Abstract
Luminous bacteria are those bacteria that carry the lux genes, genes that code for proteins involved in light production. Many luminous bacteria emit light at high, easily visible levels in laboratory culture and in nature, and the phenomenon of light emission has generated interest in these bacteria for over 125 years. Luminous bacteria are especially common in ocean environments where they colonize a variety of habitats, but some species are found in brackish, freshwater, and terrestrial environments. This chapter, which begins with an historical perspective, summarizes current understanding of the biochemistry and genetics of bacterial light emission, the taxonomy and phylogenetics of light-emitting bacteria, the evolutionary origins and hypothesized physiological and ecological functions of bacterial luminescence, the distributions and activities of these bacteria in nature, their symbiotic interactions with animals and especially with marine fishes, and the quorum sensing regulatory circuitry controlling light production at the operon level. This chapter concludes with information on the isolation, cultivation, storage, and identification of luminous bacteria.
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction and Historical Perspective
Luminous bacteria are those bacteria that carry lux genes, at a minimum luxA and luxB, the genes coding for bacterial luciferase, either as vertically inherited genes or genes naturally acquired by horizontal transfer. Most of the currently known luminous bacteria express the lux genes and produce light at high, readily visible levels in laboratory culture (Fig. 13.1 ) or in nature. Not all lux gene-carrying bacteria, however, produce levels of light visible to the human eye. To date, luminous bacteria have been found in only three closely related Gammaproteobacteria families, Vibrionaceae, Enterobacteriaceae, and Shewanellaceae, and most species are members of Vibrionaceae. Most luminous bacteria are facultatively aerobic, but two, Shewanella hanedai (Jensen et al. 1980) and Shewanella woodyi (Makemson et al. 1997), are respiratory. Additional and detailed information on the metabolism, physiology, morphology, and ecology of these bacterial groups and individual species can be found in Baumann and Baumann (1981), Baumann et al. (1984), Farmer and Hickman–Brenner (1992), Boemare et al. (1993), Forst et al. (1997), and Urbanczyk et al. (2007). Bacterial luminescence is one of several evolutionarily distinct forms of bioluminescence, an attribute of a wide diversity of eukaryotic organisms (Hastings 1995; Widder 2010).
The ability of certain bacteria to produce light has been known since 1875, when Pflüger (1875) related the luminescence coming from the slime of fish to bacteria present in the slime (Harvey 1957; Robertson et al. 2011). Many earlier observations suggest the presence of luminous bacteria and knowledge of their existence. During the 1700s and 1800s, various animal products (such as meats, fish, and eggs), the decaying bodies of marine and terrestrial animals, and even human wounds and corpses, were reported to emit light (Harvey 1940, 1952). Many years before those observations and long before bacteria and the oxygen dependence of bacterial luminescence were known, Boyle (1668) demonstrated that the “uncertain shining of fish,” the light coming from decaying fish, required air. Indeed, encounters with luminous objects and substances extend back to the beginnings of recorded history in Greece and China (Harvey 1957), and they continue in modern times to be causes of concern and wonder. Many of these encounters can be attributed to the saprophytic or pathogenic growth of luminous bacteria on or in marine and terrestrial animals.
According to Harvey (1940), J. F. Heller in 1854 was the first to give a name, Sarcina noctiluca, to an organism suspected to be responsible for luminescence. Following Pflüger’s work in 1875, other scientists working in the late 1800s and early 1900s isolated and named luminous bacteria, including “Bacterium lucens,” “Micrococcus phosphorescens,” “Micrococcus pflügeri,” “Bacillus phosphorescens,” and “Bacterium phosphoreum” (Neush 1879; Ludwig 1884; Fischer 1887; Molisch 1912; Dahlgren 1915; Zobell 1946; Harvey 1952, 1957; Robertson et al. 2011). Particularly notable among early researchers of bacterial luminescence was Martinus W. Beijerinck, a founder of general microbiology, who carried out research on the physiology of light-emitting bacteria and who coined the name Photobacterium, a genus within which he grouped all luminous bacteria (Beijerinck 1889a, b, 1891, 1916; van Iterson et al. 1940; Robertson 2003; Robertson et al. 2011). The recent revival and phylogenetic characterization of strains isolated by Beijerinck and stored in the 1920s (Figge et al. 2011) provide a direct link to the origins of general microbiology and the first studies of luminous bacteria.
Following these early studies at the end of the nineteenth and the beginning of the twentieth century, luminous bacteria were isolated from various habitats, the chemistry of bacterial light production and the culture requirements for growth and luminescence of the bacteria were characterized, and they were placed taxonomically as microbial systematics developed (e.g., Zobell and Upham 1944; Farghaly 1950; Johnson 1951). In the latter half of the twentieth century and continuing to date, taxonomic efforts have paralleled the growth of microbiology, incorporating the tools and knowledge developing from advances in biochemistry, physiology, and genetics (Baumann and Baumann 1977, 1981; Farmer and Hickman–Brenner 1992; Hastings and Nealson 1977, 1981; Hendrie et al. 1970; Nealson and Hastings 1992; Singleton and Skerman 1973). Presently, over 25 species of luminous bacteria are validly described (Table 13.1 ). Marine luminous species are found in Aliivibrio, Photobacterium, Vibrio (Vibrionaceae), and Shewanella (Shewanellaceae), and terrestrial light-producing species are members of Photorhabdus (Enterobacteriaceae) (Dunlap and Kita–Tuskamoto 2006; Urbanczyk et al. 2007, 2008; Ast et al. 2009; Dunlap 2009; Yoshizawa et al. 2009a, b; 2010a, b). Current understanding of the systematic relationships of luminous bacteria, as well as recent descriptions of new species, has utilized phylogenetic analysis of multiple, functionally independent housekeeping genes, including the 16S rRNA gene, gyrB, pyrH, and recA, among others (e.g., Ast and Dunlap 2005; Thompson et al. 2005; Ast et al. 2007b, 2009; Urbanczyk et al. 2007). Particularly useful for resolving the separate species status of closely related luminous bacteria is sequence analysis of the lux genes, luxCDABE, found to date in all luminous bacteria, due to their relatively rapid sequence divergence compared to most housekeeping genes (Ast and Dunlap 2004, 2005; Dunlap et al. 2004; Ast et al. 2007a; Urbanczyk et al. 2007). The description of several new species in the past few years (Table 13.1 ) suggests that many more species of luminous bacteria remain to be discovered. The advents of whole genome sequencing, metagenomics, and single-cell genomics and their application to luminous bacteria will undoubtedly provide additional insight into the systematics of luminous bacteria, the evolution of the bacterial luminescence system, and many other aspects of the biology of these bacteria.
Biochemistry of Bacterial Luminescence
Light emission in bacteria is catalyzed by luciferase, a heterodimeric protein of approximately 80 kD, composed of α (40 kDa) and β (37 kDa) subunits. Bacterial luciferase mediates the oxidation of reduced flavin mononucleotide (FMNH2) and a long-chain aliphatic (fatty) aldehyde (RCHO) by O2 to produce blue-green light according to the following reaction:
In the luminescence reaction, binding of FMNH2 by the enzyme is followed by interaction with O2 to form a flavin-4a-hydroperoxide. Association of this complex with aldehyde forms a highly stable intermediate, the slow decay of which results in oxidation of the FMNH2 and aldehyde substrates and the emission of light. Quantum yield for the reaction has been estimated at 0.1–1.0 photons. The reaction is highly specific for FMNH2, and the aldehyde substrate in vivo is likely to be tetradecanal. FMNH2 is provided by the activity of an NAD(P)H-flavin oxidoreductase (flavin reductase). Synthesis of the long-chain aldehyde is catalyzed by a fatty acid reductase complex composed of three polypeptides, an NADPH-dependent acyl protein reductase (r, 54 kDa), an acyl transferase (t, 33 kDa), and an ATP-dependent synthetase (s, 42 kDa). The complex has a stoichiometry of r4s4t2–4, and its activity is essential for the production of light in the absence of exogenously added aldehyde. Luciferases from different species of luminous bacteria exhibit substantial amino acid residue and nucleotide sequence identity (Meighen and Dunlap 1993; Dunlap et al. 2007), consistent with a common evolutionary origin of luminescence in bacteria. For references and detailed information on the biochemistry of bacterial light production, the reader is directed to reviews by Hastings (1995), Lee et al. (1990), Hastings et al. (1985), Meighen (1988; 1991), Meighen and Dunlap (1993), and Wilson and Hastings (1998).
Species and Phylogeny of Luminous Bacteria
Over 25 species of luminous bacteria are validly described at this time (Table 13.1 ). Taxonomically, luminous bacteria are members of six of genera in three Gammaproteobacteria families: Vibrionaceae, Enterobacteriaceae, and Shewanellaceae (Fig. 13.2 ) To date, no luminous strains belonging to other families have been reported. Most luminous species are members of Aliivibrio, Vibrio, and Photobacterium in Vibrionaceae. Detailed phylogenetic analysis has shown that most extant luminous members of Vibrionaceae acquired their luxCDABE genes vertically, with only a few cases of acquisition by intraspecies horizontal transfer from members of Vibrionaceae, whereas luminous members of Enterobacteriaceae and Shewanellaceae apparently acquired their lux genes by horizontal transfer from members of Vibrionaceae (Urbanczyk et al. 2008). These considerations, and others described below, suggest that the luxCDABE-based luminescence system of bacteria arose just once evolutionarily, apparently in an ancestor of Vibrionaceae (Urbanczyk et al. 2008).
Phylogeny of luminous bacteria. The analysis, parsimony implemented in PAUP*, is based on sequences of the 16S rRNA and gyrB genes. Luminous species (in boldface) are found in three families, Vibrionaceae, Shewanellaceae, and Enterobacteriaceae. These families contain many more nonluminous species than shown here. Also, recently identified luminous bacteria, for example, Vibrio “beijerinckii” (proposed name) (Figge et al. 2011) and Candidatus Photodesmus katoptron (Hendry and Dunlap 2011), and additional species whose descriptions are underway, are not shown
Several new species of luminous bacteria have been described in the past few years (Table 13.1 , Fig. 13.2 ). These include Aliivibrio sifiae (Ast et al. 2009; Yoshizawa et al. 2010a), Photobacterium kishitanii (Ast et al. 2007a), Photobacterium aquimaris (Yoshizawa et al. 2009b), Candidatus Photodesmus katoptron (Hendry and Dunlap 2011), Vibrio azureus (Yoshizawa et al. 2009a), and Vibrio sagamiensis (Yoshizawa et al. 2010b). Recent studies have also revealed the presence of luminous strains of species not previously reported as luminous, that is, Vibrio campbellii (Lin et al. 2010), Vibrio vulnificus (Urbanczyk et al. 2008), and Vibrio damsela (Urbanczyk et al. 2008). With respect to V. campbellii, a recent genomic analysis has revealed strain ATCC BAA-1116 (aka BB120), previously classified as Vibrio harveyi and studied intensively for quorum sensing control of luminescence and other cellular functions in this species (e.g., Bassler et al. 1993; Waters and Bassler 2005; Long et al. 2009), is actually a member of V. campbellii (Lin et al. 2010). Furthermore, newly recognized clades, for example, Aliivibrio “thorii” and Vibrio “beijerinckii” (Ast et al. 2009; Figge et al. 2011), have been identified, and formal description of these and other new species is under way. In most cases, taxonomic identification has followed cultivation-based detection of light emission; most of the bacteria listed in Table 13.1 grow and emit light in laboratory media. However, several luminous bacteria, bioluminescent symbionts of anomalopids (flashlight fish) and ceratioids (deep-sea anglerfish), are known that have not been cultured; these bacteria are members of Vibrionaceae but are divergent from known species of luminous bacteria (Haygood 1990, 1993; Haygood and Distel 1993). Very recently, luminous bacteria symbiotic with the anomalopid fish Anomalops katoptron were characterized phylogenetically and assigned Candidatus status as a new Vibrionaceae genus and species, Photodesmus katoptron (Hendry and Dunlap 2011).
Most luminous strains isolated from natural habitats group taxonomically as members of well-recognized species that typically are considered to be luminous (Table 13.1 ). However, luminescence often is not a uniformly consistent phenotype of even these luminous species (e.g., Wollenberg et al. 2011) or their genera. Nonluminous species of Vibrio are well known and are more common than luminous strains (e.g., Baumann and Baumann 1981), and several nonluminous species of Photobacterium and Aliivibrio have been found, some of which apparently lack lux genes (Dunlap and Ast 2005; Urbanczyk et al. 2011a). Furthermore, strains of Photorhabdus luminescens symbiotic with entomopathogenic nematodes have been found that do not produce light and lack genes necessary for light production (Akhurst and Boemare 1986; Forst and Nealson 1996). In addition, strains luminous on primary isolation often become dim or dark in laboratory culture (Nealson and Hastings 1979, 1992; Akhurst 1980; Silverman et al. 1989), and some species that grow well in laboratory culture at room temperature, that is, A. logei and S. hanedai, typically produce readily visible light only when grown at cooler temperatures. In some cases, that is, luminous bacteria infecting crustaceans (Giard and Billet 1889b) and strains of A. fischeri symbiotic with the Hawaiian sepiolid squid, Euprymna scolopes (Boettcher and Ruby 1990), the bacteria produce a high level of light in their natural habitat but produce little or no light when grown in laboratory culture.
Adding to this complexity, V. cholerae, generally considered to be a nonluminous species, has many luminous strains (e.g., Kaeding et al. 2007; Zo et al. 2009), and many of the nonluminous strains of this species carry lux genes that apparently are not expressed in laboratory culture (Palmer and Colwell 1991; Ramaiah et al. 2000). In addition, bacteria identified as related to V. harveyi and V. cincinnatiensis carry the lux genes but have been found to have lux gene mutations that result in a dark phenotype (O’Grady and Wimpee 2008). Furthermore, luminous strains of three other species generally known as nonluminous, Vibrio vulnificus, Vibrio chagasii, and Photobacterium damselae, recently were identified (Urbanczyk et al. 2008). This substantial variation in the incidence of luminous strains within a species has implications for understanding the evolutionary origins of bacterial luminescence and its patterns of inheritance, as described in sections that follow. It should be noted that the presence of luminescence strains has likely been overlooked in many species. Routine use of cooler temperatures (10–20 °C) for growth and examination, and utilization of conditioned media, inducers, and luciferase substrates (Fidopiastis et al. 1999), along with the application of probes for luxA and other lux genes (Wimpee et al. 1991), and full sequence-based characterization of the lux operons of new luminous bacteria will undoubtedly lead to a more complete understanding of the species diversity of bacteria able to produce light. A further complication in gaining a more comprehensive understanding of the diversity of luminous bacteria is that descriptions of new species of luminous bacteria often lack detailed information on the lux genes, relationships to type strains, or detailed phylogenetic analysis (e.g., Yoshizawa et al. 2009b, 2010b). In this regard, characterization of multiple newly isolated strains, the use of multiple independent loci, and the use of type strains and other key strains are imperatives in new species descriptions for revealing the otherwise hidden species diversity of luminous bacteria (e.g., Ast et al. 2009).
The Bacterial Luminescence Operon
The genes coding for the α- and β-subunits of bacterial luciferase, luxA and luxB, respectively, are part of the lux operon, luxCDABE, which is present in the genomes of all luminous bacteria examined to date as a conserved, contiguous, and coordinately expressed set of genes (Fig. 13.3 ). The luxC, luxD, and luxE genes, respectively, code for the r, s, and t polypeptides of the fatty acid reductase complex that synthesizes and recycles aldehyde substrate for luciferase. The lux operons of most bacteria also contain luxG, which codes for a flavin reductase (Lin et al. 1998; Meighen and Dunlap 1993; Nijvipakul et al. 2008; Swartzman et al. 1990a). The absence of luxG from the lux operon of Ph. luminescens apparently is compensated for by the activity of a flavin reductase activity coded for by an Escherichia coli fre-like gene, homologs of which are found in various luminous bacteria (Zenno et al. 1992, 1994; Zenno and Saigo 1994). Several species of Photobacterium bear an additional lux operon gene, luxF, between luxB and luxE. The luxF gene, coding for a nonfluorescent flavoprotein, is apparently specific to Photobacterium, as it is not present in the lux operons of Aliivibrio, Photorhabdus, Shewanella, or Vibrio species (Fig. 13.3 ), but it has been secondarily lost in Photobacterium leiognathi (Ast and Dunlap 2004). The LuxF protein might function in the luminescence system by scavenging an inhibitory side product of the luciferase reaction (Moore and James 1995), but it is not necessary for light production even in those Photobacterium species that normally carry this gene (Kaeding et al. 2007). In the examined species of luminous Photobacterium, the lux operon genes are followed, without a transcriptional stop or other regulatory sites, by genes involved in the synthesis of riboflavin, ribEBHA, the products of which presumably function in generating FMNH2, a substrate of luciferase (Lee and Meighen 1992; Lee et al. 1994; Lin et al. 2001; Sung and Lee 2004; Ast et al. 2007b). We refer to this gene arrangement as the Photobacterium lux-rib operon (Fig. 13.3 ). Strains of P. phosphoreum lack one of the rib genes, ribE; the gene presumably was lost in the divergence from an ancestral Photobacterium that gave rise to this species. The presence of genes for synthesis of riboflavin as part of the lux operon might facilitate light production by ensuring coordinate synthesis of luciferase and substrates for the enzyme. In this regard, the lux operon of V. campbellii (previously classified as V. harveyi; Lin et al. 2010) contains ribB, coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase, a key enzyme in riboflavin synthesis (referred to originally as luxH; Swartzman et al. 1990b) as the final gene, as does the lux operon of Candidatus Photodesmus katoptron (Hendry and Dunlap 2011). Furthermore, although ribB is not part of the lux operon of A. fischeri, its expression nonetheless is under the same quorum sensing control as the lux genes (Callahan and Dunlap 2000).
Genes of the lux operons of luminous bacteria. Shown are the lux genes and the organization of lux operons for those bacteria for which complete lux operon sequence data are available. Contiguous genes of the luminescence operons of luminous bacteria are aligned to highlight commonalities and differences. Four distinct types of lux operons are evident based on commonalities of gene content, organization, and sequence similarity, (1) the Aliivibrio/Shewanella type, with luxI/luxR regulatory genes; (2) Photobacterium type, with ribEBHA genes; (3) the Vibrio/Candidatus Photodesmus type, with neither regulatory nor additional linked genes; and (4) the Photorhabdus type, composed of just the core luxCDABE genes
In addition to presence of ribEBHA genes in Photobacterium as part of the lux-rib operon, genes upstream of the lux operon contribute to luminescence and also show genus and species differences. In Photobacterium mandapamensis, for example, the lux-rib operon is preceded by lumQ and lumP, which form the lumazine operon. The function of lumQ is not yet known, although it might code for a DNA binding protein (Lin et al. 1995). LumP, a 21 kDa fluorescent accessory protein referred to as lumazine protein, functions to shift the emission wavelength of luciferase from blue-green (495 nm) to blue (475–486 nm) and enhance the intensity of light emission (Lee 1993; O’Kane et al. 1985,1991; Petushkov et al. 1996). LumP, which has been isolated from P. phosphoreum and strains called P. leiognathi (which actually are P. mandapamensis, see below) and also purified from P. kishitanii, contains a noncovalently bound fluorophore, 6,7-dimethyl-8-ribityllumazine, the immediate biosynthetic precursor of riboflavin (O’Kane et al. 1985; Sato et al. 2010; Small et al. 1980). In P. leiognathi lumP is not found, although approximately 200 nucleotides of the P. leiognathi luxC–lumQ intergenic region can be aligned to the P. mandapamensis lumP gene (Fig. 13.4 ; Ast et al. 2007b). The activity of the LumP protein apparently accounts for the blue-shifted luminescence of P. mandapamensis compared to P. leiognathi, one of the diagnostic traits distinguishing these two species (O’Kane et al. 1985, 1991; Lee 1993; Petushkov et al. 1996; Ast and Dunlap 2004; Kaeding et al. 2007). The genes flanking the P. leiognathi and P. mandapamensis lux-rib operons are homologous to a single contiguous region in nonluminous P. angustum (Lin et al. 1993, 1995, 1996a, b, 2001; Ast et al. 2007b).
