting of wild edible fungi for a long time. A new challenge is the development of a science-based production of mushrooms in forests, sustaining the development of a mycosilviculture. For a review on the trends in this topic, see Savoie and Largeteau (2011).
Today, most of the mushrooms recognized as cultivated are saprophytic species. Some of them can be produced in forests on inoculated wood logs or other substrates, but this outdoor culture is dependent on local climatic conditions, and hence they are generally cultivated indoors. FAOSTAT (2011) indicates that the world mushroom production in 2010 was about 6.0 million tons, with significant progress in the past 20 years (2.1 million tons in 1991, 4.2 million tons in 2000) which shows the increasing interest for edible and medicinal mushrooms as an agricultural crop. Marshall and Nair (2009) reported 12 species that are commonly grown for food and/or medicinal purposes, across tropical and temperate zones, including the common mushroom (Agaricus bisporus), shiitake (Lentinula edodes), oyster (Pleurotus sp.), straw mushroom (Volvariella volvacea), lion’s head or pom pom (Hericium erinaceus), ear (Auricularia auricula), reishi (Ganoderma lucidum), maitake (Grifola frondosa), winter (Flammulina velutipes), white jelly (Tremella fuciformis), nameko (Pholiota nameko), and shaggy mane mushrooms (Coprinus comatus). One can add at least three other species, the pavement mushroom (Agaricus bitorquis), the almond mushroom (Agaricus subrufescens), and blewit (Lepista nuda). Commercial markets are dominated by A. bisporus, L. edodes and Pleurotus spp., which represent three quarters of mushrooms cultivated globally.
Understanding the ecology of mushrooms in their natural environment is the main requirement for efficient development of a cultivation process.
-
Cultivated mushrooms have two kinds of saprophytic lifestyles. Most of them are primary decomposers that can be cultivated on pasteurized or sterilized lignocellulosic substrates. The others are leaf-litter secondary decomposers cultivated on composts prepared from various agricultural wastes including manures. The cultivation substrates are both physical supports and nutrient sources for the mushrooms, which need to be able to degrade them with appropriate sets of enzymes. The choice of the agricultural by-products and their processing before cultivation is directed by local availabilities, and there are many projects attempting to optimize the bioconversion of these wastes by mushroom cultivation. The reader is directed to the proceedings of the International Conferences on Mushroom Biology and Mushroom Products (Savoie et al. 2011) for examples of experiments on various cultivation substrates.
-
The fruiting-bodies are reproductive differentiated parts of macroscopic fungi, and the bottlenecks to large development of mushroom technologies are due to a lack of knowledge with regard to two major parts of the biology of most of the potentially interesting species: (i) life cycle and genetics, and (ii) factors and mechanisms responsible for fruiting induction and fruiting body development.
-
As plant crops are, cultivated mushrooms are susceptible to a variety of viral, bacterial and fungal diseases as well as various pests. Studies concerning the mechanisms involved in host–pathogen interactions are intended to improve the control of pests and microbially induced diseases of mushrooms (Gaze and Fletcher 2007; Largeteau and Savoie 2010).
-
The quality of the final products is the final challenge for the mushroom growers. The mushrooms should have a good quality and a long storage shelf life. Food quality is defined by the degree to which it meets consumers’ expectations, which are mainly taste and nutritional/healthful profiles for mushrooms.
Growers and other participants in the mushroom industry are faced with these different challenges. The use of natural biodiversity and molecular genetic approaches for breeding might contribute significantly to achieving a successful outcome of these challenges. The information discussed in the rest of this chapter deals mainly with saprophytic edible mushrooms belonging to the genus Agaricus. Agaricus bisporus (Lange) Imbach, the button mushroom, is arguably the most studied mushroom species.
II. Wild Germplasms for Mushroom Breeding
Crop wild relatives and local varieties are the elements of agricultural biodiversity most likely to contain the necessary novel, unique, and high level of genetic diversity needed to sustain innovations in breeding programs. This assertion developed for plants is also true for mushrooms but it implies a robust taxonomic and phylogenetic knowledge and a preservation of the wild resources.
A. Phylogeny for Identifying New Interesting Species or Varieties
Agarics are popular fungi, picked and consumed in many countries. However, their determination remains difficult even for expert mycologists in Europe. Tropical or subtropical species are less well-known than temperate ones. A review of the twentieth-century literature on Agaricus L.: Fr. emend Karst finds a diversity of opinion on the circumscription of natural infrageneric groups and on the relationships of species within and among the proposed groups. The group of species most closely related to the economically important, cultivated species A. bisporus (Lange) Imbach is no exception. This situation is changing because of recent progress in the classification facilitated by molecular characterization and phylogeny. The genus Agaricus has been shown to be monophyletic (Vellinga et al. 2011). Eight sections are recognized in the subgenus Agaricus: Agaricus, Arvenses, Bivelares, Chitonioides, Minores, Sanguinolenti, Spissicaules, and Xanthodermatei (Parra 2008; Zhao et al. 2011). The sections Bivelares (Kauffman) L.A. Parra and Xanthodermatei Singer have been phylogenetically reconstructed by analyzing DNA sequences from the ITS1+2 region of the nuclear rDNA (Challen et al. 2003; Kerrigan et al. 2006, 2008), and other sections are under investigation. Such a taxonomic and phylogenetic project is valuable for the development of the cultivation of new Agaricus species or varieties.
It is noteworthy that some important traits are shared by all or almost all the species of certain sections, and not by the others. For example, the toxic species mostly belong to Agaricus section Xanthodermatei (Kerrigan et al. 2006), and the more popular edible species belong to four sections, but only those of two sections (Bivelares with A. bisporus and A. bitorquis; Arvenses with A. subrufescens) can be easily cultivated on compost. Similar data are noted for odors, volatile components, and for certain macro-chemical reactions. Agaricus section Xanthodermatei comprises a group of species allied to A. xanthodermus and generally characterized by sporophores having phenolic odors, transiently yellowing discolorations in some parts of the sporophore, and Schaeffer’s reaction negative (Kerrigan et al. 2006). Certain odors appear to be synapomorphic characters and are crucial for taxonomy (Parra 2008). It can be hypothesized that these traits and their underlying secondary metabolism have been conserved over dozens of million years because they were implicated in crucial biological processes such as spore dissemination or sporophore defence (Callac et al. 2005). We therefore predict that phylogeny will be very helpful to detect species of nutritional, biochemical or medicinal interest. Work is in progress to derive supported hypotheses about phylogenetic relationships and trends in character evolution within the genus Agaricus, by sequencing nuclear genes and comparing synapomorphic characters linked to the production of secondary metabolites implicated in mushroom adaptation and/or having potential interest for human industry. A new approach in mycology tends to resolve phylogenetic relationships at the infra-genus level, and to use phylogenies as tools for interpreting adaptive evolution and predicting the potentialities of this phylum to contain individuals with valuable properties. This concept of useful phylogenetic systematics has to be developed, and Agaricus are good models.
The geographic origins of the species and their dominant climatic conditions could be another interesting way to find specific adaptations to climate. A recent study attempting to compare temperate and tropical Agaricus species (Zhao et al. 2011) showed that classifying the species into climatic groups is not so easy. Among the cultivated species, the geographical range of some temperate species such as A. bisporus and A. bitorquis extends into tropical areas and, reciprocally, the tropical species A. subrufescens exists also in Europe (Zhao et al. 2011). This intraspecific diversity might also be a source of interesting traits to be selected for scientific studies of biodiversity or the production of new cultivars of edible mushrooms.
B. Lack of Diversity in the Cultivated Strains
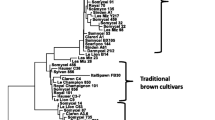
Despite the economic importance of A. bisporus and its long history of cultivation since the eighteenth century, few efforts have been made in terms of breeding and strain improvement. As reported in the previous edition of this volume (Horgen and Castle 2002) the first cross-bred A. bisporus strains (Horst-U1 and U3) were developed in The Netherlands in the 1980s, and no new hybrids with a different genetic background have been developed since then. As a result, all currently grown cultivars are assumed to be related to a limited number of traditional genotypes, and A. bisporus appears to be nearly a monolineage crop. This has been observed during the past 30 years on sets of about 20 strains per study by using different markers: isoenzymes (Royse and May 1982a), RFLP (Loftus et al. 1988), RAPD (Khush et al. 1992; Moore et al. 2001; Staniaszek et al. 2002), and ISSR (Guan et al. 2008). In a recent study with 75 cultivated genotypes provided by European spawn makers from 1990 to 2005 and maintained under liquid nitrogen, the combination of the allelic patterns obtained with 14 SSR loci made it possible to identify 13 distinct genotypes (Foulongne-Oriol et al. 2011c). Six groups were identified which corresponded to the five ancestral lineages and the hybrids Horst-U1 or Horst-U3. The ancestral lineages had been previously defined by phenotypes as “off-white”, “small white”, “white”, “brown”, “small brown”, and “golden white” (Royse and May 1982b; Foulongne-Oriol et al. 2009). Thirty-three cultivars showed the same genetic profile as U1/U3 hybrid strains. Using a mitochondrial marker, it was possible to separate them into two subgroups that correspond to either U1 or U3 as expected (Sonnenberg et al. 1991). In a parallel project, Sonnenberg et al. (2011) generated single nucleotide polymorphic markers (SNP’s) from analysis of the genomes of the two parental homokaryons of the hybrid Horst U1, and selected 600 markers evenly distributed over the whole genome. In two traditional commercial white varieties cultivated before the release of HU1, 46 % and 50 % of all 600 SNP markers showed both alleles present in the hybrid that had been obtained by crossing these two varieties. All of the nine present-day white commercial strains show a striking similarity to Horst U1. Taking into account an error of 1 % in SNP marker scoring, the authors consider these varieties as identical to Horst U1 and identical to each other. All these studies show the narrow genetic variability among the cultivars of A. bisporus. An exception is a hybrid developed during the 1980s in China (Wang et al. 1995). Despite the striking genetic similarity of the present-day commercial varieties to the first released hybrid Horst U1, phenotypic differences can be seen in the scaling, pinning, or the size of mushrooms, as observed by growers.
The lack of diversity in the cultivated strains of the button mushroom is considered an important risk for this culture, and efforts have been made during the past 30 years to overcome this problem.
Agaricus subrufescens Peck (syn. A. blazei Murrill sensu Heinemann, A. rufotegulis Nauta or A. brasiliensis Wasser, M. Didukh, Amazonas & Stamets), is a cultivated mushroom whose cultivation is developing in various countries since 25 years. For taxonomy and synonymy of this taxon we followed Kerrigan (2005); Arrillaga and Parra (2006); Ludwig (2007); and Cappelli (2011). Kerrigan (2005) and Wasser et al. (2002) agreed that the name A. blazei Murrill sensu Heinemann was inappropriate. The homonyme A. subrufescens Ellis & Everh is posterior as this has been corrected in Index Fungorum. Because of its particular fragrance and taste, this basidiomycete popularly known as “the almond mushroom” is now considered as one of the most important culinary–medicinal biotechnological species, with rising demand in consumption and production worldwide (for reviews see Largeteau et al. 2011b; Wisitrassameewong et al. 2012a). The cultivation of the almond mushroom started on a commercial scale in the 1980s in Japan, after the isolation and study of one Brazilian isolate, from the region of Piedade, São Paulo State. The majority of the strains spread over the world most probably came from the culture originally sent from Piedade to Japan, as no further discovery in nature was reported until January 2001, when the species was found growing naturally on a heap of mown grass at Embrapa Florestas, Colombo, State of Paraná, Brazil (Amazonas 2005). The mushroom was for a long time considered as endemic. Consequently, few commercial cultivars are currently available. Brazilian and Japanese authors have investigated the genetic polymorphism among cultivated strains, mainly by using RAPD markers, and they showed a high genetic homogeneity (see Largeteau et al. 2011b). In each country, the strains currently cultivated probably derived from a single or very few sporophores, because the growers select the best strains. However, with the increasing interest in this mushroom, a new hybrid was recently patented in USA (Kerrigan and Wach 2008) and work is in progress to improve the genetic diversity of A. subrufescens for the development of its cultivation under various conditions.
As in plants, the progressive loss of genetic diversity in cultivated lines (genetic erosion) resulting from man’s selection of the best Agaricus strains, or the absence of initial diversity due to a specific history of the cultivated species, raises the issue of the sanitary and economic risks related to a mono-crop. Wild types are important sources of breeding material to restore genetic variability as well as to improve the characteristics of commercially cultivated varieties.
C. Agaricus Collections, a Source of Diversity
1. Collections of Genetic Resources for Agaricus spp.
A prerequisite for breeding is the availability of genetically diverse source materials. In the 1980s, there appear to have been fewer than 20 independent lines of A. bisporus in mainstream culture collections worldwide, including those of commercial laboratories (Kerrigan 1996). There was concern about losing genetic diversity forever, and at the end of the 1980s, a few researchers decided to support collection and conservation of the germplasm of A. bisporus. Two major collections, the ARP (Agaricus Resource Program, Kerrigan 1996) in the USA, and the CGAB (Collection du Germoplasme des Agarics à Bordeaux, Callac et al. 2002) at INRA-Bordeaux have been developed. They now contain hundreds of wild isolates originating from various habitats and numerous geographical origins, representing a source of genetic diversity.