Region upstream of the lux operon in Photobacterium. (a) The lux-rib operon is preceded in P. mandapamensis by lumQ and lumP. (b) For the lux-rib 1 operon of P. leiognathi, lumQ is present upstream and lumP is not found, although approximately 200 nucleotides of the P. leiognathi luxC–lumQ intergenic region can be aligned to the P. mandapamensis lumP gene sequence. (c) The region upstream of the lux-rib 2 operon of P. leiognathi contains lumQ and a transposase gene. For details, see the text and Ast et al. (2007b). The regions flanking the lux-rib operons of other Photobacterium species remain to be defined, but preliminary information for some species indicates differences from the arrangement shown here (Urbanczyk et al. unpublished data)
In the examined Aliivibrio species, regulatory genes, luxI and luxR, which control transcription of the lux operon, precede or flank the luxCDABEG genes (Fig. 13.3 ). The luxI gene codes for an acyl-homoserine lactone (acyl-HSL) synthase (Schaefer et al. 1996), and luxR codes for a receptor protein that interacts with acyl-HSL to activate transcription of the lux operon (Engebrecht et al. 1983), as described in more detail below. In Aliivibrio fischeri, luxI is the first gene of the lux operon, and luxR, upstream of luxI, is divergently transcribed (Fig. 13.3 ). The same gene arrangement is present in Shewanella hanedai, and this identity together with the high degree of lux gene sequence similarity in S. hanedai and A. fischeri has led to the suggestion that S. hanedai acquired its lux operon by horizontal transfer from A. fischeri or the ancestor of A. fischeri (Urbanczyk et al. 2008), as described in more detail below. In Aliivibrio salmonicida, a bacterium that requires exogenous addition of aldehyde to produce a high level of light (Fidopiastis et al. 1999), two luxR genes, homologous to A. fischeri luxR, flank the lux operon; a luxI gene also is present, divergently transcribed from the downstream luxR (Fig. 13.3 ; Nelson et al. 2007; Hjerde et al. 2008). Very recently, the same arrangement of luxI and luxR genes as in A. salmonicida was identified in Aliivibrio logei (Manukhov et al. 2011). In contrast to A. salmonicida, however, A. logei does not require exogenous aldehyde to produce a high level of light (Manukhov et al. 2011); sequence comparison of the two operons identified mutations in luxD of A. salmonicida that presumably account for the exogenous aldehyde requirement of this species. Genes flanking the lux operons of other luminous Aliivibrio species (Table 13.1 ) apparently have not yet been identified.
The arrangement of genes flanking the lux operons of the examined Vibrio species differs substantially from that in Photobacterium and in Aliivibrio (Fig. 13.3 ). First, regulatory genes controlling transcription of the lux operon are not part of and are not adjacent to the lux operon in those Vibrio species examined; specifically, a luxR gene, which is not homologous to A. fischeri luxR, is not physically associated with the lux operon in V. campbellii (V. harveyi); the role of luxR Vh in the phosphorelay cascade controlling luminescence in this species is outlined below. Conservation of luxCDABE as a unit might reflect a need for close interaction of luciferase and fatty acid reductase proteins, based on coordinate regulation, to facilitate substrate generation necessary for efficient light production. However, it is not obvious what led to the genus-specific differences in the presence of genes flanking and contiguous with the lux operons of luminous members of Aliivibrio, Photobacterium, and Vibrio, three closely related genera of Vibrionaceae.
Genomes of Luminous Bacteria
The genomes of luminous Vibrio and Photobacterium species are similar in structure, overall size, and organization to other members of Vibrionaceae, with two chromosomes of unequal size and an overall size of approximately 4.5–5.4 Mb (Egan et al. 2005; Okada et al. 2005; Ruby et al. 2005; Vezzi et al. 2005; Reen et al. 2006; Lauro et al. 2009; Ast et al. 2007a; Urbanczyk et al. 2011a, b; Urbanczyk et al. unpublished data). The significance of this organization for members of Vibrionaceae is not yet known, but differences between the two replicons suggest that each chromosome carries out substantially different roles in the cell. More of the core (essential) genes are found on the large chromosome, whereas the small chromosome contains mostly lineage specific genes. Furthermore, gene content and position appear to be more highly conserved on the large chromosome than on the small chromosome (Reen et al. 2006). The small chromosome in members of Vibrionaceae nonetheless contains some essential genes (Reen et al. 2006), which might guarantee retention of the small chromosome during cell division (Egan et al. 2005). The origin of the small chromosome in Vibrionaceae remains unknown but has been hypothesized in V. cholerae to have originated from a plasmid that accumulated additional genes, including some genes transferred from the large chromosome (Egan et al. 2005). The apparent ubiquity of two chromosomes of unequal size in Vibrionaceae suggests that the small chromosome may have arisen in the ancestral lineage leading to Vibrionaceae.
In view of the different predicted roles for the large and small chromosomes in Vibrionaceae, it is significant that in luminous species for which complete sequences of both chromosomes are available, the lux operons are all located on the small chromosome. These species include A. fischeri (Ruby et al. 2005; Mandel et al. 2009), A. salmonicida (Hjerde et al. 2008), and V. campbellii (Lin et al. 2010). This pattern indicates that the lux genes have an accessory function, that is, they are not part of the core genome, a view that is consistent with the many nonluminous Vibrionaceae species and many nonluminous strains of luminous species, as noted above. Which chromosome, large or small, carries the lux genes is not yet known in Photobacterium species. A single chromosome is characteristic of Enterobacteriaceae and Shewanellaceae, and the lux genes in Photorhabdus and luminous Shewanella are chromosomal.
The genomes of luminous bacteria analyzed to date have been found to carry multiple rRNA operons. Specifically, the genome of A. fischeri carries 12 rRNA operons (Ruby et al. 2005), the P. kishitanii genome has eight or more (Ast et al. 2007a), the P. mandapamensis genome has six or more (Urbanczyk et al. 2011b), and the Ph. luminescens genome has seven (Duchaud et al. 2003; Wilkinson et al. 2009). A high copy number of rRNA operons may be an adaptation for a copiotrophic lifestyle and for rapid response to nutrient availability (Klappenback et al. 2000; Lauro et al. 2009). Taken together, the relatively large genome size and multiple rRNA operons of luminous bacteria and other members of Vibrionaceae may be adaptations for rapidly utilizing a wide range of different nutrients under feast-or-famine conditions.
Evolutionary Origin and Function of Bacterial Luminescence
The origin of bacterial luminescence has been of interest since the early days of microbiology. The natural presence of genes necessary for producing light defines the luminous bacteria. The necessary genes, luxA and luxB, encoding the luciferase subunits, luxC, luxD and luxE, for the fatty acid reductase subunits, and luxG, encoding a flavin reductase, are consistently found together as a cotranscribed unit, luxCDABEG. The reason for this conservation of lux genes as a unit is not known, but it might relate to efficient light production; the contiguous presence of these genes as an operon might help promote the coordinated production of luciferase and substrates for luciferase, long-chain aldehyde and reduced flavin mononucleotide (FMNH2). The conservation of these genes as a unit in nearly all luminous bacteria examined suggests that the lux operon arose just once in the distant past. Supporting this view, phylogenetic analysis demonstrates that the individual lux genes of different bacterial species are homologous, as was suggested by the high levels of amino acid sequence identities of the inferred Lux proteins. This homology implies that the bacterial luxCDABEG genes arose one time in the evolutionary past. The use by luciferase of oxygen as a substrate implies that this enzymatic activity originated after oxygenic photosynthesis by ancestors of modern-day cyanobacteria began to increase the level of O2 on Earth, approximately 2.4 billion years ago, during the Great Oxidation Event. A marine origin for bacterial luminescence (Palmer and Colwell 1991; Dunlap 2009) seems likely because most species of luminous bacteria are marine (Table 13.1 ).
Seliger (1987) proposed that bacterial luminescence arose under ecological selection for light emission. A flavoprotein catalyzing fatty acid α-oxidation reactions with low chemiluminescent quantum yields is postulated to have mutated under hypoxic conditions to accept FMNH2 as the flavin cofactor, generating a fortuitously high fluorescence yield, termed “protobioluminescence,” via the 4a-hydroxy-FMNH product. This flavin-dependent, aldehyde-oxidizing protoluciferase produced sufficient light and with an appropriate emission spectrum, to be detected by phototactic organisms. Ingestion by visually cueing animals of particles colonized and made luminous by these early luminous bacteria presumably enhanced their reproduction by bringing them into the animal’s nutrient-rich digestive system, ensuring the emitter’s survival and thereby possibly leading to selection for more intense light output (Widder 2010). It is possible that early evolutionary steps leading to protoluciferase involved oxygen detoxification activity that permitted early anaerobic organisms to survive an increasingly aerobic environment (McElroy and Seliger 1962; Rees et al. 1998). An alternative hypothesis for the evolution of bacterial luciferase for DNA repair (Czyż et al. 2003) has been called into question (Walker et al. 2006).
A single gene was hypothesized to encode bacterial protoluciferase (O’Kane and Prasher 1992). Although a single-subunit protoluciferase, monomer or dimer, presumably would have differed somewhat from the modern-day luciferase α-subunit and therefore might have produced light, the inability of either of the extant α- or β-subunits alone to produce light in vitro or in vivo (Li et al. 1993) argues against the single-gene hypothesis. Alternatively, bacterial protoluminescence may have arisen following a gene duplication event that is postulated to have created luxB from luxA (Baldwin et al. 1979; O’Kane and Prasher 1992; Meighen and Dunlap 1993). Based on amino acid sequence identities, a tandem duplication of the ancestral luxA gene, followed by sequence divergence in the duplicated gene, is thought to have given rise to luxB, leading to the formation of the heterodimeric luciferase present in extant luminous bacteria. Similarly, a tandem duplication of luxB followed by loss of approximately 300 nucleotides coding for N-terminus amino acids is thought to have given rise to luxF in a luminescent ancestor of Photobacterium; this gene apparently was later secondarily lost in P. leiognathi (Baldwin et al. 1979; O’Kane and Prasher 1992; Meighen and Dunlap 1993; Ast and Dunlap 2004; Dunlap 2009).
Although the evolutionary origin of luxA and other bacterial luminescence genes remains obscure (Dunlap and Kita–Tuskamoto 2006), the conserved gene content and gene order of the lux operon in bacteria, luxCDABEG, and the high levels of lux gene and Lux protein amino acid sequence identities among luminous bacteria (e.g., Meighen and Dunlap 1993) leave little doubt of the homology of all presently known bacterial lux operons. Furthermore, the general congruence of phylogenies based on lux genes and other protein coding genes (and the 16S rRNA gene) (Urbanczyk et al. 2008) suggests that the lux operon is ancestral at least to Aliivibrio, Photobacterium, and Vibrio, and possibly to Vibrionaceae. The association of the fatty acid reductase genes, luxCDE, with luxA might have predated the luxA to luxB gene duplication event. Alternatively, the presence of ERIC sequences flanking luxA and luxB in Ph. luminescens (Meighen and Szittner 1992) might mark an insertion of the luxAB genes into the fatty aldehyde reductase operon during the evolution of the bacterial luminescence system. The origins and evolution of other luminescence genes are not well understood (O’Kane and Prasher 1992).
The evolution of bacterial luminescence system also involved recruitment of regulatory and other genes to the lux operon. The lux operons of certain Aliivibrio species contain two regulatory genes, luxR and luxI (Fig. 13.3 ), the protein products of which mediate a population density-responsive autoinduction, that is, quorum sensing. Recruitment of regulatory genes to the lux operon during evolution of Aliivibrio presumably enhanced quorum sensing control of luminescence (Dunlap 2009). Furthermore, as mentioned above, luminous Photobacterium strains carry genes involved in the synthesis of riboflavin, the ribEBHA genes, as part of the lux-rib operon (Lee et al. 1994; Ast et al. 2007b). Recruitment of the rib genes to the lux operon likely happened in an ancestor of Photobacterium, since other luminescent bacteria contain the rib genes elsewhere in the genome and not associated with the lux operon (e.g., Callahan and Dunlap 2000). An interesting exception to that general pattern is the presence of ribB initially named (luxH) as the last gene of the lux operon in V. campbellii (previously classified as V. harveyi; Lin et al. 2010).
The production of light consumes a substantial amount of energy, through the synthesis of Lux proteins and through their activity (Dunlap and Greenberg 1991). This energetic cost, which may explain the fact that luciferase synthesis is regulated in most luminous bacteria, suggests that activity of the luminescence system plays an important role in the physiology and ecology of luminous bacteria. Most attention to what that role might be has focused on oxygen. One consideration is that, as noted above, the light-emitting reaction might have arisen evolutionarily as a detoxification mechanism, removing oxygen and thereby allowing an organism that is otherwise anaerobic to survive. Related to this possibility is that luciferase, as an oxidase, might function as a secondary respiratory chain that is active when oxygen or iron levels are too low for the cytoplasmic membrane-associated electron transport system to operate. This activity would allow cells expressing luciferase to reoxidize reduced coenzyme even when oxygen levels are low (Hastings and Nealson 1981; Hastings 1983; Nealson and Hastings 1992). Consistent with this view, growth of cytochrome-deficient luminous bacteria is dependent on induction of luciferase, limitation for iron stimulates light production, low oxygen levels promote the luminescence of some luminous bacteria, and luciferase synthesis can be induced under anaerobic conditions (Eberhard et al. 1979; Haygood and Nealson 1985; Makemson 1986; Makemson and Hastings 1982; Nealson and Hastings 1977, 1979). As an alternative to the electron transport system, the activity of luciferase in reoxidizing reduced coenzyme could permit cells of luminous bacteria in low oxygen habitats, such as in animal gut tracts, to continue to transport and metabolize growth substrates, thereby continuing to gain energy through substrate-level phosphorylation. Furthermore, light production presumably facilitates dissemination of luminous bacteria. The feeding of animals on luminous particles (decaying tissues, fecal pellets, and moribund animals infected by luminous bacteria), to which they are attracted, would bring the bacteria into the animal’s nutrient-rich gut tract for additional rounds of reproduction followed by dispersal (Hastings and Nealson 1981; Nealson and Hastings 1992), and recent evidence supports this possibility (Zarubin et al. 2012). Alternatively, the function of the bacterial lux system might be to generate a halotolerant flavodoxin, with light emission an incidental consequence (Kasai 2006). Future studies may test and possibly provide additional support for these and other proposed functions for luminescence, such as a physiological role for luciferase activity in bioluminescent symbioses, but it is not yet clear what factors, physiological or ecological, actually select for the retention and expression of this energetically expensive enzyme system.
Horizontal Acquisition of the Bacterial lux Genes
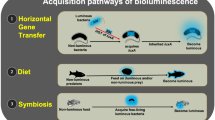
Inheritance of the lux genes has been shown to be primarily vertical. However, some instances of acquisition by horizontal transfer have been identified (Ast et al. 2007; Urbanczyk et al. 2008). In the instances identified, horizontal acquisition of lux genes within Vibrionaceae has been found to be limited to species within the same genera, and no instance of the horizontally transferred genes replacing vertically inherited lux operons has been reported. In contrast to the proposal that horizontal gene transfer drives bacterial speciation (e.g., Gogarten et al. 2002; Ochman et al. 2000), horizontal acquisition of lux genes apparently has not led to phylogenetic divergence of the recipients (Urbanczyk et al. 2008). The predominant pattern of vertical inheritance of the lux genes, together with the fact that most species of luminous bacteria are members of Vibrionaceae, leads to the hypothesis that these genes arose in an ancestor of Vibrionaceae. The scattered incidence of luminous members in Vibrionaceae, with many nonluminous species and many species with nonluminous strains, indicates that the lux genes have been lost from many descendants of this putative ancestor (Urbanczyk et al. 2008; Dunlap 2009).
In Photobacterium, many strains of P. leiognathi carry two intact and apparently functional lux-rib operons in their genomes (Ast et al. 2007b). This situation represents an unusual case of natural merodiploidy in bacteria, the presence of two or more copies of the same gene or genes in the genome of a bacterium, because of the large number of genes involved and because the second operon did not arise by tandem duplication of the first. The two lux-rib operons are distinct in sequence and genomic location. One operon, lux-rib 1, is in the ancestral chromosomal location of the lux-rib operon in P. leiognathi and related bacteria. The other, lux-rib 2, is located elsewhere in the genome and is present in many but not all strains of P. leiognathi; it is flanked by genes coding for transposases, which suggests it can transfer between strains. Phylogenetic analysis indicates that the lux-rib 1 and lux-rib 2 operons are more closely related to each other than either is to the lux and rib genes of other bacterial species (Ast et al. 2007b). This finding rules out interspecies horizontal transfer as the origin of the lux-rib 2 operon in P. leiognathi; instead, lux-rib 2 apparently arose in the distant past within a lineage of P. leiognathi that either has not yet been sampled or has gone extinct.
Merodiploidy of the lux-rib operon in P. leiognathi also is the first instance of merodiploid strains of a bacterium having a nonrandom geographic distribution; strains bearing a single lux-rib operon are found over a wide geographic range, whereas lux-rib merodiploid strains have been found only in coastal waters of Honshu, Japan (Ast et al. 2007b; Urbanczyk et al. 2012b). The presence of multiple copies of each of the lux and rib genes might provide opportunities for sequence divergence and selection that could lead to the evolution of new gene functions in one or the other of the duplicate genes.
The P. leiognathi lux-rib 2 operon has also been found in two strains of P. mandapamensis, which also carry a normal P. mandapamensis lux-rib operon, and in a strain of P. damselae, a species not previously known to be luminous (Urbanczyk et al. 2008). Furthermore, evidence has been obtained indicating horizontal acquisition of the lux genes by a recently recognized species, P. aquimaris (Yoshizawa et al. 2009b; Urbanczyk et al. 2012a).
With respect to S. hanedai and S. woodyi, comparison of genes flanking the lux operons suggested that these species had acquired lux genes from a member of Aliivibrio (Kasai et al. 2007), a possibility confirmed through phylogenetic analysis (Urbanczyk et al. 2008). In Photorhabdus species as well, the luxCDABE genes may have been acquired by horizontal gene transfer (Forst et al. 1997), possibly from an ancestor of V. harveyi (Meighen 1999). Differences between ecologically distinct strains of Ph. luminescens in the DNA flanking the lux operon (Meighen and Szittner 1992) raise the possibility that lateral transfer of the lux genes to this species occurred more than once (Forst et al. 1997). However, phylogenetic analysis of the Photorhabdus lux genes in the context of Vibrionaceae and Shewanellaceae sequences did not find support either for or against horizontal acquisition of the lux genes by Ph. luminescens (Urbanczyk et al. 2008). It is possible that substantial sequence divergence of the lux genes has occurred since their transfer to Ph. luminescens, thereby making problematic the identification of their source species.
Instances of the horizontal acquisition of lux genes have been identified also in Vibrio (Urbanczyk et al. 2008). The only known luminous strain of the human pathogen V. vulnificus (Oliver et al. 1986) apparently acquired its lux genes from V. harveyi, and in V. chagasii, a species not previously known to be luminous (Thompson et al. 2003), two luminous strains were identified and through phylogenetic analysis were shown to have acquired their lux genes apparently from V. harveyi and V. splendidus, respectively (Urbanczyk et al. 2008). A mechanism for these transfers, however, has not been proposed.
It should be noted that most species of Vibrionaceae lack the lux genes and therefore are nonluminous. Also, most strains of some luminous species, such as V. cholerae, are nonluminous. The low incidence of luminous species in the family suggests that the lux genes have been lost over evolutionary time from many of the lineages that have given rise to extant species. It also seems likely that nonluminous variants of luminous species can arise frequently through loss of one of more the core genes of the lux operon, luxCDABE (e.g., Wollenberg et al. 2011). The scattered incidence of lux genes in Vibrionaceae presumably relates to different ecologies of the different species. It is not clear, however, how having and expressing lux genes contributes to the lifestyle of most luminous bacteria, because there are no obvious ecological differences between luminous and nonluminous species except in the case of those species that are bioluminescent symbionts of fish and squids.
Habitats and Ecology of Luminous Bacteria
The luminous Aliivibrio, Photobacterium, Vibrio, and Shewanella species occur in the marine environment, whereas Photorhabdus species are terrestrial. Vibrio cholerae also occurs in brackish environments and freshwater, although strains of this species also commonly occur in coastal seawater (e.g., Kaeding et al. 2007; Urbanczyk et al. 2008).
Marine
Luminous bacteria are globally distributed in the marine environment (Table 13.1 ) and have been isolated from seawater, sediment, and suspended particulates from a wide variety of locations (Baumann and Baumann 1981; Harvey 1952; Zobell 1946). They also commonly colonize marine animals as saprophytes, commensal enteric symbionts, and parasites (Baumann and Baumann 1981; Harvey 1952; Kozukue 1952; Makemson et al. 1997; Makemson and Hermosa 1999; Meighen and Dunlap 1993; Ruby and Morin 1979; ZoBell 1946). They can also be isolated from inanimate surfaces and macroalgae (Makemson et al. 1992). A few species of luminous bacteria establish bioluminescent symbiosis with marine fish and squids (Dunlap 2009; Dunlap et al. 2007; Hastings and Nealson 1981; Haygood 1993; Ruby 1996; Ruby and Morin 1978; Visick and Ruby 2006; Urbanczyk et al. 2011a, b). In seawater, the incidence of luminous bacteria generally is low (from 0.01–40 cells per ml; Nealson and Hastings 1992), with higher numbers in coastal seawater and lower numbers in open ocean and deeper waters (Ruby and Nealson 1978; Ruby et al. 1980). Possibly reflecting this variation, metagenomic analyses of different marine waters have identified the presence of genes related to luxA (Martín Cuadrado et al. 2007) and conversely showed an absence of bacterial lux genes (Nealson and Venter 2007). Therefore, the geographic distribution of luminous bacteria in the plankton varies substantially.
In contrast to their generally low incidence in seawater, luminous bacteria can attain very high numbers in saprophytic, commensal, parasitic, and symbiotic associations with animals (up to 1011 cells per ml in symbiotic habitats; Ruby and Nealson 1976; Dunlap 1984; Nealson and Hastings 1992; Visick and Ruby 2006). For example, luminous bacteria can be readily isolated by enrichment from the muscle tissue and skin of marine fish (e.g., Budsberg et al. 2003; Ast and Dunlap 2005) (Fig. 13.5 ), and Photobacterium iliopiscarium, a nonluminous species closely related to P. phosphoreum and P. kishitanii, has been isolated from the intestines of several species of cold-water fish and from spoiled packaged fish (Ast and Dunlap 2005; Flodgaard et al. 2005; Onarheim et al. 1994; Urakawa et al. 1999). Saprophytic, commensal, parasitic, and symbiotic habitats have the potential to make substantial contributions to the density and distribution of luminous bacteria in seawater, sediments, and marine snow (Reichelt et al. 1977; O’Brien and Sizemore 1979; Ruby and Morin 1979; Haygood et al. 1984; Nealson et al. 1984; Ramesh et al. 1987; Ruby and Lee 1998; Visick and Ruby 2006), which in turn presumably serve as environmental sources of these bacteria for recolonization of animals. As commensal enteric symbionts of fish, luminous bacteria may contribute significantly to the digestion of crustacean prey through the activity of chitinase (Spencer 1961; Baumann and Schubert 1984; Ramesh and Venugopalan 1989). It should be noted that luminous bacteria coexist with and presumably carry out metabolic activities similar to nonluminous bacteria in these different habitats. Luminous bacteria in general, however, show little specificity when forming opportunistic saprophytic and enteric associations with marine animals such as mussels and clams. This lack of specificity can be attributed to the steady influx of bacteria from the water column, which presumably would prevent selection for specialization (Preheim et al. 2011). The exception to this general lack of specificity is bioluminescent symbiosis, in which the luminous bacteria able to colonize this kind of habitat typically are present as single species.
Saprophytic growth of luminous bacteria. Luminous bacteria have colonized this slice of fish meat, which was photographed in the dark by the light the bacteria produce. Growth of luminous bacteria in and on surfaces of animal tissues is common in nature. This attribute is one means by which luminous bacteria from the environment can be enriched for and isolated
In contrast to their associations with marine animals, luminous bacteria apparently do not commonly colonize the surfaces of marine algae. Agar digestion is often observed among nonluminous Vibrio species and other marine bacteria (e.g., Humm 1946), and various attempts, successful (Makemson et al. 1992) and otherwise, have been made to isolate light-emitting bacteria from algal surfaces. To date, however, only one luminous strain, provisionally identified as a member of V. harveyi, that has the ability to digest agar has been isolated from algae (Fukasawa et al. 1987). The uniqueness suggests that the single known isolate of agar-digesting luminous bacteria might have acquired either the genes for agar digestion or the lux genes by horizontal gene transfer.
The distributions and numbers of individual species of luminous bacteria tend to correlate with certain environmental factors (Baumann and Baumann 1981). Primary among these factors are temperature and depth (Ruby and Nealson 1978; Yetinson and Shilo 1979; Ruby et al. 1980; Ramaiah and Chandramohan 1987), salinity (Yetinson and Shilo 1979; Feldman and Buck 1984), and nutrient limitation and sensitivity to photooxidation (Shilo and Yetinson 1980; Makemson and Hastings 1982; Haygood and Nealson 1985a). Temperature, along with being an important environmental factor, can influence whether luminous bacteria from environmental samples are detected. For example, Shewanella hanedai, which is psychrotrophic, grows and produces light at low temperature (e.g., 4–15 °C) and grows but does not produce light at room temperature (24 °C). Therefore, incubation of platings of environmental samples at lower temperatures may reveal the presence of other luminous species with naturally temperature-sensitive luminescence systems. Temperature relationships would appear to be species-specific, however. For example, S. woodyi (found in squid ink and seawater in the Alboran Sea near Gibraltar; Makemson et al. 1997), a species closely related to S. hanedai, grows and produces light at room temperature.
Studies of the distribution and density of luminous bacteria in the marine environment traditionally have used visual detection of luminescent colonies arising from seawater spread on nutrient-containing agar plates to identify the presence of these bacteria. However, there are several kinds of luminous bacteria that can be missed with this method. One kind is bacteria that are physiologically cryptic for luminescence, producing visible light in culture only in response to the addition of inducers or other substances to the growth medium (Boettcher and Ruby 1990; Fidopiastis et al. 1999; Nelson et al. 2007) or that require growth at lower than typical room temperatures for light production. Another kind is bacteria with incomplete or defective lux operons (O’Grady and Wimpee 2008). Furthermore, enzyme assay and antibody methods have detected luciferase in several Vibrio spp. that do not produce visible light in culture (Nealson and Walton 1978; Makemson and Hastings 1986; Kou and Makemson 1988). Similarly, luxA-based DNA probes and PCR amplification of lux gene sequences have identified lux gene-containing bacteria from seawater that do not produce light in culture (Potrikus et al. 1984; Palmer and Colwell 1991; Lee and Ruby 1992; Wimpee et al. 1991; Ramaiah et al. 2000; Grim et al. 2008). These studies demonstrate that bacteria carrying the lux genes are more abundant in the marine environment and more phylogenetically diverse than is revealed by analysis of strains isolated on the basis of the production of readily visible levels of light. A counterpoint to this view, however, is the apparently low incidence of lux gene sequences in metagenomic databases (Martín Cuadrado et al. 2007; Nealson and Venter 2007), which suggests that luminous Photobacterium, Vibrio, and Aliivibrio, and presumably nonluminous members of these genera as well, represent a very small fraction of the microscopic plankton.
Freshwater
Luminous strains of V. cholerae can be isolated from freshwater and brackish estuarine waters (Desmarchelier and Reichelt 1981; West and Lee 1982; West et al. 1983; Palmer and Colwell 1991; Ramaiah et al. 2000; Table 13.1 ), as well as from coastal seawater (e.g., Urbanczyk et al. 2008). The first such strain, isolated in 1893 by F. Kutscher from the Elbe River in Germany (Harvey 1952), was named “Vibrio albensis” and later was synonymized with V. cholerae (Reichelt et al. 1976). This species also infects freshwater crustaceans; Thulis and Bernard in 1786 described the luminescence of a freshwater crustacean (possibly the common amphipod Gammarus pulex, which apparently was infected with luminous bacteria) from a river in southern France (Harvey 1957). Yasaki (1927) reported the isolation of luminous bacteria from strongly luminous specimens of the freshwater shrimp, Xiphocaridina compressa, in Lake Suwa, Japan. Initially characterized as Microspira phosphoreum, the bacterium was later redescribed as Vibrio yasakii (Majima 1931). A bacterium responsible for this “light disease of shrimp” was isolated more recently from freshwater shrimp in Lake Biwa, Japan, and identified as non-O1 V. cholerae (Shimada et al. 1995). In addition to V. cholerae in freshwater habitats, strains of P. phosphoreum have been isolated from migrating salmon in the Yukon River, Alaska (Budsberg et al. 2003; Ast and Dunlap 2005); presumably, their association with fish slime protected these marine bacteria from osmotic lysis.
Terrestrial
Luminous bacteria in the terrestrial environment have been noticed mostly as parasites of insects that cause the infected animal to luminesce. Observations of luminous midges, caterpillars, mole crickets, mayflies, and ants, among other infected insects, have been reported from the 1700s into modern times (Harvey 1952; Haneda 1950). As described and summarized by Harvey (Harvey 1952; Harvey 1957), other early reports of terrestrial luminescence attributable to luminous bacteria include luminous mutton, veal, eggs of chickens and lizards, human corpses, and battlefield wounds. Many, and perhaps all, of the observations of luminous insects result from colonization by members of the genus Photorhabdus, of which three species are currently described, Ph. luminescens, Ph. temperata, and Ph. asymbiotica (Fischer–Le Saux et al. 1999; Table 13.1 ). Two of the three Photorhabdus species occur as the mutualistic symbionts of soil nematodes of the family Heterorhabditidae (Table 13.1 ) (Akhurst and Dunphy 1993; Forst and Nealson 1996; Forst et al. 1997; Gerrard et al. 2006; Kuwata et al. 2008; Waterfield et al. 2009). They are carried in the intestine of the infective juvenile stage of the nematode and participate in a lethal infection of insect larvae. When the nematode enters the insect, via the digestive tract or other openings, and penetrates the insect’s hemocoel, the bacteria are released into the hemolymph, where they use its constituents for growth. The bacteria elaborate a variety of extracellular enzymes that presumably break down macromolecules of the hemolymph. Proliferation of the bacteria leads to death of the insect, and its carcass becomes luminous. The bacteria also produce various extracellular and cell surface-associated factors pathogenic for the insect, as well as bacteriocins and hydroxystilbene and anthraquinone antibiotics, which apparently inhibit the growth of other microorganisms in the insect cadaver and combat scavenging organisms, such as nematodes and amoeba (Akhurst 1982; Sicard et al. 2007; Waterfield et al. 2009). Crystalline protein inclusion bodies of unknown function are also produced (Bintrim and Ensign 1998). The nematodes feed on the bacteria or products of bacterial degradation of the hemolymph enabling them to develop and sexually reproduce (Boemare et al. 1997; Forst et al. 1997). Completion of the nematode life cycle involves reassociation with the bacteria and the emergence from the insect cadaver of the nonfeeding infective juveniles, carrying the bacteria in their intestines. Cells of Ph. luminescens presumably are present in soil, but association with the nematode apparently is important for their survival and dissemination. Luminescence of the infected insect larva might function to attract nocturnally active animals to feed on the glowing carcass, thereby increasing the opportunities for the bacterium and the nematode to be disseminated. However, luminescence in Ph. luminescens, which is stimulated in laboratory culture by exogenous aldehyde, is not required for successful symbiosis with the nematode; not all strains of Ph. luminescens produce luminescence (Akhurst and Boemare 1986; Forst and Nealson 1996; Schmidt et al. 1989). Furthermore, bacteria in the genus Xenorhabdus, which are symbiotic with entomopathogenic nematodes in the family Steinernematidae, are ecologically very similar to Photorhabdus, except that they do not produce light (Akhurst and Dunphy 1993). The similarities between the lifestyles and activities of Photorhabdus and Xenorhabdus are postulated to be a case of ecological convergence (Forst and Nealson 1996).
Human clinical infections have yielded P. asymbiotica, introduced apparently by spider and insect bites (Farmer et al. 1989; Peel et al. 1999). Luminous battlefield wounds are intriguing in this regard because luminescence apparently was a sign that the wound would heal well (Harvey 1957). Indeed, luminous bacteria will grow and produce light on living mammalian tissue (Johnson 1988). Perhaps antibiotic-producing, nonpathogenic Photorhabdus strains promoted wound healing by preventing the growth of putrefying, pathogenic bacteria. On the other hand, the human pathogenicity of P. asymbiotica suggests that this species might have killed rather than healed if introduced into wounds. The recent description of P. asymbiotica and P. temperata and the presence of genetically distinct subspecies within Ph. luminescens and P. temperata (Fischer Le Saux et al. 1999; Tailliez et al. 2010) indicate that additional diversity, possibly at the species level, may exist in this genus.
Along with terrestrial Photorhabdus species, marine luminous bacteria might have been responsible for some of the early reports of luminous meats and eggs, especially if brine was used in their preparation or they otherwise were exposed to seawater. Haneda (1950), following the observation by Molisch (1925) of luminous bacteria growing on beef, demonstrated that luminous bacteria could be isolated from certain samples of beef, pork, and chicken meat. These meats might have contained enough salt to support the growth of marine species, and Haneda cultured the bacteria in media containing 0.5 % salt. However, whether these bacteria were terrestrial (i.e., Photorhabdus), from brackish water (i.e., V. cholerae), or marine in origin is not known.
Parasitism of Marine Invertebrates
Most of the commonly encountered marine luminous bacteria are not known to be highly invasive or virulent in animals. Many or perhaps all luminous species, however, can act as opportunistic pathogens upon entering an animal’s body through lesions resulting from injury or stress. First noted in marine animals apparently by Viviani in 1805 (Harvey 1957), infections of marine crustaceans by luminous bacteria are common, causing the infected animal to luminesce (Giard 1889; Giard and Billet 1889; Inman 1926). Luminous bacteria inhabit the gut tract and colonize external surfaces of marine crustaceans (Inman 1926; Baross et al. 1978; O’Brien and Sizemore 1979; Lavilla–Pitogo et al. 1992); many are chitinolytic (Spencer 1961; Baumann and Schubert 1984). The bacteria enter the hemocoel of the animal through lesions in the gut or carapace, developing luminescence and killing the animal within a few days. The species of luminous bacteria infecting isopods and amphipods commonly encountered in coastal environments have not been identified in recent times, but they exhibit characters consistent with members of the genera Aliivibrio, Photobacterium, and Vibrio (Hastings and Nealson 1981; P. Dunlap, unpubl. data). Nonluminous bacteria undoubtedly cause similar infections that go unnoticed due to the lack of light production.
As opportunistic pathogens of marine crustaceans, luminous bacteria and their nonluminous relatives have had a profoundly deleterious effect on commercial prawn mariculture (Owens and Busico–Salcedo 2006; Haldar et al. 2011). The development of intensive monoculture of Penaeus monodon, the giant tiger prawn, and other penaeids during the 1980s led to a dramatic increase in disease and death of the animals due to luminous bacteria. Shrimp hatchery rearing ponds can become heavily infested with luminous bacteria, with shrimp larvae developing “luminescent vibriosis,” a pathogenic state responsible for massive mortalities. The problem continues in grow-out ponds, where the infection localizes to the hepatopancreas in juveniles, limiting the growth of the animals and further increasing losses to mortality (Lavilla–Pitogo and de la Peña 1998). Primarily responsible are strains of V. harveyi, though other luminous and nonluminous Vibrio species have been identified (Lavilla–Pitogo et al. 1990; Karunasagar et al. 1994; Lavilla–Pitogo and de la Peña 1998; Suwanto et al. 1998; Leano et al. 1998; Austin and Zhang 2006).
Parasitism of Vertebrates
In contrast to the situation with marine invertebrates, luminous bacteria apparently only rarely infect vertebrate animals. The ability of P. asymbiotica to infect humans has been mentioned above. Vibrio harveyi has been identified in fish disease, and recently, A. salmonicida (a pathogen of salmonids and cod) has been shown to contain a lux operon (Nelson et al. 2007). Clinical strains of V. vulnificus and V. cholerae typically are nonluminous, but a luminous strain of V. vulnificus has been isolated from a lethal human infection (Oliver et al. 1986; Kaeding et al. 2007), and luminous strains of V. cholerae have been isolated from humans suffering from cholera (Jermoljewa 1926). Furthermore, Weleminsky (1895) demonstrated that a nonluminous clinical isolate of V. cholerae developed luminescence apparently by passage through pigeon’s blood (Harvey 1952). Vibrio cholerae strains that are luminous or that contain the luxA gene are present in relatively high percentages in freshwater and estuarine environments (West and Lee 1982; West et al. 1983; Palmer and Colwell 1991; Ramaiah et al. 2000). However, O1 or O139 serotypes of V. cholerae, which are responsible for life-threatening cases of human diarrheal disease, do not include the light-producing or luxA gene–containing strains (Palmer and Colwell 1991; Ramaiah et al. 2000; Grim et al. 2008).
Bioluminescent Symbiosis
A special attribute of a few of the luminous bacteria is the ability to form highly specific, luminescence-based mutualisms, called bioluminescent symbiosis, with certain marine fish and squids (Table 13.2 ). Early work is reviewed in detail by Harvey (1952), Buchner (1965), Herring and Morin (1978), and Hastings and Nealson (1981). In these associations, the animal cultures a dense population of luminous bacteria in a tissue complex called a light organ, providing them with nutrients and oxygen for reproduction and light production. The animal in turn uses the bacterial light for luminescence displays associated with sex-specific signaling, predator avoidance, seeing and attracting prey, or schooling. In most of the bacterially bioluminescent fish, the light organs are associated with the gastrointestinal tract; in others, they are subocular (anomalopids), mandibular (monocentrids), or escal (ceratioids). In squids, the bacterial light organs are found as bilobed structures within the mantle cavity, associated with the ink sac. Accessory tissues associated with the light organ, that is, shutter, lens, and reflector, direct and focus the light the bacteria produce. The light organs open to the external environment, either directly or via the intestinal tract or mantle cavity, allowing the excess bacterial cells to be released from the animal’s light organ into the environment as the light-organ population reproduces. In the cases studied, the members of each new host generation of the animal acquire their symbiotic bacteria from the environment. These associations typically are highly specific at the animal family–bacterial species level; members of a family of fish or squid often all harbor the same individual bacterial species as their symbiont (Harvey 1922, 1952; Okada 1926; Harms 1928; Kishitani 1930; Yasaki 1928; Haneda 1938, 1950; Ahrens 1965; Buchner 1965; Hastings 1971; Morin et al. 1975; Herring 1977; Herring and Morin 1978; Nealson 1979; McFall–Ngai 1983; McFall–Ngai and Dunlap 1983; Haygood et al. 1984; Nealson et al. 1984; Dunlap and McFall–Ngai 1987; Wei and Young 1989; McFall Ngai and Morin 1991; McFall Ngai and Ruby 1991; Ruby and Asato 1993; Graf and Ruby 1998; Wada et al. 1999; Woodland et al. 2002; Sasaki et al. 2003; Jones and Nishiguchi 2004; Sparks et al. 2005; Dunlap et al. 2009; Charkrabarty et al. 2011; Dunlap and Nakamura 2011). The bacteria are housed extracellularly, and in most cases they are known to not be obligately dependent on the host for their reproduction, as they colonize a variety of other habitats (Baumann and Baumann 1981; Hastings and Nealson 1981; Visick and Ruby 2006). Bioluminescent symbiosis appears to be a unique kind of symbiosis; the bacterial metabolic product needed by the host animal is light, used in bioluminescence displays, rather than a bacterially produced nutrient or enzymatic activity needed for host nutrition (Claes and Dunlap 2000).
Luminous bacteria might also form symbioses with pyrosomes and salps; little is known, however, and the topic remains controversial (Harvey 1952; Buchner 1965). Pyrosome zooids bear a pair of simple photophores that contain intracellular bacteroids, but the involvement of the bacteroids in pyrosome luminescence has been both discounted and supported (Galt 1978; Herring 1978; Mackie and Bone 1978; Haygood 1993). Although the bacteroids have not been cultured, the presence of bacterial luciferase in photophores is consistent with a bacterial origin for pyrosome luminescence (Leisman et al. 1980). A similar proposal for luminous myctophid and stomiiform fish, that the luminescence of the fish’s photophores is produced by symbiotic luminous bacteria (Foran 1991), however, has been conclusively refuted (Haygood et al. 1994).
The following information focuses primarily, although not exclusively, on bioluminescent symbiosis in fish. Detailed information on the bioluminescent mutualism of A. fischeri with the sepiolid squid Euprymna scolopes can be found in the chapter by K. Visick (Chap. 20, “Vibrio fisheri: Squid Symbiosis,” Vol. 1).
Patterns of Host Affiliation
Six species of luminous bacteria form bioluminescent symbioses with fish and squids, A. fischeri, A. “thorii,” A. wodanis, P. kishitanii, P. leiognathi, and P. mandapamensis. Their currently known host affiliations are listed in Table 13.2 . There are over 460 species of bacterially luminous marine fish, in 21 families of seven teleost orders, and several species of squid in two families of two cephalopod orders (Dunlap et al. 2007; Herring and Morin 1978; Nelson 2006) (Fig. 13.6 ). The most numerous of these symbiotic bacteria, due to the exceptional abundance of their host animals in the marine environment, are likely to be P. kishitanii and P. leiognathi. The hosts of P. kishitanii are fish of diverse families in deep-sea habitats worldwide, many of which are abundant, and the hosts of P. leiognathi are primarily fish of the family Leiognathidae, which are abundant in shallow coastal waters of Southeast Asia and South Asia (Tiews and Caces Borja 1965; Kühlmorgen–Hille 1974; Herring and Morin 1978; Cohen et al. 1990; Orlov and Iwamoto 2006; Dunlap et al. 2007; Dunlap et al. 2009). The bioluminescent symbionts of deep-sea fish previously were thought to be P. phosphoreum, but detailed phylogenetic analyses of the phosphoreum species group identified P. kishitanii as the species occurring in light organs of deep-sea fish. Despite extensive testing, no bonafide member of P. phosphoreum has been found in light-organ symbiosis (Ast and Dunlap 2005; Ast et al. 2009; Dunlap and Ast 2005; Dunlap et al. 2007; Kaeding et al. 2007; Urbanczyk et al. 2007). Similarly, strains of bacteria from light organs of the sepiolid squids Sepiola affinis and Sepiola robusta, previously identified as A. logei (V. logei) (Fidopiastis et al. 1998), were recently identified based on detailed phylogenetic criteria as three entities, A. fischeri; A. “thorii”, a newly recognized bacterial clade; and A. wodanis, a previously described species newly recognized as a bioluminescent symbiont; apparently no bonafide member of A. logei has been found in light-organ symbiosis (Ast et al. 2009). In addition to these bacteria, strains identified as V. harveyi have been found in the light organ of larval leiognathid fish (Dunlap et al. 2008); it is not yet known if V. harveyi is present as an incidental, transient colonizer of the nascent light organ of this fish, as a pathogen, or, possibly, as an actual symbiont (Dunlap et al. 2008).