The aim of the Agaricus Resource Program (ARP) was to encourage the discovery, acquisition, preservation, characterization, and distribution of novel germplasm of A. bisporus and other closely related species of Agaricus. Meanwhile, with the contribution of some European mycological societies, mycologists, and collaborations with scientists from North America, Greece, Mexico, and more recently Thailand and China, French mushroom scientists had gradually constituted another collection, CGAB (Callac et al. 2002). Some specimens are both in ARP and CGAB, but the sum of the two collections represents more than 800 wild specimens, mostly collected either on cypress or spruce litter, or on manure, but also in sandy semi-arid habitats. Their distribution mainly covers Europe, the Mediterranean region, and North America. Some wild specimens have also been isolated in other areas such as Asia and are in laboratory collections of universities and research institutes. For instance, wild A. bisporus strains were collected from the Tibetan Plateau, and are in the collection of the Sichuan Academy of Agricultural Sciences (Wang et al. 2008a). The known geographic range of A. bisporus extends from the boreal region of Alaska (Geml et al. 2008) to the equatorial climate of Congo (Heinemann 1956), and from coastal dunes to mountains of more than 3,000 m elevation (Largeteau et al. 2011a), but few living specimens from extreme habitats are available in culture collections. The geographic and ecological diversity of the strains in collections is a positive point for the objective of biodiversity preservation and valorization. An efficient germplasm resource base is available for the commercial Agaricus strain development industry, while enabling scientific study of this natural resource.
For the other cultivated and potentially cultivable Agaricus species, germplasms are scarce. The Agaricus subrufescens germplasm suffered from the controversy concerning its taxonomy, and probably from the fact that it is a relatively rare species in Northern countries. In addition to the cultivated strains with their low genetic diversity presented above, the collection of genetic resources for this organism is becoming enriched with a small number of North American, European (Kerrigan 2005), and more recently Asian isolates (Wisitrassameewong et al. 2012b). Because of the new interest in this mushroom, work is in progress to increase the number of specimens in collections to study their genetic diversity.
2. Genetic Diversity in Collections
The diversity of Agaricus species is to be discovered with investigation in new areas. A recent project showed that approximately 50 distinct species were harvested from a small area of northern Thailand in a few days, most of them probably being novel species. Only about one-third of tropical species belong to the classical sections of the Agaricus based on temperate species (Zhao et al. 2011). This diversity indicates that Agaricus is a species-rich genus in the tropics as well as in temperate regions, with potentialities for identification of new cultivable species with culinary or medicinal interests. An interspecific genetic diversity is expected to be revealed in the next few years.
At the intraspecific level, due to the wild germplasms of the other Agaricus species being less developed than for A. bisporus, there is little information on their intraspecific genetic diversity.
Some projects on ITS and other taxonomic markers have used several specimens of the same species. That is the case, for instance, with A. bitorquis or A. cupressicola, for which three genotypes based on ITS sequences were identified, with five isolates of each species analyzed (Kerrigan et al. 2008). It is noteworthy that ITS1+2 sequences have been shown to be informative for the intraspecific diversity in Agaricus spp. With regard to A. subrufescens, Kerrigan (2005) did not distinguish geographical populations (with the exception of the Hawaiian samples) by sequence characters within the strains from North America, South America, or Europe which he studied.
Enrichment of the germplasm of these species is promising for an increasing availability of genetic diversity.
There have been several works using different markers showing the genetic diversity in collections of A. bisporus (see Horgen and Castle 2002). Recently, microsatellite markers and SNPs used to demonstrate the close relationship between the cultivated strains of A. bisporus (see II-B) were also useful for evaluating the available genetic diversity in collections. In 19 wild accessions on average for 29 % of all 600 SNP markers, both alleles of the cultivar strain used as reference were found by Sonnenberg et al. (2011) whilst the percentage was about 50 % for non-hybrid cultivars. Using 33 SSR markers, Foulongne-Oriol et al. (2009) observed a significant higher polymorphism among 20 wild isolates than among seven cultivars representing the six morphotype lineages assumed to represent all or almost all the genetic variability available among the traditional cultivars and the hybrid Horst U1. This clearly shows that wild accessions are distantly related to commercial varieties. But in clustering analysis, relatedness of cultivars with wild strains originating from France was observed (Foulongne-Oriol et al. 2009), in agreement with the hypothesis that most of the cultivars are probably derived from a native European ancestral population (Xu et al. 1997).
Germplasm collections are often a result of historical events and arbitrary decisions, collecting missions, and specific research programmes, resulting in over-representation of certain materials, whereas other types of material can be under-represented. The A. bisporus germplasm is no exception. As a consequence, five known major populations appearing reproductively isolated from each other had been defined by Kerrigan (2004): three in North America (western Canada, coastal California, Sonoran Desert), one in the Middle East, and one in Europe. Inside the European population, the subpopulation in Greece and Crete is genotypically distinctive whilst retaining European characteristics (Callac et al. 2002). This subpopulation, as well as four different French local populations, is probably over-represented in the European germplasm. However, the genetic diversity inside these different levels of populations is interestingly high.
In a fine-scale genetic analysis of diversity, Xu et al. (2002) monitored French samples from one field (50 × 70 m) containing horse manure as substrate and having frequent human disturbance, and from another site (20 × 30 m) associated with Monterey cypress trees and without human disturbance, over a 2-year period. There were high levels of genetic variation, and their results demonstrated limited evidence for vegetative clonality of A . bisporus in nature. The largest potential genet was found in about 1 m2. Genetic drift within a population and gene flow among neighboring populations could contribute to gene and genotype changes over years. However, the significant differentiation between the two sites located about 450 km apart suggests that long-distance gene flow was relatively limited and that a high biodiversity might be preserved in situ in local populations for their further use.
Besides, hybridizations between cultivar-like strains and elements of other populations have been truly demonstrated on California Coast samples (Kerrigan et al. 1998) whereas other populations such as that of Alberta were proven to be poorly contaminated by cultivar-like genotypes (Xu et al. 1997). When new sources of breeding material are needed to restore genetic variability in cultivars, one can collect new strains in the wild. Because of the risk of introgression in wild populations and to the dependence on climatic conditions for the collection in natural populations, the preservation of the genetic diversity in perennial germplasm is a challenge, both for mycologists and the commercial Agaricus strain development industry.
3. Phenotypic Diversity in Collections
Studies on wild germplasm have produced data on phenotypic diversity for morphological traits and behavioral traits. Trait diversity in wild A. bisporus was reviewed by Kerrigan (2004). The main characteristics that have relevance to economic development of the A. bisporus cultivation are cap color, post-harvest quality reaching consumer expectation, temperature tolerance, disease resistance, and differences in cultivation characteristics such as time of fruiting or number and weight of fruiting-bodies. Some recent data on this diversity are presented here.
The cap in A. bisporus is variably fibriollo-squamulose and color varies from white to dark brown, with a diversity that could be used in breeding programs.
The majority of the current button mushroom sales throughout the world are white mushrooms, while the wild specimens are mainly brown with many gradations. Pileus color indicated by brightness (L parameter measured by a chromameter) in a sample of 418 isolates studied in our research group ranged from 52 % to 93 % of the reference white color (see Kerrigan 2004). The percentage of isolates exhibiting white cap (L > 88) in this sample was lower than 4 %, and it may vary from 0 % in a French population to about 10 % in a Greek population (Callac et al. 2002). Using samples from an open site and from a site under cypresses (the same two sites studied for fine-scale genetic analysis of diversity described in paragraph C-2 above), it was observed that all the 21 isolates found under cypress had a brown cap color (L < 60), whilst 2 out of 16 isolates from the open site had a cream cap color (80 < L < 88), and 9 had light brown pilei (80 < L < 88). The colors were measured after cultivation in climatic rooms protected from daylight (Callac et al. 2005), showing the genetic origin of this trait.
Both the available diversity and knowledge concerning genetic control (see III-B) meet the requirements for breeding programs on this easily measurable trait. In breeding programs of research institutes or spawn companies, white mushrooms derived from commercial strains are crossed with brown wild strains. The objectives are either to obtain white hybrid of A. bisporus with given characteristics of the wild strains, or to introgress brown color and other wild traits in a white commercial strain, as recently described for instance in a United States Patent (Robles and Lodder 2009).
Cap color diversity in other cultivated Agaricus species is less well-documented. Agaricus bitorquis is one of the three species of the section Bivelares reported by Callac et al. (2005) which should have definitely lost putative ancestral alleles determining the brown color, whereas cap color of A. subrufescens isolates has been reported to vary from cream to brownish-gold in the few studied specimens (Kerrigan 2005; Llarena Hernández et al. 2011). Hybrids we obtained between brown and cream strains exhibited various colors (unpublished data). Further work is necessary for a better evaluation of the cap color diversity in A. subrufescens, and to know whether genetic determinants of the color are homologous to those of A. bisporus.
Shelf life performance and susceptibility to discoloration after harvest are other quality traits to be taken into account in addition to cap color, because they affect the commercial value of mushrooms. Mushroom discoloration is a post-harvest stress disorder caused by senescence processes and by mechanical damage as a consequence of the enzyme-catalyzed oxidation of phenols into quinones. By comparing 2-day post-harvest mushrooms with freshly harvested mushrooms, 20 genes with increased expression levels have been identified (Eastwood et al. 2001), showing a probable genetic determinism of post-harvest stress disorders in addition to effects of culture and storage conditions (Burton 2004). A collection of A. bisporus strains was screened for their bruising sensitivity in order to analyze the phenotypic variation in susceptibility to discoloration after mechanical damages among strains (Weijn et al. 2011). The results indicated that some brown wild strains showed less bruising sensitivity than white commercial lines. Breeding programs for improving insensitivity are in progress based on the use of this diversity (Gao et al. 2011).
Disease resistance in A. bisporus has recently been reviewed (Berendsen et al. 2010; Largeteau and Savoie 2010), with significant reports of work on susceptibility diversity in many cultivars and wild strains. We will not deal here in detail with the different diseases. For instance, information is available on the wide range of sensitivity to Lecanicillium fungicola in the wild lines of A. bisporus, and work published recently states the genetic bases of this trait and its interest for breeding programs (Kerrigan 2004; Largeteau et al. 2004, 2005; Sonnenberg et al. 2005; Foulongne-Oriol et al. 2011d). We tested 450 strains of A. bisporus from CGAB for their susceptibility to an isolate of L. fungicola in experiments with controlled inoculation of the pathogen, as in Juarez del Carmen et al. (2002). Between 20 % and 35 % of diseased mushrooms were recorded for commercial hybrids used as controls, and Fig. 1.1 shows the large diversity and the interesting potential of some strains on the left bottom part of the figure producing high yields with low rates of affected mushrooms. The exploitation of strains which are less affected or show fewer or milder symptoms than the commercial strains when exposed to pathogens is an objective shared by breeding companies and research institutes or universities.
There are numerous published and unpublished pieces of work in which diversity in the yield parameters of Agaricus mushrooms has been recorded. Examples of diversity in the germplasm of A. subrufescens have recently been reported, with yield of the better wild strains from temperate countries being 500 % that of some presently cultivated strains originating from Brazil (Llarena Hernández et al. 2011; Zied et al. 2011). This opens the possibility of improvements in hybrids that are already exploited (Kerrigan and Wach 2008). For A. bisporus, the standards of yield obtained with the current hybrids when cultivated under controlled conditions are close to the maximum level, and some secondary components of the complex determinants of the yield have to be selected.
Ecological and physiological adaptation to specific environmental conditions is an important trait for the ability of the strains to colonize the substrate and to produce fruiting bodies during cultivation under various conditions. Adaptation to the composts used as cultivation substrate is a behavioral trait with economic interest for the diversification of production areas. Using the same two sites studied for the fine-scale genetic analysis of diversity (Xu et al. 2002) and for the correlation of cap color with the habitat (Callac et al. 2005), Savoie et al. (1996) observed a significant difference between the two populations in their ability to colonize and degrade mushroom compost. The population from the cypress litter was less efficient than that from the open area exposed to horse manure on a field. This is a second illustration, after the cap color, of the fact that knowledge of the population may help to select samples of strains where the chance to find a given trait is increased.
Temperature tolerance is a useful commercial trait related to the yield parameters, for which geographical variation should be expected. However, in a project considering high temperature tolerance for both mycelial growth rate and fruiting ability in A. bisporus, Largeteau et al. (2011a) observed that the phenotypes correlated neither with climate/microclimate nor with habitat. Strains from the same sub-populations had contrasted phenotypes. This illustrates the limit of the prediction of the frequency of an interesting trait based on geographical origin. However, an intercontinental difference was observed. The ability to produce mature fruiting bodies at 25 °C taken as a whole appeared higher in North American populations than in European ones. This difference could result from the different history of the two continental populations.
III. Genetic Improvement of Agaricus Strains
A. Various Reproducing Systems Limiting or Facilitating Breeding Strategies
As in other fungi, there are different processes of reproduction in A. bisporus that can be classified into three groups: vegetative reproduction (which produces through mitosis a mycelium theoretically genetically identical to the original), sexual reproduction through meiosis, and other processes sometimes grouped under the name of parasexual reproduction. An understanding of reproduction gives basic information for other research, for the management of natural resources, and for breeders. A. bisporus is an interesting model for addressing these questions, because different sexual life cycles can occur in this species.
1. The Three Life Cycles of A. bisporus
Agaricus bisporus var. bisporus has a multiallelic unifactorial system of sexual intercompatibility (Miller and Kananen 1972), the locus MAT (Xu et al. 1993) having 14 alleles (Kerrigan et al. 1994; Imbernon et al. 1995), and its life cycle is amphithallic, i.e., pseudohomothallic (= secondary homothallic) or heterothallic (Fig. 1.2), according to the ploidy level of the spores, which can be respectively heterokaryotic (n + n) or homokaryotic (n) (Lange 1952; Kuhner 1977). In this variety, most of the basidia are bisporic and produce heterokaryotic spores which confer upon it a predominant pseudohomothallic life cycle (Raper et al. 1972).