Bacterially luminous fish. Shown are a few of the more than 460 species of fish that host luminous bacteria as bioluminescent symbionts. Counterclockwise from the top, the fish are Physiculus japonicus (Gadiformes: Moridae) (photo by A. Fukui), host of P. kishitanii and, less commonly, A. fischeri; Eubleekeria jonesi (Perciformes: Leiognathidae) (photo by P.V. Dunlap), host of P. leiognathi; Acropoma japonicus (Perciformes: Acropomatidae) (photo by A. Fukui), host of P. mandapamensis and, less commonly, P. leiognathi; Chlorophthalmus nigromarginatus (Aulopiformes: Chlorophthalmidae), host of P. kishitanii; Monocentris japonicus (Beryciformes: Monocentridae) (photo by P.V. Dunlap), host of A. fischeri; and Aulotrachichthys prosthemius (Beryciformes: Trachichthyidae) (photo by A. Fukui), host of P. kishitanii
Species Specificity, Cosymbiosis, and Symbiont: Host Codivergence
Previously, bioluminescent symbioses were characterized as species specific, with the light organ of each animal thought to harbor a single, pure culture of bacteria and with the members of each family of fish or squids thought to all harbor the same bacterial species as their symbiont (Hastings and Nealson 1981; Nealson and Hastings 1992; Dunlap and Kita–Tsukamoto 2006). This pattern of specificity still generally holds, but several deviations from a strict host family–bacterial species specificity have been identified. On the one hand, individual light organs of certain squid and fish have been found to harbor two bacterial species, a situation termed cosymbiosis. In contradicting a strict one-to-one relationship, cosymbiosis requires the mechanism by which the host might select its symbiotic bacteria, such as surface recognition, to respond to features common to both bacterial species or to distinct features of each (Ast et al. 2009; Fidopiastis et al. 1998; Dunlap et al. 2007; Dunlap et al. 2008; Kaeding et al. 2007; Dunlap et al., unpubl. data). On the other hand, different host species and genera within a family have been found to harbor different species of bacteria (Table 13.2 ). One example of this breakdown of host family level bacterial specificity is the presence in light organs of Acropoma hanedai of P. kishitanii, whereas P. mandapamensis is present as the primary symbiont in light organs of Acropoma japonicum. Another is the presence in different species of Coelorinchus of P. kishitanii or A. fischeri (Dunlap et al. 2007; Kaeding et al. 2007; Wada et al. 2006). These discrepancies suggest that a strict genetically based host selection of a specific symbiotic bacterium (McFall Ngai and Morin 1991) may not be operative in bioluminescent symbiosis or may not be operative for all bacterially luminous animals.
Consistent with this possibility is the lack of codivergence, that is, cospeciation, between host and symbiont lineages (Fig. 13.7 ). Genetic selection might reasonably lead to a codivergence, as reported for squid-symbiotic bacteria (Nishiguchi et al. 1998), but instead of congruent host and symbiont phylogenies, detailed phylogenetic analysis based on homologous genes, suitable numbers of strains, and a diversity of hosts reveals instead that the patterns of symbiont affiliations for fish and squids are strikingly noncongruent (Dunlap et al. 2007). Furthermore, phylogenetically distantly related hosts, for example, bacterially luminous aulopiforms, most gadiforms, and certain beryciforms, all harbor the same bacterial species, P. kishitanii, whereas some closely related hosts, such as the acropomatid fish A. hanedai and A. japonicum, as noted above, harbor distinct species, P. kishitanii and P. mandapamensis, respectively (Dunlap et al. 2007) (Fig. 13.7 ).
Host affiliations of symbiotic luminous bacteria. Families of bacterially luminous squids and fish are listed on the left, with lines to the corresponding bacterial species on the right that have been isolated from light organs of these animals. Different members of individual families of fish and some squids often harbor different species of bacteria, in some cases within the light organ of the same host specimen. Some of the bacteria, for example, A. fischeri, P. kishitanii, P. mandapamensis, are found in light organs of a diversity of fish and squids. These attributes highlight the lack of strict family level bacterial species specificity and the lack of phylogenetic congruence between host and symbiont in bioluminescent symbiosis (Dunlap et al. 2007; Kaeding et al. 2007). The question mark for the link from Leiognathidae to V. harveyi reflects the single instance that this bacterial species has been isolated from light organ symbiosis (Dunlap et al. 2008)
An alternative hypothesis to account for the observed patterns of symbiont–host affiliation in bioluminescent symbiosis is environmental congruence. This hypothesis, first outlined by Hastings and Nealson (1981), links the differing environmental distributions of different species of luminous bacteria, that is, where each species is most abundant, with the environmental distribution of its host animal (Dunlap et al. 2007; Hastings and Nealson 1981; Kaeding et al. 2007; Dunlap et al. 2008). Temperature, which influences the presence and relative numbers of the different species of luminous bacteria in the marine environment, may be the key environmental factor; deeper, colder dwelling hosts harbor the more psychrotrophic luminous species found in those habitats, P. kishitanii, for example, as their bioluminescent symbiont, whereas shallower and warmer dwelling hosts harbor the more mesophilic luminous species found in those habitats, A. fischeri and P. leiognathi, for example. An important further consideration is the ontogenetic ecology of the host. Early life history stages of these animals, for example, eggs, larvae, and juveniles, often are distributed in the environment differently from adults. The key factor therefore may be where in the environment the animal is when it is developmentally ready to initiate symbiosis. The luminous bacterial species most abundant in and adapted to the conditions of those habitats presumably would the ones most likely to initiate symbiosis (Dunlap et al. 2007; Kaeding et al. 2007). Information about early life history stages of bacterially luminous animals, especially fish, is very limited, but evidence is beginning to accumulate that supports the environmental congruence hypothesis (Dunlap et al. 2008). Nonetheless, some form of host selection must be occurring because to date only luminous bacteria, and only certain species of luminous bacteria, have been found in light organs of fish and squid. Most likely, a combination of environmental congruence and some level of selection are operative.
The luminous bacteria symbiotic with two other groups of fish, the flashlight fish, family Anomalopidae, and bacterially luminous deep-sea anglerfish, in order Lophiiformes, present a possible contrast to the apparent lack of strict species specificity in bioluminescent symbiosis. Microscopic analysis showing the presence of masses of bacterial cells within the light organs, assays specific for bacterial luciferase, and other studies convincingly demonstrate the bacterial nature of light emission in these fish (Bassot 1966; Harvey 1922; Haygood et al. 1984; Kessel 1977; Leisman et al. 1980; Munk et al. 1998), but the bacteria from light organs of these fish have not been grown in laboratory culture despite numerous attempts (Hastings and Nealson 1981; Haygood 1993; Hendry and Dunlap 2011; Herring and Morin 1978). The inability to grow these bacteria in the laboratory suggests that they have lost the ability to reproduce outside the host light organ and are therefore might be obligately dependent on their hosts (Haygood 1993; Haygood and Distel 1993). An obligate relationship presumably would lead to a very high degree of specificity and possibly also to codivergence between the host fish and its bacteria. 16S rRNA gene sequence analysis of the symbiotic bacteria of two ceratioids, representing different families of lophiiformes, places these bacteria as members of Vibrionaceae and possibly as a new bacterial species in each fish (Haygood and Distel 1993; Hendry and Dunlap 2011). Analysis of the luxA and 16S rRNA genes of anomalopid symbionts suggested these bacteria are members of Vibrio and that different genera of the fish harbor bacteria that differ at greater than the strain level (Haygood 1990; Wolfe and Haygood 1991). Consistent with and extending these findings, a recent detailed multilocus analysis classified the bacteria symbiotic with the anomalopid fish Anomalops katoptron as a new Vibrionaceae genus and species, Candidatus Photodesmus katoptron, which is closely related to Vibrio (Hendry and Dunlap 2011).
Symbiont Acquisition
In the few cases studied, bacterially luminous squid and fish have been found to acquire their symbiotic luminous bacteria from the environment with each new host generation. The sepiolid squid E. scolopes acquires cells of A. fischeri from seawater shortly after hatching from the egg as aposymbiotic juveniles; bacteria other than the native symbiont fail to colonize the light organ or do so less effectively (Wei and Young 1989; McFall Ngai and Ruby 1991). In fish, the symbiotic bacteria are acquired later, following development of larvae and inception of light organ formation (Wada et al. 1999; Dunlap et al. 2008; Dunlap et al. 2009; Dunlap et al. 2012) (Fig. 13.8 ), which is consistent with acquisition of the bacteria from the environment. Also consistent with environmental acquisition, symbiotic bacteria apparently are not present on eggs within the ovary of anomalopid fish (Haygood 1993).
Developing light organ. This electron micrograph of a section of the light organ of the fish Nuchequula nuchalis (Leiognathidae) (micrograph prepared by Sasha Meschinchi, Microscopy and Imaging Laboratory, University of Michigan) shows tubules of the nascent light organ. Some tubules are empty, whereas some are filled or becoming filled with bacteria
Quorum Sensing Control (Autoinduction) of Bacterial Luminescence
In many luminous bacteria, luciferase synthesis and luminescence are regulated in a population density-responsive manner. At low population density, very little luciferase is synthesized, and consequently, little light is produced, whereas at high population density, luciferase levels are induced 100–1,000-fold and light levels increase by 103–106-fold. Population density–responsive induction of luciferase synthesis and luminescence is controlled in part by the production and accumulation in the cell’s local environment of small signal molecules, called autoinducers (acyl-homoserine lactones, AHLs, and other low molecular weight compounds), which function via regulatory proteins to activate transcription of the lux operon. Originally called autoinduction and discovered through study of the pattern of luminescence and luciferase production of V. harveyi in batch culture (Nealson et al. 1970), this gene regulatory mechanism is now referred to as quorum sensing to reflect its relationship with population density (Fuqua et al. 1994; Greenberg 1997; Hastings and Greenberg 1999; Miller and Bassler 2001).
Over the past 30 years, there has been a very substantial accumulation of information on how two luminous bacteria, V. harveyi (a key strain recently was reclassified as V. campbellii; Lin et al. 2010) and A. fischeri, regulate luminescence by quorum sensing. Studies in these two bacteria established a base of knowledge that led to the discovery of biochemically and genetically homologous quorum sensing systems in a wide variety of nonluminous bacteria; quorum sensing controls many cellular activities other than light production, particularly the production of extracellular enzymes and other extracellular factors thought to be adaptive for bacteria at high population density and in association with animal and plant hosts (Fuqua et al. 1996; Dunlap 1997; Bassler, et al. 1997; Swift et al. 1999; Callahan and Dunlap 2000; Waters and Bassler 2005; Dunlap and Kita–Tuskamoto 2006; Higgins et al. 2007). Quorum sensing therefore is not only not unique or even special to luminous bacteria, it is also widespread and evolutionarily conserved across a diversity of bacteria. As a signal–response mechanism by which bacteria can assess their local population density, quorum sensing might have arisen evolutionarily as a diffusion sensor or efficiency sensor (Redfield 2002; Hense et al. 2007), mediating whether or not cells produce extracellular enzymes and other factors for obtaining nutrients. Substantial attention has been placed recently on the definition and correct usage of terms for quorum sensing and other chemically mediated bacterial interactions (Platt and Fuqua 2010; Stacy et al. 2012). This form of genetic regulation has been studied in detail in A. fischeri and V. harveyi but remains poorly understood in other luminous bacteria (Meighen 1999). It should be noted also that some luminous bacteria express luminescence constitutively during growth in batch culture (Katznelson and Ulitzur 1977; P. Dunlap, unpubl. data) and therefore apparently do not use a quorum sensing system to control luminescence. An overview of quorum sensing in V. harveyi and A. fischeri is provided here, and more information on this topic is provided in the chapters by B. Bassler (Chap. 22, “Quorum Sensing,” Vol. 2) and K. Visick (Chap. 20, “Vibrio fisheri: Squid Symbiosis,” Vol. 1).
Early Studies of Quorum Sensing Control of Luminescence. In A. fischeri and V. harveyi, expression of the lux operon, which initially is low in early exponential phase cultures, induces strongly as cultures attain the high cell densities associated with late exponential to early stationary phases of growth (Hastings and Greenberg 1999). Early analyses of the “phases of luminescence” in culture (e.g., Baylor 1949; Farghaly 1950) were followed by the demonstration that luciferase synthesis is inducible and that complete medium contained a compound inhibitory to induction (Nealson et al. 1970; Eberhard 1972). During growth, cells of A. fischeri and V. harveyi were found also to release into the medium species-specific positively acting secondary metabolites, called autoinducers. These compounds accumulate in the growth medium in a population density-dependent manner, and once they attain threshold concentrations, they induce luciferase synthesis (Nealson et al. 1970; Eberhard 1972; Nealson 1977; Nealson and Hastings 1979; Ulitzur and Hastings 1979; Rosson and Nealson 1981).
Analysis of quorum sensing attained benchmarks of progress in the 1980s with the identification of autoinducer signal molecules and lux regulatory genes. The first autoinducer, 3-oxo-hexanoyl-HSL (3-oxo-C6-HSL), and the first lux regulatory genes, luxI (encoding 3-oxo-C6-HSL synthase; Schaefer et al. 1996) and luxR Af (encoding acyl-HSL receptor/transcriptional activator), were identified in A. fischeri (Eberhard et al. 1981; Engebrecht et al. 1983; Engebrecht and Silverman 1984), followed by identification of 3-hydroxybutanoyl-HSL and a nonhomologous regulatory gene, luxR Vh, in V. harveyi (Cao and Meighen 1989; Martin et al. 1989; Showalter et al. 1990).
Quorum Sensing Regulatory Circuitry. Ongoing progress since the 1980s has substantially deepened understanding of the quorum sensing genetic regulatory circuitry controlling luminescence in V. harveyi and A. fischeri. The two regulatory systems are strikingly different. In V. campbellii (previously classified as V. harveyi; Lin et al. 2010), three chemically distinct autoinducers are produced, 3-hydroxybutanoyl-HSL (harveyi autoinducer-1, HAI-1), (2 S,4 S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate (V. harveyi autoinducer-2, AI-2Vh), and (S)-3-hydroxytridecan-4-one (cholerae autoinducer, CAI-1) (Cao and Meighen 1989; Cao and Meighen 1993; Chen et al. 2002; Higgins et al. 2007) (Fig. 13.9 ). Synthesis of HAI-1 is dependent on LuxM (Bassler et al. 1993), synthesis of AI-2Vh is catalyzed by LuxS (Schauder et al. 2001), and synthesis of CAI-1 is catalyzed by CqsA (Kelly et al. 2009; Wei et al. 2011). Each of these molecules is specifically recognized by a different cytoplasmic membrane-associated two-component histidine kinase receptor, LuxN (Bassler et al. 1993; Freeman et al. 2000), LuxPQ (Bassler et al. 1994b), and CqsS (Henke and Bassler 2004), respectively (Fig. 13.9 ). When concentrations of the autoinducers are low, such as at low population density or in habitats in which the autoinducers diffuse away rapidly from cells, that is, seawater, the receptor proteins function as kinases, transferring phosphate to LuxU, a histidine-phosphotransfer protein. LuxU then transfers the phosphate to LuxO, a DNA binding response regulator, the expression of which is subject to repression by LuxT (Bassler 1999; Bassler et al. 1994a; Cao et al. 1995; Freeman and Bassler 1999a, b; Surete et al. 1999; Lilley and Bassler 2000; Lin et al. 2000; Miyamoto et al. 2003; Waters and Bassler 2006). LuxO ∼ P, together with sigma factor σ54, then activates expression of genes coding for five small quorum regulatory RNAs (Qrr), Qrr1 through Qrr5 (Lenz et al. 2004; Tu and Bassler 2007). The Qrr RNAs bind and destabilize the luxR Vh transcript, blocking production of LuxRVh protein, the transcriptional activator of the lux operon (Showalter et al. 1990; Swartzman et al. 1992), and thereby preventing activation of lux operon transcription. Conversely, once autoinducer concentrations have attained high levels in the cell’s local environment, they bind to their receptors, causing the receptors to switch from kinases to phosphatases, leading to the dephosphorylation of LuxO. With the resulting cessation of qrr gene transcription, luxR Vh message is produced and translated, and LuxRVh activates lux operon transcription. Negative autoregulation of LuxRVh adds additional complexity to this system (Chatterjee et al. 1996; Miyamoto et al. 1996), as does the negative autoregulation of LuxO and posttranscriptional control of LuxO by Qrr sRNAs (Tu et al. 2010; Fig. 13.9 ). The complexity of this regulatory system apparently benefits V. campbellii by allowing a fine tuning of its quorum sensing response to differences in the various habitats the bacterium colonizes (Waters and Bassler 2005; Ng and Bassler 2009; Tu et al. 2010).
Regulatory circuitry controlling luminescence in V. campbellii. The expression of lux operon, and of other quorum sensing-regulated genes, in V. campbellii (previously classified as V. harveyi), is coordinated by three chemically distinct autoinducers, HAI-1, AI-2Vh, and CAI-1, that modulate the phosphorylation state of luxU. The synthesis of each autoinducer is catalyzed by a different protein, LuxM, LuxS, and CqsA, and each is recognized by a different cytoplasmic membrane-associated two-component histidine kinase receptor, LuxN, LuxPQ, and CqsS, respectively. Low concentrations of the autoinducers lead to phosphorylation of LuxO and via quorum regulatory RNAs to the destabilization the luxR Vh transcript, thereby blocking lux operon transcriptional activation by LuxRVh. High concentrations of the autoinducers reverse the phosphorylation cascade, allowing formation of LuxRVh and activation of lux operon transcription. Arrows indicate positive interactions or transcriptional activation, whereas bars indicate blocking of transcription. See the text for details and references (Redrawn from Tu et al. (2010))
Quorum sensing control of luminescence in A. fischeri involves a very different regulatory system. A population density-dependent accumulation of the autoinducer 3-oxo-hexanoyl-homoserine lactone (3-oxo-C6-HSL), a membrane-permeant molecule, triggers induction when it reaches a critical concentration (Fig. 13.10 ). Synthesis of 3-oxo-C6-HSL is catalyzed by LuxI, an acyl-homoserine lactone synthase. The regulatory genes, luxR Af and luxI, are directly linked to the lux operon (Figs. 13.2 , 13.10 ). The luxR Af gene, which is upstream of the lux operon and divergently transcribed from it, encodes a transcriptional activator protein, LuxRAf, which associates with 3-oxo-C6-HSL, forming a complex that binds at a site in the lux operon promoter and that facilitates the binding of RNA polymerase, thereby activating transcription of the genes for light production, luxICDABEG. Because luxI is a gene of the lux operon, increased transcription leads to increased synthesis of 3-oxo-C6-HSL in an autocatalytic, positive feedback manner. The result is a rapid and strong induction of luciferase synthesis and luminescence (Engebrecht et al. 1983; Engebrecht and Silverman 1984; Kaplan and Greenberg 1985; Eberhard et al. 1991; Schaefer et al. 1996; Stevens and Greenberg 1997).
Regulatory circuitry controlling luminescence in A. fischeri. The expression of the lux operon, and of other quorum sensing-regulated genes, in A. fischeri is mediated primarily by the concentration of AI-1, which forms a complex with LuxRAf. Synthesis of AI-1 is dependent on LuxI, and the AI-1/LuxRAf complex activates luxICDABEG transcription. Together with cAMP, the CRP protein activates expression from the luxR Af promoter, increasing synthesis of LuxRAf and potentiating the system to induce strongly once sufficient AI-1 has accumulated. Increased expression from the lux operon promoter leads to a stimulation of AI-1 synthesis in an autocatalytic, positive feedback manner, resulting in a rapid and strong induction of luciferase synthesis. A second autoinducer, AI-2Af, interacts with LuxRAf, interfering with the interaction between AI-1 and LuxRAf. The hypothesized AI-2Af/LuxRAf complex is thought to be transcriptionally less effective and functions to delay the onset of AI1/LuxRAf activation of luxICDABEG transcription. See the text for details and references. Figure provided by K. Dougan, University of Michigan
Other regulatory factors modulate quorum sensing in A. fischeri. GroEL is necessary for production of active LuxRAf (Adar et al. 1992; Adar and Ulitzur 1993; Dolan and Greenberg 1992), and 3’ 5’-cyclic AMP (cAMP) and cAMP receptor protein (CRP) activate transcription of luxR and thereby potentiate the cell’s response to 3-oxo-C6-HSL (Dunlap and Greenberg 1985; 1988; Dunlap 1989; Dunlap and Kuo 1992). LuxRAf/3-oxo-C6-HSL also negatively autoregulates luxR Af expression (Dunlap and Greenberg 1988; Dunlap and Ray 1989), and a second autoinducer, octanoyl-HSL, synthesis of which is catalyzed by AinS, interacts with LuxRAf apparently to delay lux operon induction (Eberhard et al. 1986; Kuo et al. 1996; Hanzelka et al. 1999) (Fig. 13.10 ). Under anaerobic conditions, Fnr contributes to lux operon expression (Müller Breitkreutz and Winkler 1993). A homolog of the V. harveyi luxO gene is carried by A. fischeri, and as with V. harveyi, LuxO in A. fischeri functions as a repressor of luminescence (Miyamoto et al. 2000, 2003), apparently in a qrr-dependent manner (Miyashiro et al. 2010). In addition, LitR, which has substantial sequence similarity to LuxRAf, positively regulates lux operon expression (Fidopiastis et al. 2002), and LexA is thought to contribute to control of luminescence (Shadel et al. 1990; Ulitzur and Dunlap 1995).
Despite the many differences in the quorum sensing regulatory systems of V. harveyi and A. fischeri, there are some commonalities. The C-terminal half of the A. fischeri AinS protein is 34 % identical to the V. harveyi LuxM protein, and the N-terminal portion of A. fischeri AinR, encoded by ainR, a gene downstream of ainS, is 38 % identical to the amino terminus of V. harveyi LuxN (Gilson et al. 1995). Whether AinR itself, possibly through interaction with C8-HSL, plays a role in lux regulation in A. fischeri (Gilson et al. 1995; Kuo et al. 1994) has not been established. The deduced amino acid residue sequence of A. fischeri LuxO is approximately 70 % identical to that of V. harveyi (Miyamoto et al. 2000), and a gene immediately downstream of luxO in A. fischeri is likely to be a homolog of V. harveyi luxU. Whether these homologies indicate overlaps in quorum sensing control, however, remains to be determined.
Another commonality between the two systems is expression of the lux operons of both species is dependent on cyclic AMP (cAMP) and cAMP receptor protein (CRP) (Ulitzur and Yashphe 1975; Chen et al. 1985; Dunlap and Greenberg 1985; 1988; Dunlap 1989; Dunlap and Kuo 1992). Consistent with this dependence, the regulatory regions upstream of the lux operons of both species contain a CRP binding site (Engebrecht and Silverman 1987; Devine et al. 1988; Miyamoto et al. 1988). Mutants of V. harveyi and A. fischeri apparently defective in adenylate cyclase and unable to produce light in the absence of added cAMP have been isolated and characterized (Ulitzur and Yashphe 1975; Dunlap 1989). Furthermore, CRP from V. harveyi has been purified and shown to be immunologically and functionally homologous to CRP of Escherichia coli (Chen et al. 1985), and the cya and crp genes of A. fischeri have been cloned and found to be highly similar in deduced amino acid residue sequence to E. coli cya and crp genes (P. Dunlap et al., unpubl. data). In V. harveyi, cAMP-dependent binding of CRP activates lux operon expression as well as expression of luxR Vh (Chatterjee et al. 2002). Studies with A. fischeri and with E. coli carrying the A. fischeri luxR–luxICDABEG genes indicate that a major effect of cAMP–CRP is to activate expression of LuxRAf while repressing transcription from the lux operon promoter (Dunlap and Greenberg 1985; 1988; Dunlap and Kuo 1992; Shadel et al. 1990), although other important lux regulatory effects have also been described (Shadel and Baldwin 1991; 1992a, b). Control by cAMP–CRP suggests that the luminescence systems of these bacteria might be part of the cellular response to stresses associated with nutrient limitation and decreasing growth rate.
Physiological Control of Luminescence. The presence of glucose can suppress bacterial luminescence; this catabolite repression effect presumably operates by modulating the levels of cAMP and CRP in the cell (Nealson et al. 1972; Meighen and Dunlap 1993; Dunlap 1997). In V. harveyi, catabolite repression by glucose in batch culture is permanent and is reversed by addition of cAMP (Nealson et al. 1972), whereas glucose repression of luminescence in A. fischeri is temporary, not reversed by addition of cAMP, and is eliminated by prior growth in the presence of glucose (Ruby and Nealson 1976). Complicating these differences from studies in batch culture are studies of A. fischeri grown in phosphate-limited chemostat culture; glucose repression of luminescence under these conditions is permanent, and it is reversible by cAMP (Friedrich and Greenberg 1983). A further complication for studies of cAMP control of luminescence in A. fischeri is the presence in this species of a novel, exceptionally potent periplasmic cyclic nucleotide phosphodiesterase specific for extracellular 3' 5'-cyclic nucleotides; activity of the enzyme enables cells to grow on exogenously supplied cAMP as a sole source of carbon and energy, nitrogen, and phosphorus (Dunlap et al. 1992; Dunlap and Callahan 1993; Callahan et al. 1995).
In addition to glucose, other physiological factors can strongly influence the amount of light produced by luminous bacteria grown in laboratory culture. Oxygen, amino acids, iron, and osmolarity have distinct effects, depending on the species studied (Harvey 1952; Coffey 1967; Nealson et al. 1970; Hastings and Nealson 1977; Makemson and Hastings 1982; Dunlap 1985; Haygood and Nealson 1985a; Hastings et al. 1987; Guerrero and Makemson 1989; Dunlap 1991). Those factors that stimulate growth rate, such as readily metabolized carbohydrates, tend to decrease light production and luciferase synthesis. They do so presumably by directing the consumption of oxygen and reducing power (FMNH2) away from luciferase (e.g., McElroy and Seliger 1962; Coffey 1967) and by indirectly or directly influencing lux gene expression (Dunlap and Greenberg 1985; Dunlap 2000). Conversely, factors that restrict growth rate, such as limitation for iron and low or high osmolarity of the growth medium, depending on the species, tend to stimulate the synthesis and activity of luciferase (Hastings and Nealson 1977; Makemson and Hastings 1982; Haygood and Nealson 1985; Hastings et al. 1987; Dunlap 1991). The mechanisms by which these factors operate, however, are not well understood, although they presumably interface with the quorum sensing control circuitry in some way.
Other Genes Subject to Quorum Sensing Regulation. Studies of V. harveyi led to the first demonstration of non-lux quorum-sensing-regulated genes. In V. harveyi, the production of the fatty acid storage product poly-β-hydroxybutyrate is controlled in a cell density-dependent manner by 3-OH-C4-HSL (Sun et al. 1994; Miyamoto et al. 1998). Along with that finding, LuxRVh has been shown to control various non-lux genes and to act as either a transcriptional activator or repressor (Chatterjee et al. 1996; Miyamoto et al. 1996; Miyamoto and Meighen 2006; Waters and Bassler 2006; Pompeani et al. 2008). In A. fischeri, proteomic analysis of mutants defective in luxR, luxI, and ainS revealed the presence of several quorum-sensing-controlled non-lux genes, components of a LuxR-dependent quorum-sensing regulon; these genes code for a variety of different of proteins, some of which apparently contribute to the ability of A. fischeri to colonize its squid host, E. scolopes (Callahan and Dunlap 2000; Qin et al. 2007). Transcript analysis confirmed and extended these results to several additional genes (Antunes et al. 2007).
Isolation, Cultivation, Storage, and Identification of Luminous Bacteria
When working with luminous bacteria, and particularly when isolating new strains from nature, the possibility that these bacteria could be pathogenic (e.g., Kaeding et al. 2007) should always be kept in mind and appropriate care to avoid infection should always be used. Additional and detailed information on the isolation, cultivation, and phenotypic characterization of luminous bacteria can be found in Nealson (1978), Baumann et al. (1984), Baumann and Baumann (1981), Farmer and Hickman–Brenner (1992), and Baumann and Schubert (1984).
Isolation
Luminous bacteria can be isolated from most marine environments, and two methods, direct plating of seawater and enrichment from marine fish skin, are effective and simple for this purpose. An easily prepared complete medium that is suitable for growing all known luminous bacteria, LSW-70, contains natural or artificial seawater, diluted to 70 % of full strength, 10 g l–1 tryptone or peptone, and 5 g l–1 yeast extract, with 15 g l–1 agar for solid medium (Dunlap et al. 2004). Sugars and sugar alcohols (i.e., glucose, glycerol) are unnecessary for good growth and luminescence and can lead to acid production and death of cultures (Hill 1928; Johnson and Shunk 1936; Dunlap et al. 1995); their use in isolation media should be avoided. For isolations from environments where high numbers of bacteria that form spreading colonies may be present, such as coastal seawater, sediment, and intestinal tracts of marine animals, the use of agar at 4 % (40 g l–1) (Baumann et al. 1984) is recommended. This harder, less moist agar limits the ability of bacteria motile on solid surfaces, for example, certain peritrichously flagellated bacteria and bacteria that move by gliding motility, to spread over the plate and cause cross contamination of colonies.
Direct plating of seawater involves simply spreading an appropriate volume, typically 10–100 μl for coastal seawater, of the sample on one or more plates and incubating at room temperature or, preferably, cooler temperatures, such as 15–20 °C. For open ocean seawater and other samples with a lower number of bacteria, larger volumes, for example, 100 ml to 1 l, can be filtered through membrane filters with a pore size of 0.2 μm or 0.45 μm to collect the bacteria. The filters are then placed, bacteria side up, on plates of the above medium. Once colonies have arisen, usually within 18–24 h at room temperature and longer for lower temperatures, the plates can be examined in a dark room. Luminous colonies can then be picked (sterile wooden toothpicks are suitable for this purpose) and streaked for isolation on fresh plates of the same medium. Use of a red light, such as a photographic darkroom light, on a variable intensity control can make the picking of luminous colonies easier; by adjusting the red light, colonies of nonluminous bacteria can be made to appear reddish, whereas luminous colonies are blue due to their luminescence. Samples collected from warm waters and incubated at room temperature are more likely to yield V. harveyi and related luminous Vibrio species, as well as A. fischeri, P. leiognathi, and P. mandapamensis, whereas cold seawater samples plated and incubated at lower temperatures are likely to yield A. logei, P. kishitanii, P. phosphoreum, and S. hanedai. It should be noted that some strains of A. logei and S. hanedai grow well but do not produce light at room temperature; attempts to isolate these and other psychrotrophic bacteria should be carried out at 15 °C. Also, some bacteria, such as A. salmonicida, may not produce visible levels of light unless aldehyde is added to the medium (Fidopiastis et al. 1999); a simple screening approach for finding these bacteria is, once the observer is dark adapted, to add a drop of decyl aldehyde to the underside of the lid of the plates used for plating environmental samples and look for previously dark colonies that then become luminous.
Enrichment from fish or squid can be made using fresh samples and sterile seawater or frozen samples with natural, unsterilized seawater (e.g., Ast and Dunlap 2005). The tissue, preferably with the skin on, is placed in a tray, skin up, covered halfway with seawater, incubated, and observed daily in the dark for luminous spots (see Fig. 13.5 ), which arise in one to a few days. These spots, colonies of luminous bacteria, can then be picked and streaked for isolation on the medium described above containing 4 % agar. From fish and squid, a variety of different species of luminous bacteria can be isolated, especially when different incubation temperatures, such as 4 °C, 15 °C, and 22 °C, are used.
Various crustaceans (e.g., gammarid and caprellid amphipods) are suitable sources for luminous bacteria, as they can become infected with luminous (and nonluminous) bacteria and develop a strong luminescence before and for several hours after dying. In a dark room, after dark adapting for 12–15 min, one can pick out the infected, luminous crustaceans from collected seaweed. In a lighted room, the exoskeleton of the animal is punctured to obtain the hemolymph, which is streaked onto LSW-70 agar plates. The plates can be incubated at ambient or cool temperatures and are observed after 12–24 h for luminous colonies, which are then picked and streaked to obtain pure cultures.
Cultivation
The easily prepared complete medium, LSW-70 (Dunlap et al. 2004), detailed above, is suitable for the growth and luminescence of all culturable luminous bacteria. Most complete marine media, whether prepared with artificial or natural seawater to supply appropriate levels of Na+, Ca2+, and Mg2+, support the growth of luminous bacteria from marine habitats. Previously, a commonly used complete medium was SWC, prepared with natural seawater diluted to 70 % or 75 %, 5 g per liter of tryptone or peptone, 3 g per liter of yeast extract and 3 ml per liter of glycerol, and with 15 g per liter of agar for solid medium. Traditionally, SWC was buffered with 50 mM Tris or HEPES, or 1 g per liter of solid calcium carbonate was incorporated into the agar medium to control acid production (Nealson 1978). Acid production during growth in SWC, which can lead to death of the cells, results, however, from the presence of glycerol, and elimination of this component avoids the problem (Hill 1928; Johnson and Shunk 1936; Dunlap et al. 1995) with no major effect on growth or luminescence. Nealson (1978) listed and compared various formulations for complete and minimal media. Artificial seawater can be prepared according to the formulation of MacLeod, as described by Nealson (1978), or for routine culture work, a commercial aquarium marine salt mix can be used. Procedures for preparing minimal media have been described by Nealson (1978). Growing marine luminous bacteria in liquid medium may require a low agitation rate, since strong aeration can cause some species to clump at low population density.
Storage
Luminous bacteria have been revived from sealed glass ampoules after more than 80 years and 28 years of storage (Figge et al. 2011; Haneda 1981). Storage of luminous bacteria on agar slants or in agar stabs is suitable for only short periods, that is, days; longer-term storage on media is not recommended, as dim and dark variants easily arise with most species, and survival can be poor. Similarly, survival under refrigeration is poor for some species. Lyophylization or storage in liquid nitrogen may be an option if appropriate equipment is available (Baumann et al. 1984). Perhaps most convenient and effective for retaining strains in their original state is storage at ultralow temperature, for example, −75 °C to −80 °C, in a cryoprotective medium. An effective cryoprotective medium for the storage of luminous bacteria is filter-sterilized double strength deep freeze medium (2X DFM), prepared with 1 % w/v yeast extract, 10 % dimethyl sulfoxide (DMSO), 10 % glycerol, and 02 M K2HPO4/NaH2PO4 (pH 70). 2X DFM, originally developed by R. Rodriquez for storing yeast, was recommended to us several years ago by E. F. DeLong. Storage using 2XDFM works well for all species of luminous bacteria (P. Dunlap, pers. obs.). For storage of a strain, a dense culture is prepared by growing the strain in a complete liquid medium (e.g., LSW-70 broth) with aeration overnight or longer, to attain a dense population, adding 0.5 ml each of the culture and 2X DFM to cryovials, briefly vortexing to mix, allowing the mixture to stand for 5 min, and then placing the vial into the ultralow temperature freezer. Commercial cell freezing containers containing isopropanol, which allow a slow rate of cooling, work particularly well. Quick freezing in an ethanol bath kept in the ultralow temperature freezer or chilled with dry ice works well, but the ethanol can cause the labeling on tubes to smear. Cultures of luminous bacteria stored in this manner retain viability apparently indefinitely when the vials are kept at constant ultralow temperature.
Identification
The taxonomy of the marine luminous bacteria and their relationships to other marine enterobacteria were established during the 1970s and 1980s through the use of an array of diagnostic physiological, biochemical, and molecular traits (Reichelt and Baumman 1973; Reichelt et al. 1976; Baumann and Baumann 1981; Baumann and Baumann 1981). Very substantial progress was made through this work in clarifying the genus and species diversity of luminous bacteria, and that work established a foundation for understanding the ecological distributions and evolutionary relationships of these bacteria. At a practical level, the use at the time of as few as 10–25 phenotypic traits allowed the identification of many of the commonly encountered species of marine luminous bacteria (Nealson 1978; Baumann and Baumann 1981; Hastings and Nealson 1981).
More recently, however, many of the entities thought to be single species based on these phenetic traits have been found to represent multiple, evolutionarily distinct lineages, that is, separate species and genera, when examined by molecular phylogenetic criteria (e.g., Ast and Dunlap 2005; Ast et al. 2009). Consequently, current methods for the rapid and accurate identification of luminous bacteria and for descriptions of new species increasingly are based on phylogenetic analysis of gene sequences, which is now inexpensive, rapid, and highly accurate. For rapid identifications, sequence analysis of just the luxA or luxB genes often is adequate for good provisional identifications and can be supplemented or replaced by analysis of the sequence of gyrB. A multilocus approach using housekeeping genes such as the 16S rRNA gene, gyrB, pyrH, recA, rpoA, and glnA has proven very effective for robustly separating closely related luminous bacteria and for revealing the rare instances in which a strain apparently has acquired lux genes horizontally. Information on primers and amplification procedures for these and other genes can be found in Ast et al. (2009), Thompson et al. (2005), Urbanczyk et al. (2008), Fischer Le Saux et al. (1999), and Peat et al. (2010). Together with housekeeping genes, sequence analysis of the lux genes has proven particularly valuable for new species descriptions and rapid identification of newly isolated strains (e.g., Ast and Dunlap 2004; Haygood 1990; Ast and Dunlap 2005; Thompson et al. 2005; Ast et al. 2007b; Urbanczyk et al. 2007; Ast et al. 2009).
Complete characterization of a new species of luminous bacteria, however, should include more than just a multigene phylogenetic analysis. Diagnostic biochemical and morphological traits, DNA hybridization analysis, determination of the mol% G + C ratio, fatty acid profile analysis, and comparative genomic analysis such as amplified fragment length polymorphism (AFLP) or repetitive extragenic polymorphic PCR (rep-PCR), in the context of the bacterium’s ecology (e.g., Ast et al. 2007a), provide a more complete description suitable for new species. Furthermore, the examination of multiple independent isolates of the new entity and the inclusion in the analysis of the type strains of all closely related and relevant species are critically important for accurate and definitive work.