The three life cycles of Agaricus bisporus. A. bisporus is an amphitallic species with a homothallic or heterothallic cycle depending on the ploidy level of the spores, which can be heterokaryotic (n + n) or homokaryotic (n) respectively. Each dominant life cycle is characteristic of a variety. A. bisporus var. bisporus is predominantly pseudohomothallic, A. bisporus var. burnettii is heterothallic, A. bisporus var. eurotetrasporus, is homothallic
More precisely, for 215 wild French isolates examined in cultivation, Callac et al. (1996) found that the percentages of bi-, tri- and tetrasporic basidia were on average 81 %, 18 %, and 1 %. The percentage of homokaryotic offspring, possibly varying between 1 % and 10 %, cannot be easily estimated for several reasons. The proportion of n-spored basidia depends not only on genetic factors but also on environmental conditions (Kerrigan and Ross 1987). The ploidy status of the spores of the three-spored basidia is unknown, and the germination rate of the homokaryotic spores and/or their viability can be lower than those of the heterokaryotic spores, because of the presence of lethal or deleterious recessive alleles. However, even with a low rate of spore germination, this is not negligible in absolute, if we consider that a single sporophore produces about one billion spores. These haploid spores give rise to unfertile homokaryons.
In the heterothallic life cycle, plasmogamy between two sexually compatible homokaryons restores a fertile heterokaryon. This is used in conventional breeding schemes. In contrast, in the pseudohomothallic life cycle, heterokaryotic spores give rise to fertile heterokarons. Most of the wild populations and all the traditional cultivated strains belong to A. bisporus var. bisporus . Consequently, the low percentage of homokaryotic offspring is a significant drawback, slowing down the breeding work (Kerrigan et al. 1992). With the enrichment of the germplasm during the past two decades, and the examination of hundreds of wild specimens, the species concept of A. bisporus (Lange) Imbach has been refined (Callac et al. 2002; Kerrigan 2007). Based on morphological, biological, and genetic studies, two new varieties of A. bisporus have been described.
A. bisporus var. burnettii Kerrigan & Callac has been described on the basis of specimens found in the Sonoran Desert of California (Callac et al. 1993). In the sporophores of this variety, most of the basidia are tetrasporic and, correlatively, its amphithallic life cycle is predominantly heterothallic (Kerrigan et al. 1994). More precisely, for 58 wild Californian isolates examined in cultivation, Callac et al. (1996) found that the percentages of bi-, tri- and tetrasporic basidia were on average 1 %, 14 %, and 85 %. This variety differs from the two other varieties by traits reflecting adaptation to dryness: smaller mean spore size and faster sporophore development. The average spore numbers per basidium, and correlatively the predominating type of life cycle, are primarily determined by the BSN locus (basidial spore number) which is linked to MAT on chromosome I (Imbernon et al. 1995, 1996; Callac et al. 1997). A. bisporus var. burnettii is known only in the population of the Sonoran Desert of California, and is completely inter-fertile with the var. bisporus. Inter-varietal hybrids (var. bisporus x var. burnettii ) have a predominantly heterothallic life cycle, because of the dominance (sometimes incomplete) of the tetrasporic allele (Bsn-t) at the BSN locus. Such hybrids make it possible to obtain large recombined homokaryotic progeny useful for breeding work, performing genetic maps, and studying the inheritance of traits of interest (see III-B)
A. bisporus var. eurotetrasporus Callac & Guinberteau was described on the basis of rare tetrasporic specimens found in France and in Greece, and belonging to the same genet (Callac et al. 2003). The life cycle of this variety is homothallic: homokaryotic sporophores produce homokaryotic spores, giving rise to fertile homokaryons (Fig. 1.2). The basidia are mainly tetrasporic as in var. burnettii, but spores have the same mean size as those of var. bisporus. This variety is interfertile with both var. bisporus and var. burnettii, and most of the basidia of such hybrids are tetrasporic. Moreover, a natural hybrid between var. bisporus and var. eurotetrasporus has been found. Tests for allelism at BSN showed that both var. eurotetrasporus and var. burnettii bear similar dominant Bsn-t alleles, but we do not know whether they are ancestral or even whether they have a common origin (Callac et al. 1998).
Segregation for the haploid fruiting ability that characterizes the var. eurotetrasporus has been studied among the homokaryotic offspring of a hybrid between var. eurotetrasporus and var. bisporus. Genetic determinants of this trait could not be detected because numerous false positive (hybridization by unexpected inoculum of A. bisporus) or negative mushrooms (contamination by a competitor due to the too slow growth rate of the homokaryon) occurred in fruiting tests. The trait was inherited by at least 24 % of the homokaryotic offspring, but the haploid sporophores were generally smaller and less vigorous than those of the parent of the var. eurotetrasporus (Couture et al. 2004). Haploid sporophores have been also obtained exceptionally in var. bisporus (Dickhardt 1985). In var. burnettii, haploid sporophores have been obtained experimentally, but they are weak.
Cytological studies have shown that karyogamy and meiosis with synaptonemal complex occur in var. eurotetrasporus as in the two other varieties (Kamzolkina et al. 2006). This homomictic process indicates that the life cycle of A. bisporus var. eurotetrasporus is homothallic in the strict sense. The persistence of sexual spores would maintain a better fitness than an asexual process (apomixis) via a more stringent screening for deleterious mutations (Bruggeman et al. 2003). Tetrasporic basidia probably also confer a better fitness than bisporic basidia, which seems to be without interest for a haploid homothallic fungus. A. bisporus var. eurotetrasporus is probably in a sympatric speciation process. Specimens of this variety were collected under cypress, and on one occasion in the company of A. bisporus var. bisporus and A. agrinferus (Kerrigan & Callac). This latter and A. subfloccosus (J.E. Lange) J. are the species most closely related to A. bisporus. These both edible and cultivable species are homothallic, and constituted of multiple non-recombining genets (Kerrigan et al. 1999, 2008). A. bisporus var. eurotetrasporus represents a source of Bsn-t alleles useful for breeding work as does the var. burnettii, but also its haploid fruiting ability is a tool for studying development of the sporophores and the genetics of traits of interest.
2. Special Features of Basidia
The pseudohomothallism in A. bisporus var. bisporus has three important characteristics that are not independent from each other: (1) the spores of the bisporic basidia receive two postmeiotic nuclei, which complement each other at the mating type locus (Sass 1929; Evans 1959; Royse and May 1982b; Summerbell et al. 1989; Kerrigan et al. 1993), (2) the parental heterozygosity is highly conserved in the heterokaryotic descendants, and (3) crossover is not frequent. What are the consequences in terms of variability recovered among heterokaryotic and homokaryotic offspring?
In the basidia, karyogamy, meiosis, and sporogenesis successively occur. In the bisporic basidia of the var. bisporus, the migration of the four haploid postmeiotic nuclei is not random: each spore receives two non-sister postmeiotic nuclei, one nucleus from each second division of meiosis. This model agrees with all studied offspring except an atypical one in which inclusion of sister nuclei has occurred (Spear et al. 1983). Evans (1959) proposed that the non-random distribution results from the spatial position of the two second divisions, whilst Kamzolkina et al. (2006) suggested it could result from their asynchronous divisions. Whatever the explanation, this process implies that the heterokaryotic spores receive the two homologous centromeres of the parental heterokaryon for each pair of chromosomes. In the absence of crossover, the heterokaryotic offspring would have the same global genotype as the parent, and 100 % of the parental heterozygosity would be conserved. However, due to the random distribution of the centromeres at the first meiotic division, the centromeres, like the alleles of any heterozygous loci located on different chromosomes, can be differently distributed among the two nuclei of the heterokaryotic descendants. Such heterokaryons can exhibit different phenotypes via epistatic effects that partly explain phenotypic variability among heterokaryotic offspring.
This has been shown experimentally in Neurospora crassa (Burton and Metzenberg 1972). If crossovers occur, alleles segregate at the second meiotic division, and as a result heterokarotic spores can be homoallelic at a locus that was heteroallelic in the parent. The probability of losing the parental heteroallelism at a given locus theoretically increases with the distance between this locus and the centromere. The fact that the heterokaryotic offspring remains heterokaryotic at MAT suggests that MAT is close to the centromere. The loss of parental heterozygosity is low in heterokaryotic offspring, suggesting that the rate of crossovers is low, but that it can also result from selection due to deleterious or lethal recessive alleles making unviable homoallelic recombined heterokaryons.
Finally, the observed variability among a heterokaryotic offspring can result from different processes: the redistribution of the centromeres in the two nuclei, the moderate loss of parental heterozygosity, heterokaryotic selection, and other possible processes (see Moquet et al. 1998). This is the genetic variability resulting from the intramictic process of the pseudohomothallic life cycle which breeders try to exploit by selecting inside monospore cultures. Heterokaryons isolated from multispore cultures could additionally result from self-cross either between homokaryon and heterokaryon or between two compatible homokaryons (plasmogamy in the heterothallic life cycle). In the latter case, half of the parental heterozygosity is theoretically lost, and inbreeding depression occurs as has been observed by Xu (1995), but in a back-cross generation.
Today, using the Bsn-t alleles of var. burnetii or var. eurotetrasporus , large homokaryotic offspring are available to develop methods based on controlled hybridization (see below III-B). The variability among the homokaryotic offspring highly depends on the rate of crossovers. In a segment of chromosome I, the rates of recombination observed among the progeny of hybrids between the different varieties have suggested that an incompletely dominant allele(s), possibly Bsn-t, could determine a high recombination rate (Kerrigan et al. 1993; Callac et al. 1997, 1998). This is consistent with the hypothesis that high and low rates of recombination are adaptive for heterothallic and pseudohomothallic isolates respectively. In A. bisporus var. bisporus, successive generations of heterokaryotic spores may permit deleterious alleles to accumulate. A low rate of recombination maintains a high level of heterozygosity and, by complementation, high viability and fitness among most offspring.
3. Outcrosses: Which Way?
Outcrosses and recombination generate variability which is required for genetic selection in breeding programs. This variability is also crucial for adaptation in nature. From the predominantly pseudohomothallic life cycle described above for A. bisporus var. bisporus, it seems difficult to perform outcrosses; as one might expect they are infrequent in the wild populations since only a small proportion of the offspring are homokaryotic. Population studies indicate that outcrossing occurs in the wild (Xu et al. 1997), but although Kerrigan found in North America some evidence of pseudoclonal lineages resulting of successive pseudohomothallic generations (Kerrigan 1990), this was less evident in European studied populations, in which outcrossing could be more frequent (Xu et al. 2002). The two main ways of outcrossing are crosses between compatible homokaryons (heterothallic life cycle) and crosses between homokaryons and heterokaryons. The latter process, also called the “Buller phenomenon” (Buller 1931) was first reported by Raper et al. (1972) in A. bisporus.
In different experiments, Callac et al. (2003, 2006, 2008, and unpublished) inoculated a standard substrate for A. bisporus cultivation simultaneously with homokaryotic mycelium from one parent and spores from a second parent (Fig. 1.3). Culture trays have consistently produced numerous sporophores that could theoretically have resulted from five different reproductive modes (pseudohomothallism, selfing or outcrossing via heterothallism, and selfing or outcrossing via the Buller phenomenon). However, genotype analysis showed that all or almost all the sporophores consistently resulted from outcrossing between the inoculated homokaryon and the inoculated heterokaryotic spores (or mycelia that grew from them), i.e., via the Buller phenomenon. Control trays inoculated with spores only or mycelium only did not produce any sporophores. The exceptions were due to contaminations by unexpected inoculum in first experiments because the air entering in the culture room was not sterilized. This method represents an easy way to get numerous hybrids in a single experiment.
Method of hybridization in A. bisporus var. bisporus using the Buller phenomenom. A homokaryon from a first parent and spores from a second parent are simultaneously inoculated in standard compost trays. Numerous hybrid sporophores are produced, each of them resulting from a cross between a heterokaryotic spore and the homokaryon
Such hybrids can receive recessive deleterious alleles, since there is no haploid step (no gametic/haploid selection) for the material coming from the parent of the spores, but this is not necessarily a disadvantage. For instance, it has been shown that such hybrids, which received recessive lethal alleles at loci tightly linked to MAT from the spores of one of the parents, were on average less susceptible to dry-bubble disease than those that did not receive these alleles (Callac et al. 2008). Such resistance linked to MAT and to a locus bearing a lethal recessive allele could not be exploited using conventional crosses between homokaryons. A QTL of disease resistance was identified in the vicinity of MAT (Foulongne-Oriol et al. 2012b). It was also shown that these hybrids systematically had the mitochondrion of the homokaryon, even though in conventional crosses between homokaryons of the two parents this mitochondrion was never inherited. This method can be used to control the effect of mitochondrial inheritance in the crosses. In these experiments, the complete absence of sporophores directly issued from the heterokaryotic spores was unexpected, and the success of this method with different parental strains suggests that the Buller phenomenon could play a role in nature.
4. Mycelium and Anastomosis
The mycelium of A. bisporus can be homokaryotic (n) or heterokaryotic (n + n), but in both cases articles of the hyphae are without clamp connection, and multinucleate with a variable number of nuclei (Saksena et al. 1976; Hou and Elliott 1978; Kamzolkina et al. 2006). The mycelium can highly resist cold or dryness. Although vegetative spores or conidia are not detected in A. bisporus, pieces of mycelium could have a role of dissemination in the field but could also contaminate another mycelium by transmitting a virus (MVX dsRNAs; Grogan et al. 2005) or by crossing with it (Callac et al. 2003). Such contaminations occur via anastomosis. Anastomosis between two mycelia permits exchange not only of nuclei but also mitochondria, cytoplasm, and any other intracellular components.
Anastomosis occurs between heterokaryons that can be genetically different and allow trophic and other exchanges, as in the following cases: (1) in experimental transplantation of sporophores on a recipient mycelium (Sinden et al. 1962), (2) in experimental co-cultivation of a homokaryotic mycelium and spores that produce many genetically different hybrid sporophores on the same compost tray (Callac et al. 2006), and (3) in certain processes of cultivation (CACing) in which mushroom spawn is added to the casing soil. Using a transgenic mycelium running in the compost while the spawn added in the casing layer was not transgenic, it was observed that the produced sporophores did not bear the transgene but exhibited the transgenic phenotype (Romaine et al. 2011; Woolston et al. 2011). In contrast, to avoid viral transmission, anastomosis is not desirable, as in the hybrid strain J10165 which exhibits cultural incompatibility with the most frequently cultivated hybrid strains (Kerrigan and Wach 2010). However, until now, genetics of the vegetative incompatibility remains unknown in A. bisporus.
Anastomosis plays a major role in the life history: it can occur between sexually intercompatible homokaryons, and thus restores a heterokaryon (plasmogamy in the heterothallic life cycle, see Fig. 1.2), between a homokaryon and a heterokaryon, in which case the resulting novel heterokaryon bears the nucleus of the homokaryon and a sexually compatible nucleus coming from the heterokaryon, and also between two heterokaryons. In the latter case, formation of a novel genetically different heterokaryon is generally neither reported nor detected, but cannot be excluded since (Xu et al. 1996) detected somatic recombinants in subcultures from both heterokaryon x heterokaryon and heterokaryon x homokaryon pairings. In A. bisporus, some processes observed in certain other agaricales (see Kues 2000) do not occur or have not been detected: for example there is no clear evidence of nuclear migration through the homokaryotic mycelium, following the Buller phenomenon. On the other hand, diploid nuclei have never been detected, except in basidia, although they could be expected to be detected through a mechanism of somatic recombination.
The heterokaryotic mycelium is homologous to a diploid organism for its genetic expression, but because it possesses individual haploid nuclei, it can play a role of gamete and crosses with a homokaryon naturally via the Buller phenomenon, or after having artificially recovered haploid status (deheterokaryotisation) in vitro, either mechanically by fragmentation of the heterokaryon (Dickhardt 1985) or biochemically by using glucanases (protoplast method; Anderson et al. 1984; Kerrigan et al. 1994). These techniques are used by breeders for recovering homokaryons.
Whatever the origin of a mycelium, germination of spores, regeneration of protoplasts, or even tissue culture from a wild sporophore that can be haploid, multilocus genotype tests using codominant markers (Kerrigan et al. 1993) are needed to know its ploidy status (n vs. n + n), because cytology cannot easily help, and other tests such as mycelium growth rate test, mating test or fruiting tests are not reliable enough (Kerrigan et al. 1994).
The ability to stimulate spore germination via volatile agents (Lösel 1964) such as isovaleric acid which would remove the C02-self-inhibitor in the spores by participation of β-methylcrotonyl-CoA carboxylase (Rast and Stauble 1970) is another property of the mycelium that is crucial for reproduction. In vitro, but also in semi-controlled condition (in the culture compost tray), the rate of spore germination can increase greatly when a mycelium is present in the neighborhood.
In conclusion, the mycelium of A. bisporus is far from a simple vegetative organ. It is treated so as to maintain isolated strains, but in the wild numerous events can occur between strains and modify the genotypes without meiosis. In fact, we found poor evidence of clonality among the studied populations in Europe, even within each site (Xu et al. 2002), with the exception of a site in Portugal where several hundreds of sporophores had the same genotype (unpublished data). Mycelium in the wild generally does not extend in an area larger than 1 m diameter. But how long this particularly resistant mycelium can be maintained in place, or how far away it can be disseminated and form new colonies, remain open questions, which we are investigating. It was illustrated above how the progress in knowledge concerning mushroom reproduction is a source of innovation for developing new breeding strategies, in addition to the conventional ones that benefit also from molecular and genomic tools.
B. Molecular Breeding
Many economically important production traits, such as yield, quality, or resistance to diseases in edible mushroom cultures are under polygenic inheritance. Selecting for such complex traits with the classical breeding method appears quite challenging. The dissection of these quantitative traits in individualized loci through QTL mapping greatly facilitates their effective manipulation in a subsequent breeding program. Therefore, the development of molecular markers and linkage maps provides efficient tools to investigate genetics of desirable traits, and offers new opportunities for breeding. Although such approaches have been extensively proven to be successful in plant or animal, the use of molecular markers in mushroom breeding is a relatively new applied science that is developing mainly with A. bisporus as a model.
1. Quantitative Genetics
The construction of a comprehensive linkage map is the first step towards understanding the genetic basis of complex traits. The genetic linkage map developed for A. bisporus by Kerrigan et al. (1993) was the first molecular-marker-based map for an edible mushroom species. This map was based on the analysis of RAPD and RFLP segregating markers in an A. bisporus var. bisporus intravarietal offspring. It was unsaturated, with fewer linkage groups (11) than the number of chromosomes (n = 13) and several unlinked markers. Another A. bisporus linkage map, based on an intervarietal var. bisporus x var. burnettii offspring, was initiated (Callac et al. 1997; Moquet et al. 1999). With only 26 markers (RAPD, CAPS) or genes spread on five linkage groups, this latter map was also far from saturation. Afterward, advances in molecular marker techniques, such as AFLP or SSR genotyping, have made it possible to enhance the level of saturation of this map. Thus, based on the same mapping progeny, the addition of hundreds of new loci (AFLP, CAPS and SSR) permitted the construction of the first comprehensive linkage map for A. bisporus (Foulongne-Oriol et al. 2010). This map was built with 324 markers, evenly spread over 13 linkage groups, each one assigned to the corresponding chromosome of A. bisporus. The map covered 1,156 cM, with an average marker spacing of 3.9 cM, and encompassed nearly the whole genome (Fig. 1.4). This reference map is a useful and adequate tool for genetic studies in A. bisporus.
Comparative mapping of quantitative trait loci involved in resistance to the three major diseases affecting Agaricus bisporus. Only the linkage groups with QTLs are represented.  QTL resistance to the bacterial blotch, Pseudomonas tolaasii (Moquet et al. 1999).
QTL resistance to the bacterial blotch, Pseudomonas tolaasii (Moquet et al. 1999).  QTL resistance to Trichoderma metabolites (Foulongne-Oriol et al. 2011c).
QTL resistance to Trichoderma metabolites (Foulongne-Oriol et al. 2011c).  QTL resistance to Lecanicillium fungicola, bubble symptom (Foulongne-Oriol et al. 2012b).
QTL resistance to Lecanicillium fungicola, bubble symptom (Foulongne-Oriol et al. 2012b).  QTL resistance to L. fungicola, spotted cap symptom (Foulongne-Oriol et al. 2012b)
QTL resistance to L. fungicola, spotted cap symptom (Foulongne-Oriol et al. 2012b)
In parallel, a phenotypic database, comprising numerous traits of interest assessed on the intervarietal derived materials, has been established (Fig. 1.5). The combination of phenotypic data along with genotypic data has permitted the genomic location of either genes or QTL. Indeed, Mendelian traits related to the reproductive mode in A. bisporus have been mapped as phenotypic markers. BSN , the primary determinant of basidial spore number, and MAT which controls the mating ability both mapped to chromosome I, but were far apart. The MAT locus was assumed to be in a centromeric position, while BSN was located in the distal portion of the chromosome (Foulongne-Oriol et al. 2010).
Cap color was first investigated as a simply inherited trait, and the PPC1 locus has been mapped on chromosome VIII (Callac et al. 1998). QTL analysis has made it possible to refine the inheritance of this trait. In addition to the major determinant PPC1 which explained 86 % of the phenotypic variability, two minor loci were found on two other chromosomes, confirming the oligogenic control of this trait. These two additional loci contributed to the variability observed for the color gradation of the cap within the brown genotypes (Foulongne-Oriol et al. 2012a). This could be interesting for further breeding purposes, with the renewed attraction of brown mushrooms for the consumer (Robles and Lodder 2009). The genetics of other quality traits related to shelf life performance and susceptibility to discoloration after harvest, such as bruising, stipe brittleness, cap scaling, or long-term storage behavior, remains poorly documented. This could be explained by the difficulty in establishing a reliable and reproducible assessment for these traits at a large population scale. Recently, thanks to progress in bruising sensibility assessment, Gao et al. (2011) identified at least three inheritance patterns, and they are mapping QTLs for this trait. With QTL mapping, various quality traits are becoming available for marker-assisted selection and studies on their biological bases.
The development of resistant cultivars is the most effective, economical, and environmentally friendly approach to the management of disease control. Bacterial brown blotch (caused by Pseudomonas tolaasii), dry bubble (caused by Lecanicillium fungicola), and green mould (caused by the fungal competitor Trichoderma aggressivum) are the most detrimental disorders affecting yield and quality of the button mushroom throughout the world. Independent QTL mapping studies investigating the genetics of the resistance to these diseases have been performed.
In one of the first QTL mapping studies in fungi, Moquet et al. (1999) described a major QTL that underlay the resistance to P. tolaasii. This QTL explained about 30 % of the phenotypic variation, and was closely linked to cap color locus PPC1 (Moquet et al. 1999). An update analysis using the saturated linkage map data made it possible to find one additional minor locus on LG I involved in P. tolaasii resistance (LOD = 2.85). These two loci together explained 38 % of the phenotypic variation (Foulongne-Oriol, unpublished data). Contrary to the major QTL, the second locus was not confirmed when using bacterial toxin to mime the infection. However, this was not surprising since toxin-induced symptoms partially reproduce living bacteria-induced symptoms (Moquet et al. 1999). The different mechanisms and traits that combine for the resistance to a disease might be linked to different QTLs, and be identified thanks to QTL analysis.
In contrast, one can decompose different traits that might be responsible for the final decreases of symptoms in a resistant cultivar. A part of the resistance to T. aggressivum in A. bisporus was indirectly assessed by the ability to counteract the growth-limiting effect of lytic enzymes and metabolites produced by Trichoderma sp. (Foulongne-Oriol et al. 2011a). The level of tolerance and the capacity of adaptation to these compounds were quantitatively inherited and under oligogenic control, with two QTL detected per trait. The QTL on LGIV was involved in the control of the two traits, suggesting a key role. The colocation with QTL related to mycelium growth in control condition makes it possible to presume that the ability to resist or adapt to Trichoderma metabolites are tightly linked to the fitness of A. bisporus strains. Even if validation in natural conditions with living T. aggressivum is needed, we show here that colocation of QTLs help us in the understanding of the host–pathogen interaction.
Complex resistance to L. fungicola was also dissected through QTL mapping (Foulongne-Oriol et al. 2012b). Bubble and spotted mushroom, which are the two symptoms typifying dry-bubble disease during the successive steps of the infection, were analyzed separately. The QTLs involved in the expression of these two symptoms were detected as expected in distinct genomic regions, except on LGI. Colocations between QTLs governing L. fungicola resistance and production traits highlighted some unfavorable linkage drag, particularly on LGI, LGII, and LGX (Foulongne-Oriol et al. 2012b). The most resistant hybrids tended to produce numerous small mushrooms early. These results emphasized the difficulty of introgressing desirable traits from wild strains while maintaining an acceptable agronomic level.
A complex picture of the resistance mechanisms in A. bisporus is provided by the comparative mapping of the QTLs controlling the resistance to three major diseases (Fig. 1.4). A majority of the QTLs was found specific to one disease, suggesting that distinct mechanisms are involved. Nevertheless, some QTLs related to multiple resistances were highlighted on LGI, LGVIII, and LG XIII. Interestingly, on LGVIII the region in the vicinity of the PPC1 locus was found to be significantly involved in the resistance against L. fungicola (during secondary infection) and P. tolaasii, two pathogens that produce spotted cap symptoms. In each case, the resistance allele was associated with the brown allele at PPC1. A shared mechanism of resistance based on melanin biosynthesis could explain such reactions. The colocation of QTL observed on LGI and LGXIII may reflect linkage or pleiotropic effects. The QTL controlling the two dry-bubble symptoms on LGI mapped in the same genomic interval as earliness (see below), and thus may be related to fitness. On LGXIII, a common genomic region was involved both in dry-bubble spotted cap symptom and adaptation to Trichoderma metabolites, but the parental origin of the resistance allele was not the same. These results suggested that selecting for multi-resistance in A. bisporus could be quite arduous. A multi-way breeding scheme could be conceived to combine all the resistance in one genotype.
As for disease resistance, inheritance and QTL analyses of parameters composing the final yield of mushrooms could be informative both for the understanding of the mushroom biology and for modifying the characteristics of the harvest (i.e., earliness, distribution of the yield between the flushes …). The complex genetic architecture of yield-related traits has been disentangled through QTL mapping (Foulongne-Oriol et al. 2012a).
As an example, earliness was also analyzed, and was found to be linked to production traits. The earliest hybrids tended to produce the highest number of smaller mushrooms. The development of new strains that cover a large range of earliness could be interesting to diversify mushroom industry outlets (Foulongne-Oriol et al. 2012a).
Work is in progress also for the analysis of the inheritance of adaptive traits such as temperature tolerance (Fructification at High Temperature 25 °C in Fig. 1.5) or ability to growth and fruit on compost. New information is expected.
2. Marker-Assisted Selection
The basic concept of marker-assisted selection (MAS) is to select on the genotype of the marker(s) tightly linked to the trait rather than to select on the phenotype (Collard and Mackill 2008; Hospital 2009). Since most of the interesting traits in edible mushrooms are only displayed at fruiting stage, early selection with molecular markers allows an accurate screening of desirable offspring without the cultivation step. Molecular markers in A. bisporus have been already used for homokaryotic spore isolation (Kerrigan 1992), cap color selection (Loftus et al. 2000; Foulongne-Oriol et al. 2011b), or mating-type compatibility design (Sonnenberg et al. 2005). A first marker-assisted backcrossing program for a polygenic trait introgression was mentioned in Sonnenberg et al. (2005). The authors used genetic markers to select not only for the trait of interest but also for the genetic background, in order to limit unfavorable linkage drag. They highlighted the difficulty of having all favourable loci in a unique genotype in the advanced generation of the breeding scheme in the case of a polygenic trait (Sonnenberg et al. 2005). To counterbalance this drawback, a promising QTL pyramiding strategy was proposed. The a posteriori assessment of the reliability and the effectiveness of multitrait marker-assisted selection in an A. bisporus breeding scheme has also been undertaken in our laboratory.