References
Adar YY, Ulitzur S (1993) GroESL proteins facilitate binding of externally added inducer by LuxR protein–containing E. coli cells. J Biolumin Chemilumin 8:261–266
Adar YY, Simaan M, Ulitzur S (1992) Formation of the LuxR protein in the Vibrio fischeri lux system is controlled by HtpR through the GroESL proteins. J Bacteriol 174:7138–7143
Ahrens G (1965) Untersuchungen am Leuchtorgan von Leiognathus klunzingeri (Steindachner). Z Wiss Zool 173:90–113
Akhurst RJ (1980) Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J Gen Microbiol 121:303–309
Akhurst RJ (1982) Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the family Heterorhabditidae and Steinernematidae. J Gen Microbiol 128:3061–3065
Akhurst RJ, Boemare NE (1986) A non–luminescent strain of Xenorhabdus luminescens. J Gen Microbiol 132:1917–1922
Akhurst RJ, Dunphy G (1993) Tripartite interactions between symbiotically associated entomopathogenic bacteria, nematodes, and their insect hosts. In: Beckage N, Thompson S, Federici B (eds) Parasites and pathogens of insects. Academic, New York, NY, pp 21–23
Antunes LC, Schaefer AL, Ferreira RB, Qin N, Stevens AM, Ruby EG, Greenberg EP (2007) Transcriptome analysis of the Vibrio fischeri LuxR–LuxI regulon. J Bacteriol 189:8387–8391
Ast JC, Dunlap PV (2004) Phylogenetic analysis of the lux operon distinguishes two evolutionarily distinct clades of Photobacterium leiognathi. Arch Microbiol 181:352–361
Ast JC, Dunlap PV (2005) Phylogenetic resolution and habitat specificity of members of the Photobacterium phosphoreum species group. Environ Microbiol 7:1641–1654
Ast JC, Cleenwerck I, Engelbeen K, Urbanczyk H, Thompson FL, De Vos P, Dunlap PV (2007a) Photobacterium kishitanii sp. nov., a luminous marine bacterium symbiotic with deep–sea fish. Int J Syst Evol Microbiol 57:2073–2078
Ast JC, Urbanczyk H, Dunlap PV (2007b) Natural merodiploidy of the lux–rib operon of Photobacterium leiognathi from coastal waters of Honshu Japan. J Bacteriol 189:6148–6158
Ast JC, Urbanczyk H, Dunlap PV (2009) Multi–gene analysis reveals previously unrecognized phylogenetic diversity in Aliivibrio. Syst Appl Micro 32:379–386
Austin B, Zhang XH (2006) Vibrio harveyi a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol 43:119–124
Baldwin TO, Ziegler MM, Powers DA (1979) Covalent structure of subunits of bacterial luciferase NH2– terminal sequence demonstrates subunit homology. Proc Natl Acad Sci USA 76:4887–4889
Bang SS, Baumann P, Baumann L (1978) Phenotypic characterization of Photobacterium logei (sp. nov.), a species related to P. fischeri. Curr Microbiol 1:285–288
Baross JA, Tester PA, Morita RY (1978) Incidence, microscopy, and etiology of exoskeleton lesions in the tanner crab, Chionoecetes tanneri. J Fish Res Board Can 35:1141–1149
Bassler BL (1999) How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol 2:582–587
Bassler BL, Wright M, Showalter RE, Silverman MR (1993) Intercellular signalling in Vibrio harveyi, sequence and function of genes regulating expression of luminescence. Mol Microbiol 9:773–786
Bassler BL, Wright M, Silverman MR (1994a) Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol 12:403–412
Bassler BL, Wright M, Silverman MR (1994b) Multiple signalling systems controlling expression of luminescence in Vibrio harveyi, sequence and function of genes encoding a second sensory pathway. Mol Microbiol 13:273–286
Bassler BL, Greenberg EP, Stevens AM (1997) Cross–species induction of luminescence in the quorum–sensing bacterium Vibrio harveyi. J Bacteriol 179:4043–4045
Bassot J-M (1966) On the comparative morphology of some luminous organs. In: Johnson FH (ed) Bioluminescence in progress. Princeton University, Princeton, NJ, pp 557–610
Baumann P, Baumann L (1977) Biology of the marine enterobacteria genera Beneckea and Photobacterium. Ann Rev Microbiol 31:39–61
Baumann P, Baumann L (1981) The marine Gram–negative eubacteria genera Photobacterium, Beneckea, Alteromonas, Pseudomonas, and Alcaligenes. In: Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG (eds) The prokaryotes. Springer, Berlin, pp 1302–1331
Baumann P, Schubert RHW (1984) Family II. Vibrionaceae Veron 1965. In: Kreig NR, Holt JG (eds) Bergey’s manual of systematic bacteriology. Williams and Wilkins, Baltimore, MD, pp 516–547
Baumann P, Baumann L, Bang SS, Woolkalis MJ (1980) Reevaluation of the taxonomy of Vibrio, Beneckea, and Photobacterium: abolition of the genus Beneckea. Curr Microbiol 4:127–132
Baumann P, Furniss AL, Lee JV (1984) Genus Vibrio Pacini 1854. In: Kreig NR, Holt JG (eds) Bergey’s manual of systematic bacteriology. Williams and Wilkins, Baltimore, MD, pp 518–538
Baylor ER (1949) Growth cycle of luminous bacteria on limited substrate. M.Sc. thesis, Princeton University
Beijerinck MW (1889a) Le Photobacterium luminosum, bacterie lumineuse de la mer du nord. Arch Néerl Sci Exactes Natur 23:401–415
Beijerinck MW (1889b) Les bactéries lumineuses dans leurs rapports avec l’oxygène. Archives Néerl Sci Exactes Nat 23:416–427
Beijerinck MW (1891) Sur l’aliment photogène et l’aliment plastique des bactéries lumineuses. Arch Néerl Sci Exactes Nat 24:369–442
Beijerinck MW (1916) Die Leuchtbakterien der Nordsee im August und September. Folia Microbiol 4:15–40
Bintrim SB, Ensign JC (1998) Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotrophic phenotypes. J Bacteriol 180:1261–1269
Boemare NE, Akhurst RJ, Mourant RG (1993) DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of Entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int J Syst Bacteriol 43:249–255
Boemare NE, Govaidan A, Brehelin M, Laumond C (1997) Symbiosis and pathogenicity of nematode –bacterium complexes. Symbiosis 22:21–45
Boettcher KJ, Ruby EG (1990) Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 17:3701–3706
Boisvert H, Chatelain R, Bassot JM (1967) Étude d’un Photobacterium isolé de l’organe lumineux des poissons Leiognathidae. Ann Inst Pasteur Paris 112:520–524
Boyle R (1668) Experiments concerning the relation between light and air in shining wood and fish. Phil Trans 2:581–600
Buchner P (1965) Endosymbiosis of animals with plant microorganisms. Wiley, New York
Budsberg KJ, Wimpee CF, Braddock JF (2003) Isolation and identification of Photobacterium phosphoreum from an unexpected niche migrating salmon. Appl Environ Microbiol 69:6938–6942
Callahan SM, Dunlap PV (2000) LuxR– and acylhomoserine– lactone–controlled non–lux genes define a quorum–sensing regulon in Vibrio fischeri. J Bacteriol 182:2811–2822
Callahan SM, Cornell NW, Dunlap PV (1995) Purification and properties of periplasmic 3’:5’–cyclic nucleotide phosphodiesterase. A novel zinc–containing enzyme from the marine symbiotic bacterium Vibrio fischeri. J Biol Chem 270:17627–17632
Cao JG, Meighen EA (1989) Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem 264:21670–21676
Cao JG, Meighen EA (1993) Biosynthesis and stereochemistry of the autoinducer controlling luminescence in Vibrio harveyi. J Bacteriol 175:3856–3862
Cao JG, Wei ZY, Meighen EA (1995) The lux autoinducer–receptor interaction in Vibrio harveyi binding parameters and structural requirements for the autoinducer. Biochem J 312:439–444
Castle PHJ, Paxton JR (1984) A new genus and species of luminescent eel (Pisces: Congridae) from the Arafura Sea, Northern Australia. Copeia 1984:72–81
Charkrabarty P, Davis MP, Smith WL, Berquist R, Gledhill KM, Frank LR, Sparks JS (2011) Evolution of the light organ system in ponyfishes (Teleostei: Leiognathidae). J Morphol 272:704–721
Chatterjee J, Miyamoto CM, Meighen EA (1996) Autoregulation of luxR the Vibrio harveyi lux–operon activator functions as a repressor. Mol Microbiol 20:415–425
Chatterjee J, Miyamoto CM, Zouzoulas A, Lang BF, Skouris N, Meighen EA (2002) MetR and CRP bind to the Vibrio harveyi lux promoters and regulate luminescence. Mol Microbiol 46:101–111
Chen PF, Tu SC, Hagag N, Wu FYH, Wu CW (1985) Isolation and characterization of a cyclic AMP receptor protein from luminous Vibrio harveyi cells. Arch Biochem Biophys 241:425–431
Chen X, Schauder S, Potier N, Dorsselaer AV, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorum–sensing signal containing boron. Nature 415:545–549
Claes MF, Dunlap PV (2000) Aposymbiotic culture of the sepiolid squid Euprymna scolopes: role of the symbiotic bacterium Vibrio fischeri in host animal growth, development, and light organ morphogenesis. J Exp Zool 286:280–296
Coffey JJ (1967) Inducible synthesis of bacterial luciferase: specificity and kinetics of induction. J Bacteriol 94:1638–1647
Cohen DM, Inada T, Iwamoto T, Scialabba N (1990) FAO species catalogue 10, gadiform fishes of the world. Food and Agriculture Organization of the United Nations, Rome
Czyż A, Plata K, Wegrzyn G (2003) Stimulation of DNA repair as an evolutionary drive for bacterial luminescence. Luminescence 18:140–144
Dahlgren U (1915) The production of light by animals. J Franklin Inst 180:513–537
Desmarchelier PM, Reichelt JL (1981) Phenotypic characterization of clinical and environmental isolates of Vibrio cholerae from Australia. Curr Microbiol 5:123–127
Devine JH, Countryman C, Baldwin TO (1988) Nucleotide sequence of the luxR and luxI genes and structure of the primary regulatory region of the lux regulon of Vibrio fischeri ATCC 7744. Biochemistry 27:837–842
Dolan KM, Greenberg EP (1992) Evidence that GroEL, not σ32, is involved in transcription regulation of the Vibrio fischeri luminescence genes in Escherichia coli. J Bacteriol 174:5132–5135
Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux–Eude C, Chandler M, Charles JF, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Médigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F (2003) The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol 21:1307–1313
Dunlap PV (1984) Physiological and morphological state of the symbiotic bacteria from light organs of ponyfish. Biol Bull 167:410–425
Dunlap PV (1985) Osmotic control of luminescence and growth in Photobacterium leiognathi from ponyfish light organs. Arch Microbiol 141:44–50
Dunlap PV (1989) Regulation of luminescence by cyclic AMP in cya–like and crp–like mutants of Vibrio fischeri. J Bacteriol 171:1199–1202
Dunlap PV (1991) Organization and regulation of bacterial luminescence genes. Photochem Photobiol 54:1157–1170
Dunlap PV (1997) N–Acyl–L–homoserine lactone autoinducers in bacteria unity and diversity. In: Shapiro JA, Dworkin M (eds) Bacteria as multicellular organisms. Oxford University Press, New York, NY, pp 69–106
Dunlap PV (2000) Quorum regulation of luminescence in Vibrio fischeri. In: Bartlett D (ed) Molecular marine microbiology. Horizon Press, Norfolk, UK, pp 3–21
Dunlap PV (2009) Bioluminescence, microbial. In: Schaechter M (ed) Encyclopedia of microbiology, 3rd edn. Elsevier, Oxford, pp 45–61
Dunlap PV, Ast JC (2005) Genomic and phylogenetic characterization of the luminous bacteria symbiotic with the deep–sea fish Chlorophthalmus albatrossis (Aulopiformes Chlorophthalmidae). Appl Environ Microbiol 71:930–939
Dunlap PV, Callahan SM (1993) Characterization of a periplasmic 3':5'–cyclic nucleotide phosphodiesterase gene, cpdP, from the marine symbiotic bacterium Vibrio fischeri. J Bacteriol 175:4615–4624
Dunlap PV, Greenberg EP (1985) Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J Bacteriol 164:45–50
Dunlap PV, Greenberg EP (1988) Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein–LuxR protein regulatory circuit. J Bacteriol 170:4040–4046
Dunlap PV, Greenberg EP (1991) Role of intercellular chemical communication in the Vibrio fischeri –– monocentrid fish symbiosis. In: Dworkin M (ed) Microbial cell–cell interactions. American Society for Microbiology, Washington, DC, pp 219–253
Dunlap PV, Kita–Tuskamoto K (2006) Luminous bacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes a handbook on the biology of bacteria, vol 3. Springer, New York, NY, pp 863–892
Dunlap PV, Kuo A (1992) Cell density–dependent modulation of the Vibrio fischeri luminescence system in the absence of autoinducer and LuxR protein. J Bacteriol 174:2440–2448
Dunlap PV, McFall–Ngai MJ (1987) Initiation and control of the bioluminescent symbiosis between Photobacterium leiognathi and leiognathid fish. Ann NY Acad Sci 503:269–283
Dunlap PV, Nakamura M (2011) Functional morphology of the luminescence system of Siphamia versicolor (Perciformes: Apogonidae), a bacterially luminous coral reef fish. J Morphol 272:897–909
Dunlap PV, Ray JM (1989) Requirement for autoinducer in transcriptional negative autoregulation of the Vibrio fischeri luxR gene in Escherichia coli. J Bacteriol 171:3549–3552
Dunlap PV, Mueller U, Lisa TA, Lundberg KS (1992) Growth of the marine luminous bacterium Vibrio fischeri on 3’ 5’–cyclic AMP correlation with a periplasmic 3’ 5’–cyclic AMP phosphodiesterase. J Gen Microbiol 138:115–123
Dunlap PV, Kita–Tsukamoto K, Waterbury J, Callahan SM (1995) Isolation and characterization of a visibly luminous variant of Vibrio fischeri strain ES114 from the sepiolid squid Euprymna scolopes. Arch Microbiol 164:194–202
Dunlap PV, Jiemjit A, Ast JC, Pearce MM, Marques RR, Lavilla–Pitogo CR (2004) Genomic polymorphism in symbiotic populations of Photobacterium leiognathi. Environ Microbiol 6:145–158
Dunlap PV, Ast JC, Kimura S, Fukui A, Yoshino T, Endo H (2007) Phylogenetic analysis of host–symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23:507–523
Dunlap PV, Davis KM, Tomiyama S, Fujino M, Fukui A (2008) Devlopmental and microbiological analysis of the inception of bioluminescent symbiosis in the marine fish Nuchequula nuchalis (Perciformes Leiognathidae). Appl Environ Microbiol 74:7471–7748
Dunlap PV, Kojima Y, Nakamura S, Nakamura M (2009) Inception of formation and early morphogenesis of the bacterial light organ of the sea urchin cardinalfish, Siphamia versicolor (Perciformes Apogonidae). Mar Biol 156:2011–2020
Dunlap PV, Gould AL, Wittenrich ML, Nakamura M (2012) Symbiosis initiation in the bacterially luminous sea urchin cardinalfish Siphamia versicolor. J Fish Biol 81:1340–1356
Eberhard A (1972) Inhibition and activation of bacterial luciferase synthesis. J Bacteriol 109:1101–1105
Eberhard A, Hinton JP, Zuck RM (1979) Luminous bacteria synthesize luciferase anaerobically. Arch Microbiol 121:277–282
Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ (1981) Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444–2449
Eberhard A, Widrig CA, McBath P, Schineller JB (1986) Analogs of the autoinducer of bioluminescence in Vibrio fischeri. Arch Microbiol 146:35–40
Eberhard A, Longin T, Widrig CA, Stranick SJ (1991) Synthesis of the lux gene autoinducer in Vibrio fischeri is positively autoregulated. Arch Microbiol 155:294–297
Egan ES, Fogel MA, Waldor MK (2005) Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol 56:1129–1138
Engebrecht J, Silverman M (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA 81:4154–4158
Engebrecht J, Silverman M (1987) Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucl Acids Res 15:10455–10467
Engebrecht J, Nealson K, Silverman M (1983) Bacterial bioluminescence, isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773–781
Farghaly AH (1950) Factors influencing the growth and light production of luminous bacteria. Comp Cell Physiol 36:165–183
Farmer JJ, Hickman–Brenner FW (1992) The genera Vibrio and Photobacterium. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd edn. Springer, Berlin, pp 2952–3011
Farmer JJ, Jorgensen JH, Grimont PAD, Akhurst RJ, Poinar GO, Pierce GV, Smith JA, Carger GP, Wilson K, Hickman–Brenner FW (1989) Xenorhabdus luminescens (DNA hybridization group 5) from human clinical specimens. J Clin Microbiol 27:1594–1600
Feldman KA, Buck JD (1984) Distribution and characterization of luminescent bacteria in a temperate estuary. Estuaries 7:93–97
Fidopiastis PM, von Boletzky S, Ruby EG (1998) A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J Bacteriol 180:59–64
Fidopiastis PM, Sorum H, Ruby EG (1999) Cryptic luminescence in the cold–water fish pathogen Vibrio salmonicida. Arch Microbiol 171:205–209
Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG (2002) LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol 45:131–143
Figge MJ, Robertson LA, Ast JC, Dunlap PV (2011) Historical microbiology: revival and phylogenetic characterization of luminous bacterial cultures of M. W. Beijerinck. FEMS Microbiol Ecol 78:463–472
Fischer B (1887) Bacteriologische Untersuchungen auf einer Reise nach West Indien. II. Ubereinen lichtentwickelnden, in Meerswasser gefunden Spaltpilz. Z hyg Infekt 2:54–95
Fischer Le Saux M, Viallard V, Brunel B, Normand P, Boemare EN (1999) Polyphasic classification of the genus Photorhabdus and proposal of new taxa P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov., and P. asymbiotica sp. nov. Int J Syst Bacteriol 49:1645–1656
Fitzgerald JM (1977) Classification of luminous bacteria from the light organ of the Australian pinecone fish, Cleidopus gloriamaris. Arch Microbiol 112:153–156
Flodgaard LR, Dalgaard P, Andersen JB, Nielsen KF, Givskov M, Gram L (2005) Nonbioluminescent strains of Photobacterium phosphoreum produce the cell–to–cell communication signal N–(3–hydroxyoctanoyl) homoserine lactone. Appl Environ Microbiol 71:2113–2120
Foran D (1991) Evidence of luminous bacterial symbionts in the light organs of myctophid and stomiiform fishes. J Exp Zool 259:1–8
Forst S, Nealson K (1996) Molecular biology of the symbiotic–pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol Rev 60:21–43
Forst S, Dowds B, Boemare N, Stackebrandt E (1997) Xenorhabdus and Photorhabdus spp. bugs that kill bugs. Ann Rev Microbiol 51:47–72
Freeman JA, Bassler BL (1999a) A genetic analysis of the function of LuxO, a two–component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol 31:665–677
Freeman JA, Bassler BL (1999b) Sequence and function of LuxU: a two–component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol 191:899–906
Freeman JA, Lilley BN, Bassler BL (2000) A genetic analysis of the functions of LuxN: a two–component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol 35:139–149
Friedrich WF, Greenberg EP (1983) Glucose repression of luminescence and luciferase in Vibrio fischeri. Arch Microbiol 134:87–91
Fukasawa S, Dunlap PV (1986) Identification of luminous bacteria isolated from the light organ of the squid, Doryteuthis kensaki. J Agric Biol Chem 50:1645–1646
Fukasawa S, Dunlap PV, Baba M, Osumi M (1987) Identification of an agar–digesting, luminous bacterium. Agric Biol Chem 51:265–268
Fukasawa S, Suda T, Kubota S (1988) Identification of luminous bacteria isolated from the light organ of the fish, Acropoma japonica. Agric Biol Chem 52:285–286
Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria the LuxR–LuxI family of cell density–responsive transcriptional regulators. J Bacteriol 176:269–275
Fuqua WC, Winans SC, Greenberg EP (1996) Census and consensus in bacterial ecosystems: the LuxR–LuxI family of quorum–sensing transcriptional regulators. Ann Rev Microbiol 50:727–751
Galt CP (1978) Bioluminescence: dual mechanism in a planktonic tunicate produces brilliant surface display. Science 200:70–72
Gerrard JG, Joyce SA, Clarke DJ, Ffrench–Constant RH, Nimmo GR, Looke DF, Feil EJ, Pearce L, Waterfield. NR (2006) Nematode symbiont for Photorhabdus asymbiotica. Emerg Infect Dis 12:1562–1564
Giard A (1889) On the phosphorescent infection of the Talitri and other crustaceans. Ann Mag Nat Hist 4:476–478
Giard A, Billet A (1889) Observations sur la maladie phosphorescente des Talitres et autres crustaces. Compt rend Soc Biol 41:593–597
Gilson L, Kuo A, Dunlap PV (1995) AinS and a new family of autoinducer synthesis proteins. J Bacteriol 177:6946–6951
Gogarten JP, Doolitte WF, Lawrence JG (2002) Prokaryotic evolution in light of gene transfer. Mol Biol Evol 19:2226–2238
Gomez–Gil B, Soto–Rodríguez S, García–Gasca A, Roque A, Vazquez–Juarez R, Thompson FL, Swings J (2004) Molecular identification of Vibrio harveyi–related isolates associated with diseased aquatic organisms. Microbiology 150:1769–1777
Graf J, Ruby EG (1998) Host–derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA 95:1818–1822
Greenberg EP (1997) Quorum sensing in Gram–negative bacteria. Am Soc Microbiol News 63:371–377
Grim CJ, Taviani E, Alam M, Huq A, Sack RB, Colwell RR (2008) Occurrence and expression of luminescence in Vibrio cholerae. Appl Environ Microbiol 74:708–715
Guerrero MA, Makemson JC (1989) The cytochromes of luminous bacteria and their coupling to bioluminescence. Curr Microbiol 18:67–73
Haldar S, Chatterjee S, Sugimoto N, Das S, Chowdhury N, Hinenoya A, Asakura M, Yamasaki S (2011) Identification of Vibrio campbellii isolated from diseased farm–shrimps from south India and establishment of its pathogenic potential in an Artemia model. Microbiology 157:179–188
Haneda Y (1938) Uber den leuchtfisch, Mulucocephalus luevis (Lowe). Jpn J Med 5:355–366
Haneda Y (1950) Luminous organisms of Japan and the Far East. In: Johnson FW (ed) The luminescence of biological systems. American Society for the Advancement of Science, Washington, DC, pp 335–385
Haneda Y (1981) Preservation and utilization of luminous bacteria as a light source. Sci Rep Yokosuka Cy Mus 28:79–83