It’s noteworthy that the successful use of molecular markers for selection is tightly linked to the recombination ability of the species (Collard and Mackill 2008). In this way, the availability of linkage maps offers also new insights into genome organization and recombination frequencies. It has been demonstrated that a higher rate of recombination occurred in the intervarietal linkage map (var. bisporus x var. burnettii) compared to the intravarietal one (Kerrigan et al. 1993; Foulongne-Oriol et al. 2010). As mentioned above, this phenomenon had been supposed to be adaptive and related to the different life cycles which typify the two A. bisporus varieties. Furthermore, the recombination ability in A. bisporus seems to be strongly impacted by the genetic background, as illustrated by a comparative linkage mapping study (Foulongne-Oriol et al. 2011b). Such variability in recombination behavior could be exploited judiciously in mushroom breeding programs. For example, a high recombination rate would greatly facilitate the introgression of a desirable trait, while limiting linkage drag. However, subsequent drawbacks would be the spreading of small unfavorable donor segments into the recipient genome, and the loss of linkage between markers used in selection and the selected trait. In this way, an acceptable compromise may be achieved.
C. Genetic Improvement of Mushrooms in the Era of Genomics
The recent sequencing of whole genomes of edible mushrooms is going to contribute hugely to our understanding of their biology, life cycles, and ecological behavior, and will contribute in the near future to crop improvements.
1. Genomics of A. bisporus
In 2007, the Joint Genome Institute, from the US Department of Energy (DOE), agreed to sequence the whole genome of A. bisporus due to the position of Agaricus spp. in forest ecosystems as a humicolous species, able to deploy a specific enzymatic pattern in comparison to other detritophilic fungi (Kerrigan 2011). Fungi which play key roles in the ecosystem process, particularly in the carbon cycle, are preferential targets supported by the Fungal Genomics Program (Grigoriev et al. 2011). Two homokaryons (haploid genome) of A. bisporus have been proposed. H97, obtained from the historically-cultivated stock Horst U1, was sequenced using the Sanger method with a depth of 8.29x. The H97 genome sequence consists of 30 Mb assembled in 29 scaffolds. A hundred of the sequenced markers located on the A. bisporus linkage maps (Foulongne-Oriol et al. 2010, 2011b) have made possible the assignment of the scaffolds to the 13 chromosomes. A few small gaps remain, but comprise less than 0.7 % of the nuclear genome. Ten thousand four hundred and thirty-eight gene models have been identified. Thanks to the genome coverage and the quality of the finished assembly, the H97 genome sequence constitutes the ‘reference’ for the species (Morin et al. 2012). The other homokaryon, JB137-s8, belonging to the tetrasporic var. burnettii, was sequenced with 454 pyrosequencing and Illumina HiSeq methods, and could be aligned on the H97 sequence. Both genomes are now in the public domain on the JGI’s portal web site (http://genome.jgi-psf.org).
Therefore, the whole-genome sequence of A. bisporus is the first to be available from the large family of Agaricaceae, and provides a solid foundation for understanding its particular nutrition mode and ecological adaptation (Morin et al. 2012). It will become a highly valuable resource for performing genomic and metabolic comparisons among fungi. Beyond its interest for fundamental knowledge, the release of the A. bisporus genome sequence opens a new era for breeding applications. First, the whole-genome sequence is a resource for new target DNA markers, especially SSRs (Labbe et al. 2011; Murat et al. 2011). The density of SSRs in the A. bisporus genome (approx. 60 SSRs per Mb, considering di, tri, and tetra motifs with at least five repeats; personal data) will provide a considerable molecular toolbox for map construction and further genetic applications. High-throughput sequencing also allows the development of single nucleotide polymorphism (SNP) markers. In A. bisporus, the sequencing of the two nuclei that constitute the Horst U1 strain has allowed the identification of more than 280,000 SNP, which corresponds to a frequency of about 0.93 %. However, less than 0.5 % of these SNP has been used for the development of 600 operational molecular markers (Sonnenberg et al. 2011). Their routine use implies high-throughput genotyping methods such as SNP microarrays. To date, these approaches have been rarely described in fungi, but are likely to increase. For example, in M. graminicola, the Diversity Arrays Technology (DArT) permitted the mapping of 1,793 markers (Wittenberg et al. 2009).
The tight relationship between linkage map and genome sequence will also make possible custom-made molecular markers tightly linked to target loci for further marker-assisted selection. It is premature to already expect concrete examples of applications in Agaricus breeding, but we can speculate that it will greatly facilitate cropping. This makes it possible to imagine that one would be able to perform some genomic selection in the next few years. It also offers a milestone towards the understanding of the molecular mechanisms that underlie traits of interest, through map-based cloning approach or candidate gene identification. Furthermore, other biotechnological tools such as transformation, reporter-gene expression, or gene-silencing are well established for A. bisporus (Burns et al. 2005, 2006; Eastwood et al. 2008), and will support subsequent functional validation of the gene(s) of interest. In combination with the genetic resources available, the whole genome sequence also offers the opportunity of performing genome-wide association studies to disentangle complex traits.
The challenge now for scientists and mushroom breeders will be to take advantage of the best of all the genome-based tools. It appears that the major limitation will be the way of implementing these new tools in research pipelines with, for example, the accessibility of a high-throughput genotyping platform, the ability to develop and use new statistical models and tools for bioinformatics, or the capacity of large-scale phenotypic screens.
2. Molecular Organization, Evolution, and Transmission of Mitochondrial Genes and Genomes in Mushrooms
Horgen and Castle (2002) have previously illustrated that the mitochondrion is a genetic component of mushrooms susceptible of being manipulated and resulting in unique and different genetic combinations involving nuclear genomes mixed with specifically chosen mitochondrion haplotypes and having potential consequences on strain performance. Improvements in the knowledge of the mitochondrial genes and genome organization, expression, and inheritance in Agaricomycetes will undoubtedly give a major contribution to the development of fungal biology and to the improvement of cultivated mushrooms.
a) Mitochondrial Genomics of Mushrooms
To date, only four complete mitochondrial genome (mtDNA) sequences have been reported and correctly annotated for the whole Agaricomycetes class: for Schizophyllum commune (49,704 bp) (GenBank Accession No: AF402141), Moniliophtora perniciosa (109,103 bp) (Formighieri et al. 2008), Pleurotus ostreatus (73,242 bp) (Wang et al. 2008b), and Trametes cingulate (91,500 bp) (Haridas and Gantt 2010). It is to be noted that the complete sequence of the mtDNA of A. bisporus (135,005 bp) is now available, in parallel with the achievement of its complete nuclear genome. The Agaricomycete mitochondrial genomes show a great variation in size from 36 kbp in Suillus cavipes (Bruns et al. 1988) to 176 kbp in Agaricus bitorquis (Hintz et al. 1985), and several Agaricomycete mtDNA possess larger size than those reported in the Ascomycota phylum (higher than 100,000 bp).
Plasmid-derived sequences are involved in the large size of the Agaricomycete mtDNA. In P. ostreatus mtDNA, Wang et al. (2008b) have described the presence of a DNA polymerase gene and a RNA polymerase gene, nearly identical to the genes harboured by a mitochondrial linear plasmid described in another P. ostreatus strain, confirming that this selfish genetic element is able to integrate and to be maintained in the mitochondrial genome. The Agrocybe aegerita mitochondrial genome has also been shown to possess two copies of a DNA polymerase gene (polB) with linear plasmid origin: one copy (Aa-polB) was putatively intact and consequently functional (Bois et al. 1999), and the other (Aa-polB P1) appeared eroded (Barroso et al. 2001). Interestingly, the regions flanking this eroded copy region (5834 nt) carried two large inverted repeats (higher than 2,421 nt), and contained identical copies of the nad4 gene (Ferandon et al. 2008). In the Agaricus genus, a linear mitochondrial plasmid, pEM, was described in isolates of A. bitorquis. The mitochondrial genome of A. bitorquis and A. bisporus were shown to possess a fragmented and potentially non-functional copy of the plasmid RNA polymerase (Robison et al. 1991), which had probably integrated the Agaricus mitochondrial genome before the divergence of A. bisporus and A. bitorquis (Robison and Horgen 1996). In addition to this remnant, RNA pol gene, the A. bisporus mitochondrial genome contains a second site of plasmid integration occupied by an intact DNA polB gene (G. Barroso, personal communication), and located near a copy of a previously reported (Jin and Horgen 1993) large inverted repeat (IR: >4,000 nt). The large (>38,000 nt) sequence separating both IR contains several typical mitochondrial genes (rps3, seven tRNAs, the LSU-rDNA, the nad3 and nad2 genes). This constitutes the second report, after A. aegerita (Ferandon et al. 2008), of the presence of a large inverted repeat located near a plasmid integration site.
Hence, mitochondrial linear plasmids appear as mobile genetic elements frequently found in mushroom mitochondria and able to integrate the mtDNA. The integrational event can be accompanied by molecular rearrangements such as large duplications of the flanking region, and is followed or not by an erosion of the integrated plasmid sequences. Finally, the Agaricomycete mitochondrial genomes seem frequently to integrate linear plasmid sequences able to promote large inverted repeats whose significance and consequences are still unknown. In this context, it will be noted that in A. bisporus as well as in A. aegerita, the duplication leads to a significant increase in the amount of mitochondrial genes such as several tRNAs in A. bisporus (G. Barroso, personal communication) or the nad4 gene in A. aegerita (Ferandon et al. 2008).
The size variations in mitochondrial genomes of agarics are mainly due to the presence of numerous large group I introns in most of the mitochondrial genes (Ferandon et al. 2010). The cox1 gene is the mitochondrial gene showing the highest number of introns. The complete sequence of the mitochondrial cox1 gene of A. bisporus has been recently achieved, and has shown that this longest cox1 gene (29,902 bp) is the largest group I intron reservoir (18 group I introns) reported to date in a eukaryote. It contains 18 out of 24 of the fungal introns described in all the fungal cox1 genes from Dikaria (Ascomycota and Basidiomycota). These long mitochondrial genes pose an appealing and still unresolved question about why and how some fungal species organize their mitochondrial genes and genomes in such an expensive manner, and what are the consequences on their biology.
mtDNA sequences are used to develop markers to follow mitochondrial genetic material during breeding experiments and strain improvement. Moreover, mitochondrial genomes, because of their rapid rate of sequence divergence, are also appealing molecules to use in the study of fungal wild populations or evolutionary biology. Two major types of mitochondrial sequences appear interesting for adding molecular markers suitable as taxonomic and/or phylogenetic tools at the species level and/or for strain fingerprinting, to the well-studied nuclear ribosomal unit. The first one is the compiled sequences of two variable domains (V6 and V9) of the SSU-rDNA, encoding the 16S RNA of the small-subunit of the mito-ribosome. Indeed, these domains mainly evolve by length mutations involving indel (insertion/deletion) sequences and, consequently, can easily lead to CAPS markers for species determination (Barroso et al. 2003). The variable domains V4, V6, and V9 of the mitochondrial SSU-rDNA have been successfully used to discriminate between closely related species in the Pleurotus and Agrocybe genera (Gonzalez and Labarère 1998, 2000). The second kind of sequence is constituted by the orthologs of the iAbi7 intron of the A. bisporus cox1 gene (Ferandon et al. 2010). Indeed, this group I intron appears widely distributed in the eukaryote kingdom, but also in the Agaricus genus (data not shown). Moreover, this mobile genetic element carries a structural gene, encoding a homing endonuclease (HE). It appears to be maintained in a functional state during the evolution course. Consequently, it can constitute a performing phylogenetic marker replacing the “barcoding region” of the cox1 gene, which is split by several large group I introns in the fungi, and this especially in the Agaricus genus.
b) Mitochondrial Effects on Mushroom Development
Influence of mitochondrial haplotype on mycelial growth of A. bisporus was studied by De La Bastide et al. (1997). Pairs of heterokaryon strains, each pair having the same nuclear genomes but a different mitochondrial genome, were produced by controlled crosses of homokaryons with wild or commercial origins. Seven genetically distinct mitochondrial DNA (mtDNA) haplotypes were evaluated in different nuclear backgrounds. The growth of heterokaryon pairs differing only in their mtDNA haplotype was compared at three temperatures similar to those utilized in commercial production facilities (18 °C, 22 °C, and 26 °C). Statistically significant differences were detected in most of the heterokaryon pairs evaluated. Some heterokaryon pairs showed differences of growth at all three temperatures of incubation, suggesting a temperature-independent difference. Others showed differences at only a single temperature, suggesting a temperature-dependent difference. The influence of some mtDNA haplotypes on growth was dependent on the nuclear genetic background, showing that mtDNA haplotype can influence growth of A. bisporus heterokaryons in some nuclear backgrounds. Combinations of mitochondrial and nuclear genomes may also affect other traits involved in agronomical performances and ageing during storage.
Ageing of biological systems is a fundamental process controlled by a complex network of molecular pathways, and mitochondria have been shown to play a crucial role in programmed cell death and ageing.
Hence, in the yeast S. Cerevisiae (for a review see Braun and Westermann 2011), different stimuli have been shown to activate distinct mitochondrion-dependent cell death pathways, and it has been demonstrated that ageing is associated with a progressive increase in mitochondrial damage, culminating in oxidative stress and cellular dysfunction. In the same way, in the filamentous fungus Podospora anserine (for review see Osiewacz 2011), a wide range of pathways have been identified that contribute to the maintenance of a population of functional mitochondria. These pathways act in a hierarchical manner, but all are limited in capacity. At the end of the life cycle, when the pathways are overwhelmed and damage has reached certain thresholds, programmed cell death brings the life of individual P. anserina to an end.
In A. bisporus sporophores, the gene of a superoxide dismutase (SOD) of the iron/manganese family usually located in mitochondria is up-regulated following harvest (Henderson et al. 2005). This renders them capable of responding rapidly to a potentially lethal level of superoxide radicals. These ageing processes linked to mitochondria might probably affect the shelf life of mushrooms, and are worth studying to produce improved strains.
Some cultivars of A. bisporus subcultured for a long time have shown phenotypic variability or vegetative decline with variation in growth which might be related to changes in mitochondrial genome organization (Jin et al. 1992). Such reports suggest that mitochondrial genetic material could be correlated with still unknown physiologically important functions of fungi and, consequently, give an important interest to the investigations on mitochondrial genomics and heredity. Identification of mitochondrial genotypes and evaluation of different nuclear–mitochondrial combinations in strain improvement programs are promising directions for the button mushroom industry.
c) Mitochondrial Inheritance in Agaricomycetes
In fungi of the Agaricomycete class, sexual mating does not rely on organs or cells specialized for sexual reproduction, but relies on a fusion between two vegetative homokaryotic hyphae (somatogamy). After cell fusion, both hyphae exchange nuclei, leading to a dikaryon with both parental nuclei formed in the existing thallus of the homokaryon. In most of the studied species, reciprocal (bidirectional) nuclear migration occurs from the junction line of the two mating colonies, since donor nuclei extensively migrate through the resident cells of each recipient homokaryon, resulting in two discrete dikaryons with the same nuclei but different cytoplasms. Agrocybe aegerita (Barroso and Labarère 1995) and L. edodes are examples (Fukuda et al. 1995). In Coprinopsis cinerea (May and Taylor 1988) and A. bitorquis (Hintz et al. 1988), mating asymmetry caused by nonreciprocal nuclear migration was described, and in A. bisporus, no nuclear migration was observed (Jin et al. 1992; Jin and Horgen 1994). In any case, it is obvious that one of the most important consequences of plasmogamy is the mixing of cytoplasmic genetic elements from different individuals. It is generally accepted that a competition between different cytoplasmic elements can lead to conflict that impairs the fitness of the cell (Billiard et al. 2010). For instance, mitochondrial mutants with an impaired contribution to the cell’s performance but possessing a more rapid replication will be able to invade. The idea that zygotes formed by the fusion of gametes with uniparental inheritance of organelles are the fittest leads to the hypothesis that processes involved in the uniparental inheritance of the cytoplasm should have been selected during the course of evolution. In accordance with this, uniparental mitochondrial inheritance, occurring at plasmogamy, has been reported in most of the studied mushrooms. However, biparental mitochondrial inheritance at plasmogamy has been observed in C. cinerea (May and Taylor 1988) or L. edodes (Fukuda et al. 1995).
Mitochondrial inheritance in A. bisporus analyzed with RFLP markers on 16 crosses obtained by combinations of 13 homokaryotic strains supported the occurrence of uniparental mitochondrial inheritance in A. bisporus , with one mtDNA haplotype usually favored in the new heterokaryon (De la Bastide and Horgen 2003). Non-parental mtDNA haplotypes were seen in heterokaryons produced from seven to 16 crosses. Evidence for the occurrence of two mtDNA haplotypes in one heterokaryotic mycelium was observed in eight out of 16 crosses, suggesting the maintenance of true heteroplasmons after three successive subculturing steps. The mating protocol described by the authors could be utilized to generate novel mtDNA haplotypes for strain improvement as an alternative to the use of the Buller phenomenon proposed above.
Mitochondrial transmission in A. aegerita was studied not only at plasmogamy, but also during vegetative growth of the resulting dikaryons, and also during sporophore differentiation (Barroso and Labarère 1995). It was demonstrated that plasmogamy between homokaryons from progeny of three wild-type strains resulted in bidirectional nuclear migration, whereas little mitochondrial migration accompanied the nuclear migration. A total of 75 % of the dikaryons from the fusion lines had both parental mitochondrial haplotypes (mixed dikaryons), and 25 % had only a single haplotype (homoplasmic dikaryons). Moreover, in some matings, there was a strong bias in favour of one parental mitochondrial haplotype.
The heteroplasmic nature of mixed dikaryons was demonstrated by isolating and subculturing apical cells in micromanipulation experiments, and also by identifying recombinant mitochondrial genomes. Conversion of heteroplasmons into homoplasmons was shown to occur (i) during long-term storage, (ii) in mycelia regenerated from isolated apical cells, and (iii) during sporophore differentiation. Homokaryons that readily accepted foreign nuclei were the most efficient homokaryons in maintaining their mitochondrial haplotype during plasmogamy, long-term storage, and sporophore differentiation.
This strongly argues for the presence of still-unknown mechanisms responsible for a non-random sorting out of mitochondrial genomes, acting at different times in the life cycle of the heteroplasmic dikaryon.
Both models, A. bisporus and A. aegerita, suggest that the mechanism responsible for the non-random retention or elimination of a given haplotype may be related to the nuclear genotype or the mitochondrial haplotype, or both in interaction. This has consequences with regard to the diffusion of mitochondrial genotypes in natural populations, and to the possibilities of strain improvements using nuclear–mitochondrial combinations.
IV. Conclusion
In the previous edition of this book, Horgen and Castle (2002) concluded their chapter with a sentence on the necessity for the mushroom industry to invest in genetic manipulation through intensive breeding programs, utilizing and adapting new technologies as they evolve to mushroom research. It “will position the industry for success in the 21st century”, they wrote. Ten years later, several results of mushroom research have been confirmed by the industry, with the development of new A. bisporus hybrids based on the utilization of both genetic diversity in germplasm and molecular approaches. The problem of monocultural practice is going to be solved with a new interest from growers in the production of more diversified strains of the button mushroom adapted to different local conditions and to the consumer demand for safe and healthy food. The recent developments in genetics and the whole genome sequence open new ways to understand fundamental biological mechanisms in the button mushroom, and also to foresee new breeding strategies. To our knowledge, the sum of the studies performed on the A. bisporus species makes up the most accomplished and integrative analysis of trait genetics in an edible mushroom. Diversification of the species is a second way for a new development of the industry. It will be reached thanks to progress in knowledge concerning the biodiversity of Agaricus species and the possibility of optimizing the development of efficient strains and culture conditions by using the findings acquired on A. bisporus.
References
Amazonas MAL de A (2005) Champignon do Brasil (Agaricus brasiliensis): nutrition, health, market demands and regulatory concerns. In: Tan Q, Chen ZM, Cao H, Buswell JA (eds) Proceedings of the 5th international conference on mushroom biology and mushroom products. Acta Edulis 12 sup, pp 111–119
Anderson JB, Petsche DM, Herr FB, Horgen PA (1984) Breeding relationships among several species of Agaricus. Can J Bot 62:1884–1889
Arrillaga P, Parra LA (2006) El género Agaricus L. en España. XI Agaricus subrufescens, primera cita para España. Bol Soc Micol Madrid 30:201–207
Barroso G, Labarère J (1995) Genetic evidence for nonrandom sorting of mitochondria in the basidiomycete Agrocybe aegerita. Appl Environ Microbiol 63:4686–4691
Barroso G, Bois F, Labarère J (2001) Duplication of a truncated paralog of the family B DNA polymerase gene Aa-polB in the Agrocybe aegerita mitochondrial genome. Appl Environ Microbiol 67:1739–1743
Barroso G, Sirand-Pugnet P, Mouhamadou B, Labarère J (2003) Secondary structure and molecular evolution of the mitochondrial small subunit ribosomal RNA in Agaricales (Euagarics clade, Homobasidiomycota). J Mol Evol 57:383–396
Berendsen RL, Baars JJP, Kalkhove SIC, Lugones LG, Wosten HAB, Bakker PAHM (2010) Lecanicillium fungicola: causal agent of dry bubble disease in white-button mushroom. Mol Plant Pathol 11:585–595
Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, Giraud T (2010) Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev Camb Philos Soc 86:421–442
Boa E (2004) Non-wood forest products 17. Wild edible fungi a global overview of their use and importance to people. FAO, Rome, ISBN 92-5-105157-7 http://www.fao.org/docrep/007/y5489e/y5489e00.htm#TopOfPage
Bois F, Barroso G, Gonzalez P, Labarère J (1999) Molecular cloning, sequence and expression of Aa-polB, a mitochondrial gene encoding a family B DNA polymerase from the edible basidiomycete Agrocybe aegerita. Mol Gen Genet 261:508–513
Braun RJ, Westermann B (2011) Mitochondrial dynamics in yeast cell death and aging. Biochem Soc Trans 39:1520–1526
Bruggeman J, Debets AJM, Wijngaarden PJ, deVisser JAGM, Hoekstra RF (2003) Sex slows down the accumulation of deleterious mutations in the homothallic fungus Aspergillus nidulans. Genetics 164:479–485
Bruns TD, Palmer JD, Shumard DS, Grossman LI, Hudspeth ME (1988) Mitochondrial DNAs of Suillus: three fold size change in molecules that share a common gene order. Curr Genet 13:49–56
Buller AHR (1931) Researches on fungi, vol IV. Longmans/Green and Co, London
Burns C, Gregory KE, Kirby M, Cheung MK, Riquelme M, Elliott TJ, Challen MP, Bailey A, Foster GD (2005) Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol 42:191–199
Burns C, Leach KM, Elliott TJ, Challen MP, Foster GD, Bailey A (2006) Evaluation of agrobacterium-mediated transformation of Agaricus bisporus using a range of promoters linked to hygromycin resistance. Mol Biotechnol 32:129–138
Burton KS (2004) Cultural factors affecting mushroom quality- cause and control of bruising. In: Romaine CP, Keil CB, Rinker DL, Royse DJ (eds) Mushroom science XVI, science and cultivation of edible and medicinal mushrooms. The Pennsylvania State University, Penn State, ISBN-1-883956-01-13, pp 397–404
Burton EG, Metzenberg RL (1972) Novel mutation causing derepression of several enzymes of sulfur metabolism in Neurospora crassa. J Bacteriol 109:140–151
Callac P, Billette C, Imbernon M, Kerrigan RW (1993) Morphological, genetic, and interfertility analyses reveal a novel, tetrasporic variety of Agaricus bisporus from the Sonoran Desert of California. Mycologia 85:335–351
Callac P, Imbernon M, Kerrigan RW, Olivier JM (1996) The two life cycles of Agaricus bisporus. In: Royse J (ed) Mushroom biology and mushroom product, proceeding of the second international conference. University Park, Pennsylvania, pp 57–66
Callac P, Desmerger C, Kerrigan RW, Imbernon M (1997) Conservation of genetic linkage with map expansion in distantly related crosses of Agaricus bisporus. FEMS Microbiol Lett 146:235–240
Callac P, Moquet F, Imbernon M, Ramos Guedes-Lafargue M, Mamoun M, Olivier JM (1998) Evidence for PPC1 a determinant of the pilei-pellis color of Agaricus bisporus fruiting bodies. Fungal Genet Biol 23:181–188
Callac P, Theochari I, Kerrigan RW (2002) The germplasm of Agaricus bisporus: main results after ten years of collection in France, in Greece, and in North America. Acta Hortic 579:49–55
Callac P, Gaubert J, Imbernon M, Guinberteau J, Desmerger C, Olivier JM (2003) Ressources génétiques chez les agarics: résultats récents et premiers essais expérimentaux d’interfécondation libre chez le champignon de Paris. Les Actes du BRG 4:331–346