Hanzelka BL, Parsek MR, Val DV, Dunlap PV, Cronan JE Jr, Greenberg EP (1999) Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol 181:5766–5770
Harms JW (1928) Bau und entwicklung eines eigenartigen leuchtorgans bei Equula spec. Z Wiss Zool 131:157–179
Harvey EN (1922) The production of light by the fishes photoblepharon and anomalops. Carnegie Institute of Washington, Washington DC
Harvey EN (1940) Living light. Princeton University Press, Princeton NJ
Harvey EN (1952) Bioluminescence. Academic, New York NY
Harvey EN (1957) A history of luminescence from the earliest times until 1900. American Philosophical Society, Philadelphia PA
Hastings JW (1971) Light to hide by: ventral luminescence to camouflage the silhouette. Science 173:1016–1017
Hastings JW (1983) Biological diversity, chemical mechanisms, and the evolutionary origins of bioluminescent systems. J Mol Evol 19:309–317
Hastings JW (1995) Bioluminescence. In: Sperelakis N (ed) Cell physiology source book. Academic, New York, NY, pp 665–681
Hastings JW, Greenberg EP (1999) Quorum sensing the explanation of a curious phenomenon reveals a common characteristic of bacteria. J Bacteriol 181:2667–2669
Hastings JW, Nealson KH (1977) Bacterial bioluminescence. Ann Rev Microbiol 31:549–595
Hastings JW, Nealson KH (1981) The symbiotic luminous bacteria. In: Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG (eds) The prokaryotes. Springer, Berlin, pp 1332–1345
Hastings JW, Potrikus CJ, Gupta SC, Kurfurst M, Makemson JC (1985) Biochemistry and physiology of bioluminescent bacteria. Adv Microb Physiol 26:235–291
Hastings JW, Makemson JC, Dunlap PV (1987) How are growth and luminescence regulated independently in exosymbionts? Symbiosis 4:3–24
Haygood MG (1990) Relationship of the luminous bacterial symbiont of the Caribbean flashlight fish, Kryptophaneron alfredi (family Anomalopidae) to other luminous bacteria based on bacterial luciferase (luxA) genes. Arch Microbiol 154:496–503
Haygood MG (1993) Light organ symbioses in fish. Crit Rev Microbiol 19:191–216
Haygood MG, Distel DL (1993) Bioluminescent symbionts of flashlight fish and deep–sea anglerfish form unique lineages related to the genus Vibrio. Nature 363:154–156
Haygood MG, Nealson KH (1985) Mechanisms of iron regulation of luminescence in Vibrio fischeri. J Bacteriol 162:209–216
Haygood MG, Tebo BM, Nealson KH (1984) Luminous bacteria of a monocentrid fish (Monocentris japonicus) and two anomalopid fish (Photoblepharon palpebratus and Kryptophaneron alfredi) Population sizes and growth within the light organs, and rates of release into the seawater. Marine Biol 78:249–254
Haygood MG, Distel DL, Herring PJ (1992) Polymerase chain reaction and 16 S rRNA gene sequences from the luminous bacterial symbionts of two deepsea anglerfish. J Marine Biol AssocUK 72:149–159
Haygood MG, Edwards DB, Mowlds G, Rosenblatt RH (1994) Bioluminescence of myctophid and stomiiform fishes is not due to bacterial luciferase. J Exp Zool 270:225–231
Hendrie MS, Hodgkiss W, Shewan JM (1970) The identification, taxonomy and classification of luminous bacteria. J Gen Microbiol 64:151–169
Hendry TA, Dunlap PV (2011) The uncultured luminous symbiont of Anomalops katoptron (Beryciformes Anomalopidae) represents a new bacterial genus. Mol Phylogenet Evol 61:834–843
Henke JM, Bassler BL (2004) Three parallel quorum–sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol 186:6902–6904
Hense BA, Kuttler C, Muller J, Rothballer M, Harmann A, Kreft JU (2007) Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol 5:230–239
Herring PJ (1977) Luminescence in cephalopods and fish. Sym Zool Soc Lond 38:127–159
Herring PJ (1978) Bioluminescence of invertebrates other than insects. In: Herring PJ (ed) Bioluminescence in action. Academic, London, pp 190–240
Herring PJ, Morin JG (1978) Bioluminescence in fish. In: Herring PJ (ed) Bioluminescence in action. Academic, London, pp 273–329
Higgins DA, Pmianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL (2007) The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450:883–886
Hill SE (1928) The influence of molds on the growth of luminous bacteria in relation to the hydrogen ion concentration, together with the development of a satisfactory culture method. Biol Bull 55:143–150
Hjerde E, Lorentzen MS, Holden MT, Seeger K, Paulsen S, Bason N, Churcher C, Harris D, Norbertczak H, Quail MA, Sanders S, Thurston S, Parkhill J, Willassen NP, Thomson NR (2008) The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 9:616
Humm HJ (1946) Marine agar–digesting bacteria of the South Atlantic coast. Duke Univ Mar Stn Bull 3:45–75
Inman OL (1926) A pathogenic luminous bacterium. Biol Bull 53:197–200
Jensen MJ, Tebo BM, Baumann P, Mandel M, Nealson KH (1980) Characterization of Alteromonas hanedai (sp. nov.), a nonfermentative luminous species of marine origin. Curr Microbiol 3:311–315
Jermoljewa S (1926) Vibrio phosphorescens beim klinischen Bilde der Cholera und sein Zusammenhang mit anderen Vibrionen Zent. Bakteriol I Abt Orig 100:170–177
Johnson FH (1951) Luminous bacteria. In: Werkman CH, Wilson PW (eds) Bacterial physiology. Academic, New York NY, pp 576–605
Johnson FH (1988) Luminescence, narcosis, and life in the Deep Sea. Vantage Press, New York
Johnson FH, Shunk IV (1936) An interesting new species of luminous bacteria. J Bacteriol 31:585–593
Jones BW, Nishiguchi MK (2004) Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 144:1151–1155
Kaeding AJ, Ast JC, Pearce MM, Urbanczyk H, Kimura S, Endo H, Nakamura M, Dunlap PV (2007) Phylogenetic diversity and co–symbiosis in the bioluminescent symbioses of Photobacterium mandapamensis. Appl Environ Microbiol 73:3173–3182
Kaplan HB, Greenberg EP (1985) Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol 163:1210–1214
Karunasagar I, Pai R, Malathi GR, Karunasagar I (1994) Mass mortality of Penaeus monodon larvae due to antibiotic–resistant Vibrio harveyi infection. Aquaculture 128:203–209
Kasai S (2006) Freshwater bioluminescence in Vibrio albensis (Vibrio cholerae biovar albensis) NCIMB 41 is caused by a two–nucelotide deletion in luxO. J Biochem 139:471–482
Kasai S, Okada K, Hoshino A, Iida T, Honda T (2007) Lateral transfer of the lux gene cluster. J Biochem 141:231–237, Tokyo
Katznelson R, Ulitzur S (1977) Control of luciferase synthesis in a newly isolated strain of Photobacterium leiognathi. Arch Microbiol 115:347–351
Kelly RC, Bolitho ME, Higgins DA, Lu W, Ng WL, Jeffrey PD, Rabinowitz JD, Semmelhack MF, Hughson FM, Bassler BL (2009) The Vibrio cholerae quorum–sensing autoinducer CAI–1: analysis of the biosynthetic enzyme CqsA. Nat Chem Biol 5:891–895
Kessel M (1977) The ultrastructure of the relationship between the luminous organ of the teleost fish Photoblepharon palpebratus and its symbiotic bacteria. Cytobiologie 15:145–158
Kishitani T (1930) Studien über die Leuchtsymbiose in Physiculus japonicus Hilgendorf, mit der Beilage der zwei neuen arten der Leuchtbacterien. Sci Rep Res Inst Tohoku Univ 5:801–823
Klappenback JA, Dunbar JM, Schmidt TM (2000) rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol 66:1328–1333
Kou YS, Makemson JC (1988) Luciferase–like protein in Vibrio cholerae. In: Abstracts of the annual meeting of the American society for microbiology, Miami Beach, FL. American Society for Microbiology. Washington DC, 37
Kozukue H (1952) Isolation of luminous bacteria from alimentary canal of symbiotic luminous fish. Tokyo Jik Med J 67:18–21
Kühlmorgen–Hille G (1974) Leiognathidae. In: Fischer W, Whitehead PJP (eds) FAO species identification sheets for fishery purposes, Eastern Indian Ocean (Fishing Area 57) and Western Central Pacific (Fishing Area 71), vol 2. Food and Agriculture Organization of the United Nations, Rome
Kuo A, Blough NV, Dunlap PV (1994) Multiple N–acyl–homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J Bacteriol 176:7558–7565
Kuo A, Callahan SM, Dunlap PV (1996) Modulation of luminescence operon expression by N–octanoyl–homoserine lactone in ainS mutants of Vibrio fischeri. J Bacteriol 178:971–976
Kuwata R, Yoshiga T, Yoshida M, Kondo E (2008) Mutualistic association of Photorhabdus asymbiotica with Japanese heterorhabditid entomopathogenic nematodes. Microbes Infect 10:734–741
Lauro FM, McDougald TT et al (2009) The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106:15527–15533
Lavilla–Pitogo CR, de la Peña LD (1998) Bacterial diseases in shrimp (Penaeus monodon) cultured in the Philippines. Fish Pathol 33:405–411
Lavilla–Pitogo CR, Baticados MCL, Cruz–Lacierda ER, de la Peña LD (1990) Occurrence of luminous bacterial disease of Penaeus monodon larvae in the Philippines. Aquaculture 91:1–13
Lavilla–Pitogo CR, Albright LJ, Paner MG, Sunaz NA (1992) Diseases in Asian aquaculture. In: Shariff IM, Subasinghe RP, Arthur JR (eds) Studies on the sources of luminescent Vibrio harveyi in Penaeus monodon hatcheries. Asian Fisheries Society, Manila, pp 157–164
Leano EM, Lavilla–Pitogo CR, Paner MG (1998) Bacterial flora in the hepatopancreas of pond reared Penaeus monodon juveniles with luminous vibriosis. Aquaculture 164:367–374
Lee J (1993) Lumazine protein and the excitation mechanism in bacterial bioluminescence. Biophys Chem 48:149–158
Lee CY, Meighen EA (1992) The lux genes in Photobacterium leiognathi are closely linked with genes corresponding in sequence to riboflavin synthesis genes. Bichem Biophys Res Commun 186:690–697
Lee KH, Ruby EG (1992) Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl Environ Microbiol 58:942–947
Lee J, Matheson IBC, Muller F, O'Kane D, Veroort J, Visser AJWG (1990) The mechanism of bacterial bioluminescence. In: Muller F (ed) Chemistry and biochemistry of flavoenzyme, vol 2. CRC Press, Boca Raton, FL, pp 109–151
Lee CY, O’Kane DJ, Meighen EA (1994) Riboflavin synthesis genes are linked with the lux operon of Photobacterium phosphoreum. J Bacteriol 176:2100–2104
Leisman G, Cohn DH, Nealson KH (1980) Bacterial origin of luminescence in marine animals. Science 208:1271–1273
Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi. Cell 118:69–82
Li Z, Szittner R, Meighen EA (1993) Subunit interactions and the role of the luxA polypeptide in controlling thermal stability and catalytic properties in recombinant luciferase hybrids. Biochim Biophys Acta 1158L:137–145
Lilley BN, Bassler BL (2000) Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma–54. Mol Microbiol 36:940–954
Lin JW, Chao YF, Weng SF (1993) The lumazine protein–encoding gene in Photobacterium leiognathi is linked to the lux operon. Gene 126:153–154
Lin JW, Yu KY, Chao YF, Weng SF (1995) The lumQ gene is linked to the lumP gene and the lux operon in Photobacterium leiognathi. Biochem Biophys Res Commun 217:684–695
Lin JW, Chao YF, Weng SF (1996a) Nucleotide sequence and functional analysis of the luxE gene encoding acyl–protein synthetase of the lux operon from Photobacterium leiognathi. Biochem Biophys Res Commun 228:764–773
Lin JW, Yu KY, Chao YF, Weng SF (1996b) Regulatory region with putA gene of proline dehydrogenase that links to the lum and lux operons in Photobacterium leiognathi. Biochem Biophys Res Commun 219:868–875
Lin JW, Chao YF, Weng SF (1998) Characteristic analysis of the luxG gene encoding the probable flavin reductase that resides in the lux operon of Photobacterium leiognathi. Biochem Biophys Res Commun 246:446–452
Lin YH, Miyamoto C, Meighen EA (2000) Cloning and functional studies of a luxO regulator LuxT from Vibrio harveyi. Biochim Biophys Acta 1494:226–235
Lin JW, Chao YF, Weng SF (2001) Riboflavin synthesis genes ribE, ribB, ribH, ribA reside in the lux operon of Photobacterium leiognathi. Biochem Biophys Res Commun 284:587–595
Lin B, Wang Z, Malanoski AP, O'Grady EA, Wimpee CF, Vuddhakul V, Alves N Jr, Thompson FL, Gomez–Hil B, Voral GJ (2010) Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA–1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep 2:81–89
Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS (2009) Quantifying the integration of quorum–sensing signals with single–cell resolution. PLoS Biol 7:e68
Ludwig F (1884) Micrococcus P flugeri, ein neuer photogener Pilz. Hedwigia 23:33–37
Lunder T, Sørum H, Holstad G, Steigerwalt AG, Mowinckel P, Brenner DJ (2000) Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from Atlantic salmon (Salmo salar) with 'winter ulcer'. Int J Syst Evol Microbiol 50:427–450
Mackie GO, Bone Q (1978) Luminescence and associated effector activity in Pyrosoma (Tunicata: Pyrosomida). Proc R Soc London 202:483–495
Majima R (1931) Studies on luminous bacteria further studies on pathogenic luminous bacteria, Microspira phosphoreum. Yasaki Sei–i–kai Japan 50:1–23
Makemson JC (1986) Luciferase–dependent oxygen consumption by bioluminescent vibrios. J Bacteriol 165:461–466
Makemson JC, Hastings JW (1982) Iron represses bioluminescence in Vibrio harveyi. Curr Microbiol 7:181–186
Makemson JC, Hastings JW (1986) Nonluminous vibrios possess proteins that share antigenic determinants with Vibrio harveyi luciferase. In: Abstr. Ann. Meet. Amer. Soc. Microbiol. Washington, DC, I–46
Makemson JC, Hermosa GV Jr (1999) Luminous bacteria cultured from fish guts in the Gulf of Oman. Luminescence 14:161–168
Makemson JC, Fulayfil N, Basson P (1992) Association of luminous bacteria with artificial and natural surfaces in Arabian Gulf seawater. Appl Environ Microbiol 58:2341–2343
Makemson JC, Fulayfil NR, Landry W, Van Ert LM, Wimpee CF, Widder EA, Case JF (1997) Shewanella woodyi sp. nov., an exclusively respiratory luminous bacterium isolated from the Alboran Sea. Int J Syst Bacteriol 47:1034–1039
Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG (2009) A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218
Manukhov IV, Khrul'nova SA, Baranova A, Zavilgelsky GB (2011) Comparative analysis of the lux operons in Aliivibrio logei KCh1 (a Kamchatka Isolate) and Aliivibrio salmonicida. J Bacteriol 193:3998–4001
Martín Cuadrado AB, López García P, Alba JC, Moreira D, Monticelli L, Strittmatter A, Gottschalk G, Rodríguez Valera F (2007) Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One 2:e914
Martin M, Showalter R, Silverman M (1989) Identification of a locus controlling expression of the luminescence genes in Vibrio harveyi. J Bacteriol 171:2406–2414
McElroy WD, Seliger HH (1962) Origin and evolution of bioluminescence. In: Kasha M, Pullman B (eds) Horizons in biochemistry. Academic, New York, NY, pp 91–101
McFall Ngai M, Morin JG (1991) Camouflage by disruptive illumination in leiognathids, a family of shallow–water, bioluminescent fishes. J Exp Biol 156:119–137
McFall Ngai MJ, Ruby EG (1991) Symbiont recognition and subsequent morphogenesis as early events in animal–bacterial mutualism. Science 254:1491–1494
McFall–Ngai MJ (1983) Adaptations for reflection of bioluminescent light in the gas bladder of Leiognathus equulus (Perciformes: Leiognathidae). J Exp Zool 227:23–33
McFall–Ngai MJ, Dunlap PV (1983) Three new modes of luminescence in the leiognathid fish Gazza minuta: discrete projected luminescence, ventral body flash, and buccal luminescence. Mar Biol 73:227–237
Meighen EA (1988) Enzymes and genes from the lux operons of bioluminescent bacteria. Ann Rev Microbiol 42:151–176
Meighen EA (1991) Molecular biology of bacterial bioluminescence. Microbiol Rev 55:123–142
Meighen EA (1999) Autoinduction of light emission in different species of bioluminescent bacteria. Luminescence 14:3–9
Meighen EA, Dunlap PV (1993) Physiological, biochemical and genetic control of bacterial bioluminescence. Adv Microb Physiol 34:1–67
Meighen EA, Szittner RB (1992) Multiple repetitive elements and organization of the lux operons of luminescent terrestrial bacteria. J Bacteriol 174:5371–5381
Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199
Miyamoto CM, Meighen EA (2006) Involvement of LuxR, a quorum sensing regulator in Vibrio harveyi, in the promotion of metabolic genes argA, purM, lysE and rluA. Biochim Biophys Acta 1759:296–307
Miyamoto CM, Graham AF, Meighen EA (1988) Nucleotide sequence of the luxC gene and the upstream DNA from the bioluminescent system of Vibrio harveyi. Nucl Acids Res 16:1551–1562
Miyamoto CM, Chatterjee J, Swartzman E, Szittner R, Meighen EA (1996) The role of lux autoinducer in regulating luminescence in Vibrio harveyi control of luxR expression. Mol Microbiol 19:767–775
Miyamoto CM, Sun W, Meighen EA (1998) The LuxR regulator protein controls synthesis of polyhydroxybutyrate in Vibrio harveyi. Biochim Biophys Acta 1384:356–364
Miyamoto CM, Lin YH, Meighen EA (2000) Control of bioluminescence in Vibrio fischeri by the LuxO signal response regulator. Mol Microbiol 36:594–607
Miyamoto CM, Dunlap PV, Ruby EG, Meighen EA (2003) LuxO controls luxR expression in Vibrio harveyi evidence for a common regulatory mechanism in Vibrio. Mol Microbiol 48:537–548
Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG (2010) A single qrr gene is necessary and sufficient for LuxO–mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567
Molisch H (1912) Leuchtende Pflanzen. Eine Physiologische Studie. Gustav Fischer, Jena
Molisch H (1925) Botanische Beobachtungen in Japan. III Über das Leuchten des Schlacht–viehfleisches in Sendai. Japan Sci Rep Tohoku Imp Univ Biol 197–103
Moore SA, James MN (1995) Structural refinement of the non–fluorescent flavoprotein from Photobacterium leiognathi at 160 A resolution. J Mol Biol 249:195–214
Morin JG, Harrington A, Nealson K, Krieger N, Baldwin TO, Hastings JW (1975) Light for all reasons: versatility in the behavioral repertoire of the flashlight fish. Science 190:74–76
Müller Breitkreutz K, Winkler UK (1993) Anaerobic expression of the Vibrio fischeri lux regulon in E coli is Fnr–dependent. J Biolumin Chemilumin 8:108
Munk O, Hansen K, Herring PJ (1998) On the development and structure of the escal light organ of some melanocetid deep sea anglerfishes (Pisces Ceratioidei). J Mar Biol Assoc UK 78:1321–1335
Nealson KH (1977) Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch Microbiol 112:73–79
Nealson KH (1978) Isolation, identification and manipulation of luminous bacteria. In: DeLuca MA (ed) Methods in enzymology, vol 57. Academic, New York, NY, pp 153–166
Nealson KH (1979) Alternative strategies of symbiosis of marine luminous fishes harboring light–emitting bacteria. Trends Biochem Sci 4:105–110
Nealson, K. H., and D. S. Walton. 1978 Luciferase in non–luminous species of Beneckea. In: Abstracts of the Annual Meeting of the American Society for Microbiology, Washington, DC American Society for Microbiology. Washington DC, 1 1–131
Nealson KH, Hastings JW (1977) Low oxygen is optimal for luciferase synthesis in some bacteria. Ecological implications. Arch Microbiol 112:9–16
Nealson KH, Hastings JW (1979) Bacterial bioluminescence its control and ecological significance. Microbiol Rev 43:496–518
Nealson KH, Hastings JW (1992) The luminous bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd edn. Springer, Berlin, pp 625–639
Nealson KH, Venter JC (2007) Metagenomics and the global ocean survey what's in it for us, and why should we care? ISME J 1:185–187
Nealson KH, Platt T, Hastings JW (1970) Cellular control of synthesis and activity of the bacterial luminescence system. J Bacteriol 104:313–322
Nealson KH, Eberhard A, Hastings JW (1972) Catabolite repression of bacterial bioluminescence, functional implications. Proc Natl Acad Sci USA 69:1073–1076
Nealson KH, Haygood MG, Tebo BM, Roman M, Miller E, McCosker JE (1984) Contribution of symbiotically luminous fish to the occurrence and bioluminescence of luminous bacteria in seawater. Microb Ecol 10:69–77
Nealson KH, Wimpee B, Wimpee C (1993) Identification of Vibrio splendidus as a member of the planktonic luminous bacteria from the Persian Gulf and Kuwait region with luxA probes. Appl Environ Microbiol 59:2684–2689
Nelson JS (2006) Fishes of the World, 4th edn. Wiley, Hoboken, NJ
Nelson EJ, Tunsjø HS, Fidopiastis PM, Sørum H, Ruby EG (2007) A novel lux operon in the cryptically bioluminescent fish pathogen Vibrio salmonicida is associated with virulence. Appl Environ Microbiol 73:1825–1833
Neush J (1879) Bactéries lumineuses sur la viande fraîsche. J Pharm Chim Paris 29:20–22
Ng WL, Bassler BL (2009) Bacterial quorum–sensing network architectures. Annu Rev Genet 43:197–222
Nijvipakul S, Wongratana J, Suadee C, Entsch B, Ballou DP, Chaiyen P (2008) LuxG is a functioning flavin reductase for bacterial luminescence. J Bacteriol 190:1531–1538
Nishiguchi MK (2000) Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl Environ Microbiol 66:3550–3555
Nishiguchi MK, Ruby EG, McFall–Ngai MJ (1998) Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in Sepiolid squid–Vibrio symbioses. Appl Environ Microbiol 64:3209–3213
O’Brien CH, Sizemore RK (1979) Distribution of the luminous bacterium Beneckea harveyi in a semitropical estuarine environment. Appl Environ Microbiol 38:933–938
O’Grady EA, Wimpee CF (2008) Mutations in the lux operon of natural dark mutants in the genus Vibrio. Appl Environ Microbiol 74:61–66
O’Kane DJ, Prasher DC (1992) Evolutionary origins of bacterial bioluminescence. Mol Microbiol 6:443–449
O’Kane DJ, Karle VA, Lee J (1985) Purification of lumazine proteins from Photobacterium leiognathi and Photobacterium phosphoreum: bioluminescent properties. Biochemistry 24:1461–1467