Callac P, Guinberteau J, Rapior S (2005) New hypotheses from integration of morphological traits, biochemical data and molecular phylogeny in Agaricus spp. In: Tan Q, Chen ZM, Cao H, Buswell JA (eds) Proceedings of the 5th international conference on mushroom biology and mushroom products, Shanghai. Acta Edulis Fungi 12 sup, pp 37–44
Callac P, Spataro C, Caillé A, Imbernon M (2006) Evidence for outcrossing via Buller phenomenon in substrate simultaneously inoculated with spores and mycelium of Agaricus bisporus. Appl Environ Microbiol 72:2366–2372
Callac P, Imbernon, Savoie JM (2008) Outcrossing via the Buller phenomenon in a substrate simultaneously inoculated with spores and mycelium of Agaricus bisporus creates variability for agronomic traits. In: Lelley JI, Buswell JA (eds) Proceedings of the 6th international conference on mushroom biology and mushroom products, GAMU, Krefeld, pp 113–119
Cappelli A (2011) Approccio al Genere Agaricus-IV. Rivista di Micologia 54:3–27
Castle AJ, Horgen PA, Anderson JB (1987) Restriction fragment length polymorphisms in the mushrooms Agaricus brunnescens and Agaricus bitorquis. Appl Environ Microbiol 53:816–822
Challen MP, Kerrigan RW, Callac P (2003) A phylogenetic reconstruction and emendation of Agaricus section Duploannulatae. Mycologia 95:61–73
Chang ST, Miles PG (1992) Mushroom biology, a new discipline. Mycologist 6:64–65
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363:557–572
Couture C, Michel A, Imbernon M, Callac P (2004) Inheritance of the haploid fruiting ability in Agaricus bisporus. In: Romaine CP, Keil CB, Rinker DL, Royse DJ (eds) Mushroom science XVI, science and cultivation of edible and medicinal mushrooms. The Pennsylvania State University, Penn State, pp 45–52
De la Bastide PY, Horgen PA (2003) Mitochondrial inheritance and the detection of non-parental mitochondrial DNA haplotypes in crosses of Agaricus bisporus homokaryons. Fungal Genet Biol 38:333–342
De La Bastide PY, Sonnenberg A, Van Griensven L, Anderson JB, Horgen PA (1997) Mitochondrial haplotype influences mycelial growth of Agaricus bisporus heterokaryons. Appl Environ Microbiol 63:3426–3431
Dickhardt R (1985) Homokaryotisation of Agaricus bitorquis (Quel.) Sacc. and Agaricus bisporus (Lange) Imb. Theor Appl Genet 70:52–56
Eastwood DC, Kingsnorth CS, Jones HJ, Burton KS (2001) Genes with increased transcript levels following harvest of the sporophore of Agaricus bisporus have multiple physiologic roles. Mycol Res 105:1223–1230
Eastwood DC, Challen MP, Zhang C, Jenkins H, Henderson J, Burton KS (2008) Hairpin-mediated down-regulation of the urea cycle enzyme argininosuccinate lyase in Agaricus bisporus. Mycol Res 112:708–716
Evans HJ (1959) Nuclear behaviour in the cultivated mushroom. Chromosoma 10:115–135
FAOSTAT (2011) Food and Agriculture Organization of the United Nations. https://www.Faostat.fao.org. Cited 10 Jan 2012
Ferandon C, ChatelSel K, Castandet B, Castroviejo M, Barroso G (2008) The Agrocybe aegerita mitochondrial genome contains two inverted repeats of the nad4 gene arisen by duplication on both sides of a linear plasmid integration site. Fungal Genet Biol 45:292–301
Ferandon C, Moukha S, Callac P, Benedetto JP, Castroviejo M, Barroso G (2010) The Agaricus bisporus cox1 gene: the longest mitochondrial gene and the largest reservoir of mitochondrial group i introns. PLoS One 5:e14048
Formighieri EF, Tiburcio RA, Armas ED, Medrano FJ, Shimo H, Carels N, Góes-Neto A, Cotomacci C, Carazzolle MF, Sardinha-Pinto N, Thomazella DP, Rincones J, Digiampietri L, Carraro DM, Azeredo-Espin AM, Reis SF, Deckmann AC, Gramacho K, Gonçalves MS, MouraNeto JP, Barbosa LV, Meinhardt LW, Cascardo JC, Pereira GA (2008) The mitochondrial genome of the phytopathogenicbasidiomyceteMoniliophthora perniciosa is 109 kb in size and contains a stable integrated plasmid. Mycol Res 112:1136–1152
Foulongne-Oriol M, Spataro C, Savoie JM (2009) Novel microsatellite markers suitable for genetic studies in the white button mushroom Agaricus bisporus. Appl Microbiol Biot 1184:1125–1135
Foulongne-Oriol M, Spataro C, Cathalot V, Monllor S, Savoie JM (2010) An expanded genetic linkage map of an intervarietal Agaricus bisporus var. bisporus x A. bisporus var. burnettii hybrid based on AFLP, SSR and CAPS markers sheds light on the recombination behaviour of the species. Fungal Genet Biol 47:226–236
Foulongne-Oriol M, Minvielle N, Savoie JM (2011a) QTL for resistance to Trichoderma lytic enzymes and metabolites in Agaricus bisporus. In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 2. pp 17–25. http://wsmbmp.org/Previous_Conference_7.html. Cited 27 Feb 2012
Foulongne-Oriol M, Dufourcq R, Spataro C, Devesse C, Rodier A, Savoie JM (2011b) Comparative linkage mapping in the white button mushroom Agaricus bisporus provides foundation for breeding management. Curr Genet 57:39–50
Foulongne-Oriol M, Rodier A, Caumont P, Spataro C, Savoie JM (2011c) Agaricus bisporus cultivars: hidden diversity beyond apparent uniformity? In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 2. pp 9–16. http://wsmbmp.org/Previous_Conference_7.html. Cited 10 Feb 2012
Foulongne-Oriol M, Rodier A, Rousseau T, Largeteau M, Savoie JM (2011d) Quantitative genetics to dissect the fungal-fungal interaction between Lecanicillium verticillium and the white button mushroom Agaricus bisporus. Fungal Biol 115:421–431
Foulongne-Oriol M, Rodier A, Rousseau T, Savoie JM (2012a) QTL mapping of yield-related components and oligogenic control of the cap color in the button mushroom Agaricus bisporus. Appl Environ Microbiol 78:2422–2434
Foulongne-Oriol M, Rodier A, Savoie JM (2012b) Relationship between yield components and partial resistance to Lecanicillium fungicola in Agaricus bisporus assessed by QTL mapping. Appl Environ Microbiol 78:2435–2442
Fukuda M, Harada Y, Imahori S, Fukumasa-Nakai Y, Hayashi Y (1995) Inheritance of mitochondrial DNA in sexual crosses and protoplast cell fusions in Lentinula edodes. Curr Genet 27:550–554
Gao W, Baars JJP, Sonnenberg ASM, Richard Visser R (2011) Inheritance pattern of bruising sensitivity trait in Agaricus bisporus. In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 1. pp 43–51. http://www.wsmbmp.org/Previous_Conference_7.html. Cited 10 Feb 2012
Gaze R, Fletcher J (2007) Mushroom pest and disease control. Academic/Elsevier, Waltham
Geml J, Laursen GA, Taylor DL (2008) Molecular diversity assessment of artic and boreal Agaricus taxa. Mycologia 100:577–589
Gonzalez P, Labarère J (1998) Sequence and secondary structure of the mitochondrial small-subunit rRNA V4, V6, and V9 domains reveal highly species-specific variations within the genus Agrocybe. Appl Environ Microbiol 64:4149–4160
Gonzalez P, Labarère J (2000) Phylogenetic relationships of Pleurotus species according to the sequence and secondary structure of the mitochondrial small-subunit rRNA V4, V6 and V9 domains. Microbiology 146:209–221
Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP, Kuske C, Magnuson JK, Martin F, Spatafora JW, Tsang A, Baker SE (2011) Fueling the future with fungal genomics. Mycology 2:192–209
Grogan H, Gaze R, Holcroft S (2005) Viral dsRNAs in Agaricus bisporus: transmisson, symptom expression and control. In: Tan Q, Chen ZM, Cao H, Buswell JA (eds) Proceedings of the 5th international conference on mushroom biology and mushroom products. Acta Edulis Fungi 12 sup, pp 363–367
Guan XJ, Xu L, Shao YC, Wang ZR, Chen FS (2008) Differentiation of commercial strains of Agaricus species in China with inter-simple sequence repeat marker. World J Microbiol Biot 24:1617–1622
Haridas S, Gantt JS (2010) The mitochondrial genome of the wood-degrading basidiomyceteTrametes cingulata. FEMS Microbiol Lett 308:29–34
Heinemann P (1956) Champignons récoltés au Congo Belge par Madame M. Goosens-Fontana. II Agaricus Fries s.s. Bulletin du Jardin Botanique de l’Etat à Bruxelles 26:1–127
Henderson J, Eastwood D, Bains N, Burton K (2005) Superoxide dismutase – mushroom under stress. In: Tan Q, Chen ZM, Cao H, Buswell JA (eds) Proceedings of the 5th international conference on mushroom biology and mushroom products. Acta Edulis Fungi 12sup, pp 61–65
Hintz W, Mohan M, Anderson JB, Horgen PA (1985) The mitochondrial DNAs of Agaricus: heterogeneity in A. bitorquis and homogeneity in A. brunnescens. Curr Genet 9:127–132
Hintz W, Anderson JB, Horgen PA (1988) Nuclear migration and mitochondrial inheritance in the mushroom Agaricus bitorquis. Genetics 119:35–41
Horgen PA, Castle AJ (2002) The application and potential of molecular approaches to mushrooms. In: Kempken F (ed) The Mycota XI agricultural applications. Springer, Berlin/Heidelberg, pp 3–17
Hospital F (2009) Challenges for effective marker-assisted selection in plants. Genetica 136:303–310
Hou HH, Elliott TJ (1978) Comparative cytology in the genus Agaricus. Mushroom Sci 10(1):51–62
Imbernon M, Callac P, Granit S, Pirobe L (1995) Allelic polymorphism at the mating type locus in Agaricus bisporus var. burnettii, and confirmation of the dominance of its tetrasporic trait. Mushroom Sci 14(1):11–19
Imbernon M, Callac P, Gasqui P, Kerrigan RW, Velcko AJ Jr (1996) BSN, the primary determinant of basidial spore number and reproductive mode in Agaricus bisporus, maps to chromosome I. Mycologia 88:749–761
Jin T, Horgen PA (1993) Further characterization of a large inverted repeat in the mitochondrial genomes of Agaricus bisporus (= A. brunnescens) and related species. Curr Genet 23:228–233
Jin T, Horgen PA (1994) Uniparental mitochondrial transmission in the cultivated button mushroom, Agaricusbisporus. Appl Environ Microbiol 60:4456–4460
Jin T, Sonnenberg AS, Van Griensven LJ, Horgen PA (1992) Investigation of mitochondrial transmission in selected matings between homokaryons from commercial and wild-collected isolates of Agaricus bisporus (= Agaricus brunnescens). Appl Environ Microbiol 58:3553–3560
Juarez del Carmen S, Largeteau-Mamoun ML, Rousseau T, Regnault-Roger C, Savoie JM (2002) Genetic and physiological variation in isolates of Verticillium fungicola causing dry bubble disease of the cultivated button mushroom, Agaricus bisporus. Mycol Res 106:1163–1170
Kamzolkina O, Volkova V, Kozlova M, Pancheva E, Dyakov Y, Callac P (2006) Karyological evidence for meiosis in the three different types of life cycles existing in Agaricus bisporus. Mycologia 98:763–770
Kerrigan RW (1990) Evidence of genetic divergence in two populations of Agaricus bisporus. Mycol Res 94:721–733
Kerrigan RW (1992) Strategies for the efficient recovery of Agaricus bisporus homokaryons. Mycologia 84:575–579
Kerrigan RW (1996) Characteristics of a large collection of wild edible mushroom germ plasm: the Agaricus resource program. Centraalbureau voor Schimmelcultures and the World Federation for Culture Collections, Veldhoven, pp 302–308
Kerrigan RW (2004) Trait diversity in wild Agaricus bisporus. In: Romaine CP, Keil CB, Rinker DL, Royse DJ (eds) Mushroom science XVI, science and cultivation of edible and medicinal mushrooms, The Pennsylvania State University, Penn State, ISBN-1-883956-01-13, pp 31–38
Kerrigan RW (2005) Agaricus subrufescens, a cultivated edible and medicinal mushroom, and its synonyms. Mycologia 97:12–24
Kerrigan RWK (2007) Lectotypification of Agaricus brunnescens. Mycologia 99:906–915
Kerrigan RW (2011) Whole-genome sequencing of the cultivated button mushroom Agaricus bisporus: history, status and applications. In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 1. pp 1–6. http://www.wsmbmp.org/Previous_Conference_7.html. Cited 01 Feb 2012
Kerrigan RWK, Ross K (1987) Basidiospore number variation in Agaricus. In: Wuest PJ, Royse DJ, Beelman RB (eds) Cultivating edible fungi. Elsevier Science Publishers, Amsterdam, pp 155–162
Kerrigan RW, Wach MP (2008) Agaricus subsrufescens mushroom plant named ‘H1X1’. US Patent USPP19, 313P3
Kerrigan RWK, Wach MP (2010) Agaricus bisporus mushroom plant named J10165 US Patent Application: 20100218294
Kerrigan RW, Baller LM, Horgen PA, Anderson JB (1992) Strategies for the efficient recovery of Agaricus bisporus homokaryons. Mycologia 84:575–579
Kerrigan RW, Royer JC, Baller LM, Kohli Y, Horgen PA, Anderson JB (1993) Meiotic behavior and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics 133:225–236
Kerrigan RW, Imbernon M, Callac P, Billette C, Olivier JM (1994) The heterothallic life cycle of Agaricus bisporus var. burnettii, and the inheritance of its tetrasporic trait. Exp Mycol 18:193–210
Kerrigan RW, Carvalho DB, Horgen PA, Anderson JB (1998) The indigenous coastal Californian population of the mushroom Agaricus bisporus, a cultivated species, may be at risk of extinction. Mol Ecol 7:35–45
Kerrigan RW, Callac P, Xu J, Noble R (1999) Population and phylogenetic structure within the Agaricus subfloccosus complex. Mycol Res 103:1515–1523
Kerrigan RW, Callac P, Challen M, Guinberteau J, Parra LA (2006 [2005]) Agaricus section Xanthodermatei: a phylogenetic reconstruction with commentary on taxa. Mycologia 97:1292–1315
Kerrigan RW, Callac P, Para LA (2008) New and rare taxa in Agaricus section Bivelares (Duploannulati). Mycologia 100:876–892
Khush R, Becker E, Wach M (1992) DNA amplification polymorphisms of the cultivated mushroom Agaricus bisporus. Appl Environ Microbiol 58:2971–2977
Kue U (2000) Life history and developmental processes in the Basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev 64:316–353
Kuhner R (1977) Variation of nuclear behaviour in the Homobasidiomycetes. Trans Brit Mycol Soc 68(Kuhner R):1–16
Labbe J, Murat C, Morin E, Le Tacon F, Martin F (2011) Survey and analysis of simple sequence repeats in the Laccaria bicolor genome, with development of microsatellite markers. Curr Genet 57:75–88
Lange M (1952) Species concepts in the genus Coprinus. Dansk Bot Ark 14:1–140
Largeteau ML, Savoie JM (2010) Microbially-induced diseases of Agaricus bisporus: biochemical mechanisms and impact on commercial mushroom production. MiniReview. Appl Microbiol Biot 86:63–73
Largeteau ML, Rodier A, Rousseau T, Juarez del Carmen S, Védie R, Savoie JM (2004) Agaricus susceptibility to Verticillium fungicola. In: Romaine CP, Keil CB, Rinker DL, Royse DJ (eds) Mushroom science XVI, science and cultivation of edible and medicinal mushrooms. The Pennsylvania State University, Penn State, ISBN-1-883956-01-13, pp 515–522
Largeteau ML, Baars JPP, Juarez del Carmen S, Regnault-Roger C, Savoie JM (2005) Wild strains of Agaricus bisporus: a source of tolerance to dry bubble disease. In: Pisabarro AG, Ramirez L (eds) IV congress genetics and cellular biology of basidiomycetes. Universidad Pública de Navarra, Pamplona, pp 77–87
Largeteau ML, Callac P, Navarro-Rodriguez AM, Savoie JM (2011a) Diversity in the ability of Agaricus bisporus wild isolates to fruit at high temperature (25 °C). Fungal Biol 115:1186–1195
Largeteau ML, Llarena-Hernandez RC, Regnault-Roger C, Savoie JM (2011b) The medicinal Agaricus mushroom cultivated in Brazil: biology, cultivation and non-medicinal valorization. Appl Microbiol Biot 92:897–907
Llarena Hernández RC, Largeteau M, Farnet AM, Minvielle N, Regnault-Roger C, Savoie JM (2011) Phenotypic variability in cultivars and wild strains of Agaricus brasiliensis and Agaricus subrufescens. In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 2. pp 38–49. http://www.wsmbmp.org/Previous_Conference_7.html. Cited 25 Jan 2012
Loftus MG, Moore D, Elliot TJ (1988) DNA polymorphisms in commercial and wild strains of the cultivated mushroom, Agaricus bisporus. Theor Appl Genet 76:712–718
Loftus M, Bouchti KL, Robles C, Van Griensven LJLD (2000) Use of SCAR marker for cap color in Agaricus bisporus breeding programs. Mushroom Sci 15:201–205
Lösel DM (1964) The stimulation of spore germination in Agaricus bisporus by living mycelium. Ann Bot 28:54I–554I
Ludwig E (2007) Pilzkompendium 2: 61. Fungicon-Verlag, Berlin
Marshall E, (Tan) Nair NG (2009) FAO Diversification booklet number 7: make money by growing mushrooms, 53 p. Food and Agriculture Organization of the United Nations. ftp://ftp.fao.org/docrep/fao/011/i0522e/i0522e00.pdf. Cited 25 Jan 2012
May G, Taylor JW (1988) Patterns of mating and mitochondrial DNA inheritance in the agaric Basidiomycete Coprinus cinereus. Genetics 118:213–220
Miller RE, Kananen DL (1972) Bipolar sexuality in the mushroom. Mushroom Sci 13:713–718
Moore AJ, Challen MP, Warner PJ, Elliott TJ (2001) RAPD discrimination of Agaricus bisporus mushroom cultivars. Appl Microbiol Biot 55:742–749
Moquet F, Guedes-Lafargue MR, Mamoun M, Olivier JM (1998) Selfreproduction induced variability in agronomic traits for a wild Agaricus bisporus. Mycologia 90(1998):806–812
Moquet F, Desmerger C, Mamoun M, Ramos-Guedes-Lafargue M, Olivier JM (1999) A quantitative trait locus of Agaricus bisporus resistance to Pseudomonas tolaasii is closely linked to natural cap color. Fungal Genet Biol 28:34–42
Morin E, Kohler A, Baker A, Foulongne-Oriol M, Lombard V, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Billette C, Coutinho PM, Doddapaneni H, Grimwood J, Hildén K, Kües U, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Lundell T, Murat C, Riley RW, Salamov AA, Schmutz J, Subramanian V, Wösten HAB, Xu JP, Eastwood DC, Foster GD, Sonnenberg ASM, Cullen D, de Vries RP, Henrissat B, Burton KS, Kerrigan RW, Challen MP, Grigoriev IV, Martin F (2012) The genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. PNAS 109(43):17501–17506
Murat C, Riccioni C, Belfiori B, Cichocki N, Labbé J, Morin E, Tisserant E, Paolocci F, Rubini A, Martin F (2011) Distribution and localization of microsatellites in the Perigord black truffle genome and identification of new molecular markers. Fungal Genet Biol 48:592–601
Osiewacz HD (2011) Mitochondrial quality control in aging and lifespan control of the fungal aging model Podospora anserina. Biochem Soc Trans 39:1488–1492
Parra LA (2008) Agaricus L. Allopsalliota Nauta & Bas. Pars 1Edizioni Candusso Alassio, Italy
Raper CA, Raper JR, Miller RE (1972) Genetic analysis of the life cycle of Agaricus bisporus. Mycologia 64:1088–1117
Rast D, Stauble EJ (1970) On the mode of action of isovaleric acid in stimulating the germination of Agaricus bisporus. New Phytol 69:557–566
Robison MM, Horgen PA (1996) Plasmid RNA polymerase-like mitochondrial sequences in Agaricus bitorquis. Curr Genet 29:370–376
Robison MM, Royer JC, Horgen PA (1991) Homology between mitochondrial DNA of Agaricus bisporus and an internal portion of a linear mitochondrial plasmid of Agaricus bitorquis. Curr Genet 19:495–502
Robles CW, Lodder SC (2009) Brown mushrooms for commercial production. US Patent 7,608,760 B2
Romaine CP, Schlagnhaufer CD, Woolston BM (2011) Strategies for the transgenic manipulation of filamentous fungi. Patent WO/2011/130247 International Application PCT/US2011/032087
Román M, Boa E (2006) The marketing of Lactarius deliciosus in northern Spain. Econ Bot 60:284–290
Royse DJ, May B (1982a) Use of isozyme variation to identify genetic classes of Agaricus brunnescens. Mycologia 74:93–102
Royse DJ, May B (1982b) Genetic relatedness and its application in selective breeding of Agaricus brunnescens. Mycologia 74:569–575
Saksena KN, Marino R, Haller MN, Lemke PA (1976) Study on development of Agaricus bisporus by fluorescent microscopy and scanning electron microcopy. J Bacteriol 126:417–428
Sass JE (1929) The cytological basis for homothallism and heterothallism in the Agaricaceae. Am J Bot 16:663–701
Savoie JM, Largeteau ML (2011) Production of edible mushrooms in forests: trends in development of a mycosilviculture. MiniReview. Appl Microbiol Biot 89:971–979
Savoie JM, Bruneau D, Mamoun M (1996) Resource allocation ability of wild isolates of Agaricus bisporus on conventional mushroom compost. FEMS Microbiol Ecol 21:285–292
Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (2011) Proceedings of the 7th international conference on mushroom biology and mushroom products. http://www.wsmbmp.org/Previous_Conference_7.html
Sinden JW, Tschierpe HJ, Hauser E (1962) Transplantation of sporophores as a new method for studying growth and nutritional factors of mushrooms. Mushroom Sci 5:250–266
Sonnenberg AS, Van Loon PC, Van Griensven LJ (1991) The occurrence of mitochondrial genotypes and inheritance of mitochondria in the cultivated mushroom Agaricus bisporus. Mushroom Sci 13:85–92
Sonnenberg, ASM, Baars JP, Hendrickx PM, Kerrigan KW (2005) Breeding mushrooms: state of the art. In: Tan Q, Chen ZM, Cao H, Buswell JA (eds) Proceedings of the 5th international conference on mushroom biology and mushroom products, Acta Edulis Fungi 12sup, 163–173
Sonnenberg, ASM, Baars JP, Hendrickx PM, Lavrijssen B, Gao W, Weijn A, Mes JJ (2011) Breeding and strain protection in the button mushroom Agaricus bisporus. In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 1. pp 7–15. http://wsmbmp.org/Previous_Conference_7.html. Cited 30 Jan 2012
Spear MC, Royse DJ, May B (1983) Atypical meiosis and joint segregation of biochemical loci in Agaricus brunnescens. J Hered 74:417–420
Staniaszek M, Marczewski W, Szudyga K, Maszkiewicz J, Czaplicki A, Qian G (2002) Genetic relationship between Polish and Chinese strains of the mushroom Agaricus bisporus (Lange) Sing., determined by the RAPD method. J Appl Genet 43:43–47
Summerbell RC, Castle ZJ, Horgen PA, Anderson JB (1989) Inheritance of restriction fragment length polymorphisms in Agaricus brunnescens. Genetics 123:293–300
Vellinga EC, Sysouphanthong P, Hyde KD (2011) The family of Agaricaceae: phylogenies and two new white-spored genera. Mycologia 103:494–509
Wang ZS, Liao JH, Li FG (1995) Studies on breeding hybrid strain As2796 of Agaricus bisporus for canning in China. Mushroom Sci 14:71–72
Wang ZS, Liao J, Li H, Wang B, Chen M, Lu Z, Guo Z (2008a) Study on the biological characteristics of wild Agaricus bisporus strains from China. In: van Gruening M (ed) Science and cultivation of edible and medicinal fungi: Mushroom Science XVII, Proceedings of the 17th congress of the international society for mushroom science. South African Mushroom Farmers Association, Pretoria, South Africa (CD-ROM). ISBN 978-0-620-40808-0
Wang Y, Zeng F, Hon CC, Zhang Y, Leung FC (2008b) The mitochondrial genome of the Basidiomycete fungus Pleurotus ostreatus (oyster mushroom). FEMS Microbiol Lett 280:34–41
Wasser SP, Didukh MY, Amazonas MAL, Nevo E, Stamets P, Eira AF (2002) Is a widely cultivated culinary-medicinal royal sun Agaricus (the Himematsutake mushroom) indeed Agaricus blazei Murill? Int J Med Mushrooms 4:267–290
Weijn A, Tomassen MMM, Bastiaan-Net S, Hendrix EAHJ, Baars JJP, Sonnenberg ASM, Wichers HJ, Mes JJ (2011) Browning sensitivity of button mushrooms. In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 1. pp 203–211. http://www.wsmbmp.org/Previous_Conference_7.html. Cited 10 Feb 2012
Wisitrassameewong K, Karunarathna SC, Thongklang N, Zhao R, Callac P, Moukha S, Férandon C, Chukeatirote E, Hyde KD (2012a) Agaricus subrufescens: a review. Saudi J Biol Sci 19:131–146
Wisitrassameewong K, Karunarathna SC, Thongklang N, Zhao RL, Callac P, Chukeatirote E, Bahkali AH, Hyde KD (2012b) Agaricus subrufescens: new to Thailand. Chiang Mai J Sci 39:281–291
Wittenberg AH, van der Lee TA, Ben M’Barek S, Ware SB, Goodwin SB, Kilian A, Visser RG, Kema GH, Schouten HJ (2009) Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS One 4:e5863
Woolston BM, Schlagnhaufer C, Wilkinson J, Larsen J, Shi Z, Mayer K, Walters DS, Curtis WR, Romaine CP (2011) Long-distance translocation of protein during morphogenesis of the fruiting body in the filamentous fungus, Agaricus bisporus. PLoS ONE 6:e28412
Xu J (1995) Analysis of inbreeding depression in Agaricus bisporus. Genetics 141:137–145
Xu J, Kerrigan RW, Horgen PA, Anderson JB (1993) Localization of the mating-type gene in Agaricus bisporus. Appl Environ Microbiol 59:3044–3049
Xu J, Horgen PA, Anderson JB (1996) Somatic recombination in the cultivated mushroom Agaricus bisporus. Mycol Res 100:188–192
Xu J, Kerrigan RW, Callac P, Horgen PA, Anderson JB (1997) Genetic structure of natural populations of Agaricus bisporus, the commercial button mushroom. J Hered 88:482–488
Xu J, Desmerger C, Callac P (2002) Fine-scale genetic analyses reveal unexpected spatial-temporal heterogeneity in two natural populations of the commercial mushroom Agaricus bisporus. Microbiology 148:1253–1262
Zhao R, Karunarathna S, Raspé O, Parra LA, Guinberteau J, Moinard M, De Kesel A, Barroso G, Courtecuisse R, Hyde KD, Guelly AK, Desjardin D, Callac P (2011) Major clades in tropical Agaricus. Fungal Divers 51:279–296
Zied DC, Pardo-Gimenez A, Savoie JM, Pardo-Gonzalez JR, Callac P (2011) “Indoor” method of composting and genetic breeding of the strains to improve yield and quality of the almond mushroom Agaricus subrufescens. In: Savoie JM, Foulongne-Oriol M, Largeteau M, Barroso G (eds) Proceedings of the 7th international conference on mushroom biology and mushroom products, vol 1. pp 424–432. http://www.wsmbmp.org/Previous_Conference_7.html. Cited 25 Jan 2012
Acknowledgments
We wish to express our gratitude to the members of our laboratory for their help throughout the work on Agaricus biology and genetics. We thank Anne Rodier and the Centre Technique du Champignon (CTC) for fruitful collaboration in the research program for several years, contributing to the progress in genetics of A. bisporus. We are grateful to Francis Martin, the JGI team, and all the Agaricus community for the sequencing of the genome and its worthwhile annotation, and to all the mushroom scientists and their groups cited in this chapter for their work contributing to the development of knowledge and applications in edible mushroom breeding and cultivation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Savoie, JM., Foulongne-Oriol, M., Barroso, G., Callac, P. (2013). 1 Genetics and Genomics of Cultivated Mushrooms, Application to Breeding of Agarics. In: Kempken, F. (eds) Agricultural Applications. The Mycota, vol 11. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-36821-9_1
Download citation
DOI: https://doi.org/10.1007/978-3-642-36821-9_1
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-36820-2
Online ISBN: 978-3-642-36821-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)