O’Kane DJ, Woodward B, Lee J, Prasher DC (1991) Borrowed proteins in bacterial bioluminescence. Proc Natl Acad Sci 88:1100–1104
Ochman H, Lawrence JG, Groisman E (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Okada YK (1926) On the photogenic organ of the knight–fish Monocentris japonicus (Houttuyn). Biol Bull 50:365–373
Okada K, Iida T, Kita–Tsukamoto K, Honda T (2005) Vibrios commonly possess two chromosomes. J Bacteriol 187:752–757
Oliver JD, Roberts DM, White VK, Dry MA, Simpson LM (1986) Bioluminescence in a strain of the human pathogenic bacterium Vibrio vulnificus. Appl Environ Microbiol 52:1209–1211
Onarheim AM, Wiik R, Burghardt J, Stackebrandt E (1994) Characterization & identification of two Vibrio species indigenous to the intestine of fish in cold sea water; description of Vibrio iliopiscarious sp. nov. Syst Appl Microbiol 17:370–379
Orlov AM, Iwamoto T (2006) Grenadiers of the World Oceans: biology, stock assessment, and fisheries. American Fisheries Society, Bethesda, MD
Ortiz Conde BA, Muir DG, Pillidge CJ, Gobius KS, Anikis MS, Powell DM, Hori H, Colwell RR (1989) Nucleotide sequences of 5 S ribosomal RNAs from three marine eubacteria Shewanella hanedai, Alteromonas colwelliana and Vibrio mediterranei. Nucl Acids Res 17:4881
Owens L, Busico–Salcedo N (2006) Vibrio harveyi: pretty problems in paradise. In: Thompson FL, Austin B, Swings J (eds) The biology of Vibrios. American Society for Microbiology Press, Washington, DC, pp 266–280
Palmer LM, Colwell RR (1991) Detection of luciferase gene sequence in nonluminescent Vibrio cholerae by colony hybridization and polymerase chain reaction. Appl Environ Microbiol 57:1286–1293
Peat SM, Ffrench–Constant RH, Waterfield NR, Marokházi J, Fodor A, Adams BJ (2010) A robust phylogenetic framework for the bacterial genus Photorhabdus and its use in studying the evolution and maintenance of bioluminescence a case for 16 S, gyrB, and glnA. Mol Phylogenet Evol 57:728–740
Peel MM, Alfredson DA, Gerrard JG, Davis JM, Robson JM, McDougall RJ, Scullie BL, Akhurst RJ (1999) Isolation, identification, and molecular characterization of strains of Photorhabdus luminescens from infected humans in Australia. J Clin Microbiol 37:3647–3653
Petushkov VN, Ketelaars M, Gibson BG, Lee J (1996) Interaction of Photobacterium leiognathi and Vibrio fischeri Y1 luciferases with fluorescent (antenna) proteins bioluminescence effects of the aliphatic additive. Biochemistry 35:12086–12093
Pflüger E (1875) Ueber die Phosphorescenz verwesender Organismen. Arch ges Physiol Men Tiere 11:222–263
Platt TG, Fuqua C (2010) What's in a name? The semantics of quorum sensing. Trends Microbiol 18:383–387
Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL (2008) The Vibrio harveyi master quorum–sensing regulator, LuxR, a TetR–type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol 70:76–88
Potrikus CJ, Greenberg EP, Hamlett NV, Gupta S, Hastings JW (1984) Hybridization of Vibrio harveyi luciferase genes to non–luminous marine bacteria. In: Abstracts of the Annual Meeting of the American Society for Microbiology, St. Louis, MO American Society for Microbiology. Washington DC, 1–42
Preheim SP, Boucher Y, Wildschutte H, David LA, Veneziano D, Alm EJ, Polz MF (2011) Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ Microbiol 13:265–275
Pujalte MJ, Garay E (1986) Proposal of Vibrio mediterranei sp. nov. A new marine member of the Genus Vibrio. Int J Syst Bacteriol 36:278–281
Qin N, Callahan SM, Dunlap PV, Stevens AM (2007) Analysis of LuxR regulon gene expression during quorum sensing in Vibrio fischeri. J Bacteriol 189:4127–4134
Ramaiah N, Chun J, Ravel J, Straube WL, Hill RT, Colwell RR (2000) Detection of luciferase gene sequences in non–luminescent bacteria from the Chesapeake Bay. FEMS Microbiol Ecol 33:27–34
Ramesh A, Venugopalan VK (1989) Role of luminous bacteria in chitin degradation in the intestine of fish. MIRCEN J 5:55–59
Ramesh A, Balakrish Nair G, Abraham M, Natarajan R, Venugopalan VK (1987) Seasonal distribution of luminous bacteria in the tropical Vellar estuary. Microbios 52:151–159
Ramaiah N, Chandramohan D (1987) Distribution and species composition of planktonic luminous bacteria in the Arabian Sea. Indian J Mar Sci 16:139–142
Redfield RJ (2002) Is quorum sensing a side effect of diffusion sensing? Trends Microbiol 10:365–370
Reen FJ, Almagro Moreno S, Ussery D, Boyd EF (2006) The genomic code: inferring Vibrionaceae niche specialization. Nat Rev Microbiol 4:697–704
Rees JF, de Wergifosse B, Noiset O, Dubuisson M, Janssens B, Thompson EM (1998) The origins of marine bioluminescence turning oxygen defense mechanisms into deep–sea communication tools. J Exp Biol 201:1211–1221
Reichelt JL, Baumman P (1973) Taxonomy of the marine, luminous bacteria. Arch Mikrobiol 94:283–330
Reichelt JL, Baumann P, Baumann L (1976) Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridization. Arch Microbiol 110:101–120
Reichelt JL, Nealson K, Hastings JW (1977) The specificity of symbiosis pony fish and luminescent bacteria. Arch Microbiol 112:157–161
Robertson LA (2003) The Delft School of microbiology, from the nineteenth to the twenty–first century. Adv Appl Microbiol 52:357–388
Robertson LA, Figge MJ, Dunlap PV (2011) Beijerinck and the bioluminescent bacteria – microbiological experiments in the late 19th and early 20th centuries. FEMS Microbiol Ecol 75:185–194
Rosson RA, Nealson KH (1981) Autoinduction of bacterial bioluminescence in a carbon–limited chemostat. Arch Microbiol 129:299–304
Ruby EG (1996) Lessons from a cooperative bacterial–animal association The Vibrio fischeri–Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624
Ruby EG, Asato LM (1993) Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol 159:160–167
Ruby EG, Lee KH (1998) The Vibrio fischeri– Euprymna scolopes light organ association current ecological paradigms. Appl Environ Microbiol 64:805–812
Ruby EG, Morin JG (1978) Specificity of symbiosis between deep–sea fish and psychrotrophic luminous bacteria. Deep–Sea Res 25:161–171
Ruby EG, Morin JG (1979) Luminous enteric bacteria of marine fish. A study of their distribution, densities, and dispersion. Appl Environ Microbiol 38:406–411
Ruby EG, Nealson KH (1976) Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica, a model of symbiosis based on bacterial studies. Biol Bull 141:574–5867
Ruby EG, Nealson KH (1978) Seasonal changes in the species composition of luminous bacteria in nearshore seawater. Limnol Oceanogr 23:530–533
Ruby EG, Greenberg EP, Hastings JW (1980) Planktonic marine luminous bacteria species distribution in the water column. Appl Environ Microbiol 39:302–306
Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP (2005) Complete genome sequence of Vibrio fischeri a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA 102:3004–3009
Sasaki A, Ikejima K, Aoki S, Azuma N, Kashimura N, Wada M (2003) Field evidence for bioluminescent signaling in the pony fish, Leiognathus elongatus. Environ Biol Fish 66:307–311
Sato Y, Shimizu S, Ohtaki A, Noguchi K, Miyatake H, Dohmae N, Sasaki S, Odaka M, Yohda M (2010) Crystal structures of the Lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7–dimethyl–8–(1_–D–ribityl) lumazine, and its analogues, riboflavin and flavin mononucleotide, at high resolution. J Bacteriol 192:127–133
Schaefer AL, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP (1996) Generation of cell–to–cell signals in quorum sensing:acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA 93:9505–9509
Schauder S, Shokat K, Surette MG, Bassler BL (2001) The LuxS family of bacterial autoinducers biosynthesis of a novel quorum–sensing signal molecule. Mol Microbiol 41:463–476
Schmidt TM, Kopecky K, Nealson KH (1989) Bioluminescence of the insect pathogen Xenorhabdus luminescens. Appl Environ Microbiol 55:2607–2612
Seliger HH (1987) The evolution of bioluminescence in bacteria. Photochem Photobiol 45:291–297
Shadel GS, Baldwin TO (1991) The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J Bacteriol 173:568–574
Shadel GS, Baldwin TO (1992a) Identification of a distantly located regulatory element in the luxD gene required for negative autoregulation of the Vibrio fischeri luxR gene. J Biol Chem 267:7690–7695
Shadel GS, Baldwin TO (1992b) Positive autoregulation of the Vibrio fischeri luxR gene. J Biol Chem 267:7696–7702
Shadel GS, Devine JH, Baldwin TO (1990) Control of the lux regulon of Vibrio fischeri. J Biolumin Chemilumin 5:99–106
Shilo M, Yetinson T (1980) Physiological characteristics underlying the distribution patterns of luminous bacteria in the Mediterranean Sea and the Gulf of Elat. Appl Environ Microbiol 38:577–584
Shimada T, Arakawa E, Itoh K, Kosako Y, Okitsu T, Yamai S, Nishino M, Nakajima T (1995) Causative agent of the so–called “light disease of shrimps” is luminescent Vibrio cholerae non–O1. Nippon Saik Z 50:863–870
Showalter RE, Martin MO, Silverman MR (1990) Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J Bacteriol 172:2946–2954
Sicard M, Hering S, Schulte R, Gaudriault S, Schulenburg H (2007) The effect of Photorhabdus luminescens (Enterobacteriaceae) on the survival, development, reproduction and behaviour of Caenorhabditis elegans (Nematoda Rhabditidae). Environ Microbiol 9:12–25
Silverman M, Martin M, Engebrecht J (1989) Regulation of luminescence in marine bacteria. In: Hopwood DA, Chater KF (eds) Genetics of bacterial diversity. Academic, London, pp 71–86
Singleton RJ, Skerman TM (1973) A taxonomic study by computer analysis of marine bacteria from New Zealand waters. J R Soc New Zealand 3:129–140
Small ED, Koka P, Lee J (1980) Lumazine protein from the bioluminescent bacterium Photobacterium phosphoreum. Purification and characterization. J Biol Chem 255:8804–8810
Smith SK, Sutton DC, Fuerst JA, Reichelt JL (1991) Evaluation of the genus Listonella and reassignment of Listonella damsela (Love et al.) MacDonell and Colwell to the genus Photobacterium as Photobacterium damsela comb. nov. with an emended description. Int J Syst Bacteriol 41:529–534
Sparks JS, Smith WL, Dunlap PV (2005) Evolution and diversification of a sexually dimorphic luminescent system in ponyfishes (Teleostei: Leiognathidae), including diagnoses for two new genera. Cladistics 21:305–327
Spencer R (1961) Chitinoclastic activity of the luminous bacteria. Nature 190:938
Stacy AR, Diggle SP, Whiteley M (2012) Rules of engagement defining bacterial communication. Curr Opin Microbiol 15(2):155–162
Stevens AM, Greenberg EP (1997) Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J Bacteriol 179:557–562
Sun W, Cao JG, Teng K, Meighen EA (1994) Biosynthesis of poly–3–hydroxybutyrate in the luminescent bacterium, Vibrio harveyi, and regulation by the lux autoinducer, N–(3–hydroxybutanoyl) homoserine lactone. J Biol Chem 269:20785–20790
Sung ND, Lee CY (2004) Coregulation of lux genes and riboflavin genes in bioluminescent bacteria of Photobacterium phosphoreum. J Microbiol 42:194–199
Surete MG, Miller MB, Bassler BL (1999) Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA 96:1639–1644
Suwanto A, Yuhana M, Herawaty E, Angka SL (1998) Genetic diversity of luminous Vibrio isolated from shrimp larvae. In: Flegel TW (ed) Advances in shrimp biotechnology. National Center for Genetic Engineering and Biotechnology, Bangkok, pp 217–224
Swartzman E, Kapoor S, Graham AF, Meighen EA (1990a) A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. J Bacteriol 172:6797–6802
Swartzman E, Miyamoto C, Graham A, Meighen E (1990b) Delineation of the transcriptional boundaries of the lux operon of Vibrio harveyi demonstrates the presence of two new lux genes. J Biol Chem 265:3513–3517
Swartzman E, Silverman M, Meighen EA (1992) The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol 174:7490–7493
Swift S, Williams P, Stewart GSAB (1999) N–Acylhomoserine lactones and quorum sensing in proteobacteria. In: Dunny GM, Winans SC (eds) Cell–cell signaling in bacteria. American Society for Microbiology Press, Washington, pp 291–313
Tailliez P, Laroui C, Ginibre N, Paule A, Pagès S, Boemare N (2010) Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein–coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int J Syst Evol Microbiol 60:1921–1937
Thompson FL, Thompson CC, Li Y, Gomez Gil B, Vandenberghe J, Hoste B, Swings J (2003) Vibrio kanaloae sp. nov., Vibrio pomeroyi sp. nov. and Vibrio chagasii sp. nov., from sea water and marine animals. Int J Syst Evol Microbiol 53:753–759
Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J (2005) Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 71:5107–5115
Tiews K, Caces Borja P (1965) On the availability of fish of the family Leiognathidae Lacepede in Manila Bay and San Miguel Bay and on their accessibility to controversial fishing gears. Phil J Fish 7:59–83
Tu KC, Bassler BL (2007) Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev 21:221–233
Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL (2010) Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum–sensing response. Mol Cell 37:567–579
Ulitzur S, Dunlap PV (1995) Regulatory circuitry controlling luminescence autoinduction in Vibrio fischeri. Photochem Photobiol 62:625–632
Ulitzur S, Hastings JW (1979) Autoinduction in a luminous bacterium: a confirmation of the hypothesis. Curr Microbiol 2:345–348
Ulitzur S, Yashphe J (1975) An adenosine 3',5'–monophosphate–requiring mutant of the luminous bacteria Beneckea harveyi. Biochim Biophys Acta 404:321–328
Urakawa H, Kita Tsukamoto K, Ohwada K (1999) Reassessment of the taxonomic position of Vibrio iliopiscarius (Onarheim et al. 1994) and proposal for Photobacterium iliopiscarium comb. nov. Int J Syst Bacteriol 49:257–260
Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV (2007) Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int J Syst Evol Microbiol 57:2823–2829
Urbanczyk H, Ast JC, Kaeding AJ, Oliver JD, Dunlap PV (2008) Phylogenetic analysis of the incidence of lux gene horizontal transfer in Vibrionaceae. J Bacteriol 190:3494–3504
Urbanczyk H, Ast JC, Dunlap PV (2011a) Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol Rev 35:324–342
Urbanczyk H, Ast JC, Dunlap PV (2011b) Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol Rev 35:324–342
Urbanczyk H, Ogura Y, Hendry TA, Gould AL, Kiwaki N, Atkinson JT, Hayashi T, Dunlap PV (2011c) Genome Sequence of Photobacterium mandapamensis svers.11, the bioluminescent symbiont of the cardinal fish Siphamia versicolor. J Bacteriol 193:3144–3145
Urbanczyk H, Furukawa T, Yamamoto Y, Dunlap PV (2012a) Natural replacement of vertically inherited lux-rib genes of Photobacterium aquimaris by horizontally acquired homologs. Environ Microbiol Rep 4:412–416
Urbanczyk H, Kiwaki N, Furukawa T, Iwatsuki Y (2012b) Limited geographic distribution of certain strains of the bioluminescent symbiont Photobacterium leiognathi. FEMS Microbiol Ecol 81: 355–363
van Iterson G, Jr. den Dooren de Jong LE, Kluyver AJ (1940) Martinus Willem Beijerinck. His life and work. Martinus Nijhoof, The Hague
Vezzi A, Campanaro S, D'Angelo M et al (2005) Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459–1461
Visick KL, Ruby EG (2006) Vibrio fischeri and its host it takes two to tango. Curr Opin Microbiol 9:632–638
Wada M, Azuma N, Mizuno N, Kurokura H (1999) Transfer of symbiotic luminous bacteria from parental Leiognathus nuchalis to offspring. Mar Biol 135:683–687
Wada M, Kamiya A, Uchiyama N, Yoshizawa S, Kita Tsukamoto K, Ikejima K, Yu R, Imada C, Karatani H, Mizuno N, Suzuki Y, Nishid M, Kogure K (2006) luxA gene of light organ symbionts of the bioluminescent fish Acropoma japonicum (Acropomatidae) and Siphamia versicolor (Apogonidae) forms a lineage closely related to that of Photobacterium leiognathi ssp. mandapamensis. FEMS Microbiol Lett 260:186–192
Walker EL, Bose JL, Stabb EV (2006) Photolyase confers resistance to UV light but does not contribute to the symbiotic benefit of bioluminescence in Vibrio fischeri ES114. Appl Environ Microbiol 72:6600–6606
Waterfield NR, Ciche T, Clarke D (2009) Photorhabdus and a host of hosts. Annu Rev Microbiol 63:557–574
Waters CM, Bassler BL (2005) Quorum sensing cell–to–cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Waters CM, Bassler BL (2006) The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767
Wei SL, Young RE (1989) Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar Biol 103:541–546
Wei Y, Perez LJ, Ng WL, Semmelhack MF, Bassler BL (2011) Mechanism of Vibrio cholerae autoinducer–1 biosynthesis. ACS Chem Biol 6:356–365
Weleminsky F (1895) Die Ursachen des Leuchtens bei Choleravibrionen. Prager med Wochenschr Bd 20:263–264
West PA, Lee JV (1982) Ecology of Vibrio species, including Vibrio cholerae, in natural waters of Kent, England. J Appl Bacteriol 52:435–448
West PA, Lee JV, Bryant TN (1983) A numerical taxonomic study of species of Vibrio isolated from the aquatic environment and birds in Kent, England. J Appl Microbiol 55:263–283
Widder EA (2010) Bioluminescence in the ocean origins of biological, chemical, and ecological diversity. Science 328:704–708
Wilkinson P, Waterfield NR, Crossman L, Corton C, Sanchez Contreras M, Vlisidou I, Barron A, Bignell A, Clark L, Ormond D, Mayho M, Bason N, Smith F, Simmonds M, Churcher C, Harris D, Thompson NR, Quail M, Parkhill J, Ffrench Constant RH (2009) Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens. BMC Genomics 10:302
Wilson T, Hastings JW (1998) Bioluminescence. Annu Rev Cell Dev Biol 14:197–230
Wimpee CF, Nadeau TL, Nealson KH (1991) Development of species–specific hybridization probes for marine luminous bacteria by using in vitro DNA amplification. Appl Environ Microbiol 57:1319–1324
Wolfe CJ, Haygood MG (1991) Restriction fragment length polymorphism analysis reveals high levels of genetic divergence among the light organ symbionts of flashlight fish. Biol Bull 181:135–143
Wollenberg MS, Preheim SP, Polz MF, Ruby EG (2011) Polyphyly of non–bioluminescent Vibrio fischeri sharing a lux–locus deletion. Environ Microbiol. doi:101111/j.1462-2920201102608x
Woodland DJ, Cabanban AS, Taylor VM, Taylor RJ (2002) A synchronized rhythmic flashing light display by schooling Leiognathus splendens (Leiognathidae: Perciformes) Mar. Mar Freshwater Res 53:159–162
Yang Y, Yeh LP, Cao Y, Baumann L, Baumann P, Tang JSE, Beaman B (1983) Characterization of marine luminous bacteria isolated off the coast of China and description of Vibrio orientalis sp. nov. Curr Microbiol 8:95–100
Yasaki Y (1927) Bacteriologic studies on bioluminescence. 1 On the cause of luminescence in the fresh water shrimp, Xiphocaridina compressa (De Haan). J Infect Dis 40:404–407
Yasaki Y (1928) On the nature of the luminescence of the knight fish, Monocentris japonicus (Houttuyn). J Exp Zool 50:495–505
Yetinson T, Shilo M (1979) Seasonal and geographic distribution of luminous bacteria in the eastern Mediterranean Sea and the Gulf of Elat. Appl Environ Microbiol 37:1230–1238
Yoshizawa S, Wada M, Kita Tsukamoto K, Ikemoto E, Yokota A, Kogure K (2009a) Vibrio azureus sp. nov., a luminous marine bacterium isolated from seawater. Int J Syst Evol Microbiol 59:1645–1649
Yoshizawa S, Wada M, Kita Tsukamoto K, Yokota A, Kogure K (2009b) Photobacterium aquimaris sp. nov., a luminous marine bacterium isolated from seawater. Int J Syst Evol Microbiol 59:1438–1442
Yoshizawa S, Karatani H, Wada M, Yokota A, Kogure K (2010a) Aliivibrio sifiae sp. nov., luminous marine bacteria isolated from seawater. J Gen Appl Microbiol 56:508–518
Yoshizawa S, Wada M, Yokota A, Kogure K (2010b) Vibrio sagamiensis sp. nov., luminous marine bacteria isolated from seawater. J Gen Appl Microbiol 56:499–507
Zarubin M, Belkin S, Ionescu M, Genin A (2012) Bacterial bioluminescence as a lure for marine zooplankton and fish. Proc Natl Acad Sci USA. doi:101073/pnas.1116683109
Zenno H, Saigo K (1994) Identification of the genes encoding NAD(P)H–flavin oxidoreductases that are similar in sequence to Escherichia coli Fre in four species of luminous bacteria Photorhabdus luminescens, Vibrio fischeri, Vibrio harveyi, and Vibrio orientalis. J Bacteriol 176:3544–3551
Zenno I, Inouye S, Saigo K (1992) Does the luxG gene in luminous bacteria code for an NAD(P)H–FMN oxidoreductase? Genetics. Life Sci Adv 11:85–91
Zenno H, Saigo K, Kanoh H, Inouye S (1994) Identification of the gene encoding the major NAD(P)H–flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J Bacteriol 176:3536–3543
Zo YG, Chokesajjawatee N, Grim C, Arakawa E, Watanabe H, Colwell RR (2009) Diversity and seasonality of bioluminescent Vibrio cholerae populations in Chesapeake Bay. Appl Environ Microbiol 75:135–146
Zobell CE (1946) Marine microbiology. Chronica Botanica Company, Waltham
Zobell CE, Upham HC (1944) A list of marine bacteria including descriptions of sixty new species. Bull Scripps Inst Oceanogr 5:239–292
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this entry
Cite this entry
Dunlap, P.V., Urbanczyk, H. (2013). Luminous Bacteria. In: Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-30141-4_75
Download citation
DOI: https://doi.org/10.1007/978-3-642-30141-4_75
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-30140-7
Online ISBN: 978-3-642-30141-4
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences