Abstract
Barley is a resilient crop plant with higher tolerance than other cereal plants for several types of abiotic stress. In this chapter, we describe the genetic components underlying barley’s response to abiotic stresses, including soil acidity, boron toxicity, soil salinity, drought, temperature, and nutrient deficiency. We describe typical symptoms observed in barley in response to these stresses. We enumerate the major qualitative trait loci (QTLs) identified so far, such as FR-H1 and FR-H2 for low-temperature tolerance. We also discuss candidate genes that are the basis for stress tolerance, such as HVP10, which underlies the HvNax3 locus for salinity tolerance. Although knowledge about barley’s responses to abiotic stresses is far from complete, the genetic diversity in cultivated barley and its close wild relatives could be further exploited to improve stress tolerance. To this end, the release of the barley high-quality reference genome provides a powerful tool to facilitate identification of new genes underlying barley’s relatively high tolerance to several abiotic stresses.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
15.1 Introduction
Barley (Hordeum vulgare ssp. vulgare) is cultivated in both high-yielding, high-input agricultural systems, and in marginal, low-input agricultural environments. Barley was domesticated 10,000 years ago in the Fertile Crescent (Badr et al. 2000); however, its domestication was not confined to a single center of origin (Allaby 2015; Zohary 1999). In fact, Poets et al. (2015) recently showed that cultivated barley results from multiple sources of wild populations. Thus, the different geographic environments to which barley and its wild relatives have been exposed have resulted in a mosaic of adaptive variants containing tolerance to abiotic stresses (Allaby 2015). Barley is thus considered to be more resilient than other crops [for further details, see Newton et al. (2011)]. Despite this resilience, barley is affected by several abiotic stresses that decrease both yield and grain quality, which in turn result in considerable economic losses (e.g., the malting industry is directly affected by the yield and quality of barley). To breed for increased abiotic stress tolerance, we must harness the wealth of genetic variation provided by cultivated barley and its wild relatives. For instance, a wild allele from one H. vulgare ssp. spontaneum donor contributed to a 30% increase in barley yield under high soil salinity conditions in the field (Saade et al. 2016).
In this chapter, we describe the geographic extent of each abiotic stress that affects barley, the general symptoms barley exhibits in response to these stresses, mechanisms of stress tolerance and their molecular bases, and future research prospects and opportunities offered by the release of the high-quality genome sequence of barley (Mascher et al. 2017). Here, we review studies of barley’s responses to single stresses although we know that, under field conditions, barley plants are often exposed to more than one stress. For example, heat stress and drought can co-occur in arid regions (Savin et al. 1996) and boron toxicity is often observed at the same time as soil salinity and alkalinity (Goldberg 1997). While work on combined stresses is beyond the scope of this review, we aim to provide a comprehensive review of the major advances in research on the genetic mechanisms affecting barley’s tolerance to abiotic stresses.
15.2 Soil Acidity
Acidity in soil affects all the world’s regions to some extent; it is a major problem in developing countries where management practices are often unable to ameliorate pH decreases (FAO 2015). It was estimated that soil acidity affects over 40% of the world’s arable land (Kochian 1995). Aluminum (Al) is the main constraint in acidic soils (Foy et al. 1978). Under acidic conditions, Al is solubilized to the toxic Al3+, which inhibits root elongation and affects absorption of several plant nutrients such as Ca, Mg, P, and K (see Zhang and Li (2016) and Foy et al. (1978) for details). Symptoms of Al toxicity in plants include root thickening and darkening; phosphorus deficiency-like symptoms in the shoots including overall stunting with small, dark green leaves and late maturity; purpling of stems, leaves and leaf veins; and yellowing and death of leaf tips.
Barley is among the most Al-sensitive of the cereal crops, although genetic variation in Al tolerance exists in barley’s germplasm (Reid et al. 1969). The differences in Al tolerance in barley genotypes have been attributed to differences in the release of organic acid anions, such as malate and citrate, from root apices (Ma et al. 2001; Ryan et al. 2001). Once these organic acid anions are released from root cells, they can bind Al3+, reducing its availability and thus toxicity (Zhou et al. 2013). The genetic mechanisms controlling the release of organic acid anions are likely associated with allelic variation at several loci.
Early research on the genetic control of Al tolerance in barley using the Dayton (Al-tolerant) and Smooth Awn 86 (Al-sensitive) genotypes detected a major locus, Alp, on the long arm of chromosome 4H (4HL) (Minella and Sorrells 1992; Reid 1971). Another early study found that a single gene, named Pht, on chromosome 4H was responsible for tolerance to high acidity in the soil (Stolen and Andersen 1978). The same chromosomal location on chromosome 4HL also included the Al tolerance locus, Alt, which was mapped using amplified fragment length polymorphism (AFLP) in an F2 progeny derived from a cross between the Yambla (moderately Al-tolerant) and WB229 (Al-tolerant) genotypes (Raman et al. 2002). Later, Minella and Sorrells (1997) showed that tolerance to low pH (Pht) and tolerance to aluminum (Alp) are controlled by the same locus. Ma et al. (2004) used F2 seedlings from a cross between the Murasakimochi (Al-tolerant) and Morex (Al-sensitive) genotypes to identify microsatellite markers associated with Al tolerance and citrate secretion on chromosome 4H. High-resolution mapping of Alp suggested that HvMATE, which encodes a multidrug and toxic compound extrusion (MATE) protein, may underlie Al tolerance (Wang et al. 2007).
In 2007, Furukawa et al. (2007) identified HvAACT1 as the gene underlying the Al tolerance locus. This gene was found to encode a citrate transport protein located on the plasma membrane and was identified by fine mapping using the F4 recombinants between the Murasakimochi and Morex genotypes. HvAACT1 is a member of the MATE family; it is constitutively expressed in the roots of the Al-tolerant barley cultivar Murasakimochi (Furukawa et al. 2007). Interestingly, although expression of HvAACT1 was not induced by Al, a good correlation was found between Al tolerance and citrate release from barley root apices; the HvAACT1 expression level was determined to be higher in Al-tolerant genotypes (Furukawa et al. 2007). Constitutive overexpression of HvAACT1 in transgenic barley and wheat plants demonstrated the ability of HvAACT1 to increase citrate efflux and Al tolerance in acidic soils (Zhou et al. 2013). Also, barley plants transformed with sorghum SbMATE showed an increased Al-dependent citrate efflux from root apices and Al tolerance when compared with null segregant plants (Zhou et al. 2014). The presence of a 1-kb insertion in the 5′ untranslated region (UTR) was later linked to the higher expression of HvAACT1 in Al-tolerant genotypes (Fujii et al. 2012). This mutation altered protein expression from root pericycle cells in sensitive genotypes to the root apices of Al-tolerant genotypes. This finding affects our understanding of the original role of HvAACT1 in facilitating the translocation of iron from roots to shoots (Fujii et al. 2012). More recently, another allele was found in HvAACT1 of the Al-tolerant Chinese genotype CXHKSL, but the mechanisms leading to this tolerance are yet to be confirmed (Ma et al. 2016).
In wheat, Al tolerance, controlled by a gene located in chromosome 4DL and expressed constitutively in root apices, depends on malate efflux from root apices (Raman et al. 2005; Sasaki et al. 2004). Indeed, the transgenic Golden Promise (Al-sensitive) barley genotype that expresses TaALMT1 under ubiquitin promoter exhibits Al-activated malate efflux and enhanced Al tolerance (Delhaize et al. 2004). Zhou et al. (2014) showed that barley lines transformed with wheat TaALMT1 were more Al-tolerant than barley lines overexpressing HvAAC1 and lines transformed with SbMATE. The closest homolog of TaALMT1 in barley is HvALMT1, which is located on chromosome 2H and is mainly expressed in stomatal guard cells (Gruber et al. 2010). In Xenopus oocytes, HvALMT1 is weakly activated by Al3+, facilitating the movement of malate and other ions across oocyte membranes. Thus, HvALMT1 has been proposed to facilitate organic anion transport to regulate turgor in stomatal guard cells and root elongation zones (Gruber et al. 2010).
Using a genome-wide association study, Zhou et al. (2016) found 22 QTLs associated with Al tolerance, including HvAAC1, but also presented unknown QTLs underlying acid soil resistance. Although the genetic mechanisms underlying Al tolerance in barley are quite well known, it is likely that publication of the barley high-quality genome sequence (Mascher et al. 2017) will facilitate the discovery of other genes related to the release of organic acid anions from root apices and new alleles of HvAACT1. In the field, barley farmers should follow effective soil management practices and use Al-tolerant germplasm to overcome the negative effects of acidic soils on yield.
15.3 Boron Toxicity
The range in which soil is considered neither deficient of boron (B) nor toxic from B, an essential plant nutrient, is quite narrow (Goldberg 1997). In arid and semiarid regions with low rainfall, soluble B is only partially leached from the subsoil, and the problem of high B concentration in the soil (B toxicity) becomes evident (Reid 2010; Ryan et al. 1998). In particular, plants in Australia (Cartwright et al. 1986) and some areas of the Middle East, such as Syria (Ryan et al. 1998), suffer from B toxicity. Symptoms of B toxicity include chlorosis and necrosis starting at the leaf tips. Brown lesions also appear in the margins of the oldest leaves and progressively affect the whole shoot (Jefferies et al. 1999). Also, the growth of shoots and roots is retarded, which results in reduced yield (Holloway and Alston 1992; McDonald et al. 2010). B toxicity in barley is managed by the use of B-tolerant genotypes that can maintain low B concentrations in the shoots (Yau and Ryan 2008). For example, Sahara 3771 (a B-tolerant Algerian landrace) is a promising source of B tolerance alleles because it can maintain growth under high B conditions. Under the same conditions, Schooner (a B-sensitive genotype) showed shoot and root biomass reduction by 79 and 51%, respectively, compared with plants grown under control conditions. Furthermore, the roots of Sahara 3771 plants had lower B concentrations than in the external solution, whereas the roots of Schooner plants had equal B concentrations to the growth solution. Hayes and Reid (2004) concluded that the mechanism responsible for B tolerance in Sahara 3771 was related to the B efflux from the roots.
Jefferies et al. (1999) used restriction fragment length polymorphism (RFLP) markers and doubled-haploid lines resulting from the cross between the Sahara (B-tolerant) and Clipper (B-sensitive) genotypes to map regions of the barley genome associated with B tolerance. They found four regions (on chromosomes 2H, 3H, 4H, and 6H) associated with different traits involved in B tolerance. Regions on chromosomes 4HL and 3HS associated with relative root length (the root length at a B concentration of 0 mg/L relative to the root length at 100 mg/L of B) accounted for 39% of the trait variation. Other regions on chromosomes 2H and 4H associated with the leaf symptom score accounted for 38% of the observed variation. The region on chromosome 4H was also associated with shoot dry mass, and together with a region on chromosome 6H, accounted for 53% of the variation observed in the B concentration in shoots. The alleles coming from Sahara conferred higher relative root length, lower leaf symptoms, higher dry mass, and lower B concentration in the shoots at the respective loci (Jefferies et al. 1999). Sutton et al. (2007) identified HvBot1 as the gene underlying the 4H QTL responsible for B tolerance in Sahara, with higher transcript levels of HvBot1 related to B tolerance (Sutton et al. 2007). Hayes et al. (2015) found that the nine Saharan accessions (Sahara 3763–Sahara 3771) from the Australian barley germplasm collection had tandemly duplicated Sahara alleles that conferred B tolerance. Genotypes carrying the duplicated (estimated four copies) Sahara allele of HvBot1 had higher gene expression in the roots (more than a 200-fold increase) (Hayes et al. 2015), where it functions as an efflux-type borate anion transporter (Sutton et al. 2007). However, more investigation of HvBot1 is needed to clarify the mechanism of B efflux (Hayes and Reid 2004).
More recently, Hayes et al. (2015) found eight synonymous single nucleotide polymorphisms (SNPs) in the HvBot1 coding regions of the Clipper and Sahara alleles, although all were silent mutations. They therefore attributed the difference between alleles solely to expression levels. Emebiri et al. (2009) used marker-assisted selection (MAS) to introgress the favorable Sahara alleles at the 4H locus to the breeding line VB9104, which is adapted to conditions in southern Australia. However, results from field experiments conducted at four locations on the generated nearly isogenic lines were inconsistent because, in some locations, genotypes with the B tolerance allele had higher yields, whereas, in other locations, these genotypes had lower yields, suggesting a significant genotype-by-environment interaction. However, malting quality was not affected, suggesting that the introgression of the B tolerance allele had no penalty on quality (Emebiri et al. 2009). Similarly, McDonald et al. (2010) found that introgressing B tolerance alleles from Sahara at the 2H and 4H loci had little or no effect on yield gain in the field and that yield gain was largely affected by site location. This recombinant inbred line (RIL) population had yields similar to or lower than their progenitors although they exhibited reduced B toxicity symptoms and reduced B concentrations in their shoots. This result could be because B-tolerant plants grow vigorously during early stages, thus using up the limited water available in arid regions and therefore affecting plants at the reproductive stage. Another reason could be that B toxicity is often accompanied by other constraints, such as high salinity. In fact, McDonald (2006) showed earlier that variation in yield in soils in which both stresses occurred simultaneously is due to salinity rather than to B toxicity. Tissue tolerance to B and the ability to overcome other soil constraints are both necessary for increasing yield under high B concentrations (McDonald et al. 2010).
The B tolerance gene underlying the 6H QTL, identified by Jefferies et al. (1999), was cloned by Schnurbusch et al. (2010). This gene, HvNIP2;1, encodes an aquaporin that belongs to the subfamily of nodulin-26-like intrinsic proteins (NIPs). NIPs are channel proteins that allow transport of water and other small solutes such as boric acid (Takano et al. 2006). The expression of HvNIP2;1 was found to be specific to the roots and to increase the permeability of the plasma membrane to B in heterologous systems (i.e., Xenopus oocytes and yeast cells). In addition, the roots of Clipper (B-sensitive genotype) plants had 15-fold more HvNIP2;1 transcripts compared with those of B-tolerant Sahara. Differences in the regulatory sequence regions, upstream of the ATG codon, may explain the differences in transcript levels of HvNIP2;1 between the two genotypes. One mechanism of barley tolerance under high B concentrations in the soil could be the reduced expression of HvNIP2;1 to limit the passive influx of B from the roots (Schnurbusch et al. 2010). This mechanism, combined with high expression of HvBot1, may increase B tolerance. In B-limited environments, higher HvNIP2;1 expression would be beneficial for plants to avoid deficiency in this nutrient (Schnurbusch et al. 2010; Takano et al. 2006). Hayes et al. (2015) screened 65 barley genotypes for differences in the coding sequence and transcription level of HvNIP2;1 and found that the open reading frame of HvNIP2;1 was highly conserved. However, Sahara genotypes presented a unique SNP in the 5′ UTR region of HvNIP2;1. This SNP, 44 bp upstream of the start codon, resulted in a shorter upstream open reading frame that translated into a short polypeptide, resulting in lower expression in Sahara (Hayes et al. (2015). Limited knowledge is available about the genes underlying the 2H and 3H QTL. HvBot2, a B-transporter gene, has been suggested as a candidate gene underlying the 3H QTL because Sahara 3771 has a deletion in the 3′ end of the coding sequence, which may disrupt the protein function. The publication of the barley high-quality genome sequence (Mascher et al. 2017) will allow advancement of the research needed to clarify the B tolerance mechanisms underlying the 2H and 3H mechanisms.
15.4 Soil Salinity
Soil salinity is a major constraint on agriculture and impacts food security and political stability. Currently, 1128 Mha, including 20% of irrigated lands (http://www.fao.org/water/en/), are estimated to be affected by soil salinity. The largest such area, of 189 Mha, is located in the Middle East (Wicke et al. 2011). These numbers are expected to increase due to climate change and poor irrigation practices (Hayes et al. 2015). Qadir et al. (2014) estimated that an annual economic loss of US$27.3 billion is due to soil salinity. Salinity tolerance is a complex polygenic mechanism (Roy et al. 2014) that enables plants to maintain growth and to produce photoassimilates under saline conditions (Munns 2002). The response of plants to salinity includes several tolerance mechanisms, namely shoot ion-independent tolerance (or osmotic tolerance), ionic tolerance and tissue tolerance (Munns and Tester 2008; Roy et al. 2014). Numerous genes have been proposed to contribute to salinity tolerance traits (Roy et al. 2014).
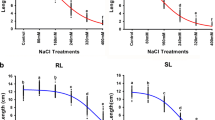
Barley is the most salt-tolerant cereal crop (Munns and Tester 2008). This distinction for barley could partially be due to salinity tolerance in its tissues. For example, evidence exists that is consistent with barley having better ability than durum wheat to compartmentalize Na+ in vacuoles (James et al. 2006). A high cytoplasmic K+/Na+ ratio in mesophyll cells would allow barley to maintain its photosynthetic capacity under saline conditions (James et al. 2006; Wu et al. 2013). Besides the ability of barley to accumulate and tolerate high levels of Na+, Na+ exclusion is still an important mechanism in barley salinity tolerance. Halophytic relatives of barley have been used in several studies to understand the differences in the mechanisms of salinity tolerance between species (Islam et al. 2007; Maršálová et al. 2016). For example, Garthwaite et al. (2005) reported that seven wild Hordeum species were better able to exclude Na+ from their shoots compared with cultivated barley, when grown on nutrient solutions with different NaCl concentrations. Hence, improving cultivated barley salinity tolerance could be done by making use of exotic germplasm. In fact, Nax3 and Nax4 loci, described below, were identified using a landrace (Sahara 3771) and a wild barley (Hordeum spontaneum) accessions.
Barley wild relatives are an important source of genes to increase salinity tolerance. For example, Saade et al. (2016) identified a wild allele on chromosome 2H that resulted in a 30% yield increase under saline conditions. Also, the overexpression of HsCBL8 gene from a H. spontaneum line native to the Qinghai–Tibet Plateau resulted in an increased salinity tolerance in rice (Guo et al. 2016).
The ability of barley to retain K+ in its cells under saline conditions is possible through high-affinity K+ transporters, such as HvHAK1 (Santa-Maria et al. 1997). In fact, HvHAK1 expression was higher in the shoots and roots of K305 (a salt-tolerant genotype) compared with I743 (a salt-sensitive genotype) (Ligaba and Katsuhara 2010). Na+ is sequestered in the vacuoles away from the cytoplasmic machinery via Na+/H+ antiporters localized on the tonoplasts (vacuolar membranes). The electrochemical gradient generated by the vacuolar H+-inorganic pyrophosphatase (V-PPase) and the vacuolar H+-ATPase (V-ATPase) provides the driving force for this movement (Rea and Poole 1993; Sze et al. 1992). Fukuda et al. (2004) studied the effects of 400 mM of mannitol (osmotic effect) and 200 mM of NaCl (ionic effect) on the expression of two V-PPases (HVP1 and HVP10), V-ATPase subunit A (HvVHA-A) and Na+/H+ antiporter (HvNHX1). One copy of each gene was detected in the barley genome. The transcript level of HvNHX1 increased fivefold in the roots of barley plants treated with 200 mM of NaCl compared with the roots of control plants, suggesting that HvNHX1 plays a role in salinity tolerance. Treatment with 400 mM mannitol increased the transcript level of HvNHX1 fourfold compared with the transcript level in control plants. HvNHX1 protein could thus be transporting ions other than Na+ into the vacuoles. In fact, AtNHX1 has a K+/H+ exchange function (Bassil et al. 2011; Venema et al. 2002) and plays an important role in turgor regulation and stomatal function through K+ uptake at the tonoplast (Barragan et al. 2012). V-PPase HVP10 exhibited a higher transcript level in roots than in shoots, whereas HVP1 exhibited higher shoot expression. HVP10 and HvVHA-A transcript levels in the roots increased in response to the addition of NaCl and decreased in response to the addition of mannitol, while the transcript level of HVP1 increased in response to the addition of both. HVP10, HvVHA-A, and HVP1 transcript levels did not change in the shoots (Fukuda et al. 2004).
HVP10 was proposed as a strong candidate underlying HvNax3, a locus on the short arm of chromosome 7H identified in a biparental population resulting from a cross between the wild barley CPI-71284-48 (H. vulgare ssp. spontaneum) and the cultivar Barque-73 (Shavrukov et al. 2010, 2013). HvNax3 was found to be associated with Na+ exclusion, explaining 10–25% difference in leaf Na+ of plants grown hydroponically under salinity (Shavrukov et al. 2013). The importance of vacuolar pyrophosphatase in improving salinity tolerance was best illustrated by Schilling et al. (2014) who found that transgenic barley genotypes expressing the arabidopsis vacuolar pyrophosphatase gene (AVP1) had a larger shoot biomass (30–42%) compared with wild-type genotypes under salinity conditions in the field. Furthermore, transgenic barley AVP1 genotypes had a higher yield as indicated by a larger number of heads per plant (16–58%), grains per plant (76–85%), average grain weight (29–43%), and grain yield per plant (79–87%) (Schilling et al. 2014). A third vacuolar Na+/H+ antiporter, HvNHX2, was isolated in barley (Vasekina et al. 2005) and increased salinity tolerance was observed when HvNHX2 was overexpressed in arabidopsis (Bayat et al. 2011) and potato plants (Bayat et al. 2010).
The role played by the so-called high-affinity potassium transporters (HKT) in the ionic component of salinity tolerance has been clearly established. In fact, TmHKT1;4-A2 and TmHKT1;5-A were suggested as candidate genes underlying two sodium exclusion loci, Nax1 and Nax2, respectively. When grown in saline fields, durum wheat genotypes carrying the TmHKT1;5-A locus had lower concentrations of leaf Na+ and 25% higher grain yield compared with near-isogenic lines lacking TmHKT1;5-A. More importantly, TmHKT1;5-A had no effect on yield in low-salinity soils (Munns et al. 2012). HKTs can be classified into two groups based on their amino acid sequences (Platten et al. 2006). Class I HKTs are more likely to be Na+-transporters whereas class II HKTs (high-affinity Na+) function as Na+/K+ symporters or Na+- or K+-uniporters (Hauser and Horie 2010; Maser et al. 2002). Furthermore, some class I HKT transporters are localized in the plasma membrane of xylem parenchyma cells in the root stele, allowing them to retrieve Na+ from the xylem and to prevent Na+ from reaching the shoot (Byrt et al. 2007). Class II HKT proteins, which are localized at the root epidermis/cortex, play an important role in plant growth under K+-limited conditions, and they may be downregulated under salinity conditions (Huang et al. 2008).
Identifying the specific functions of HKT has been challenging and controversial, especially when heterologous systems are used (Haro et al. 2005). In barley, two HKT proteins have been characterized: HvHKT1;5 (class I) and HvHKT2;1 (class II) (Hauser and Horie 2010). Mian et al. (2011) reported that HvHKT2;1 was predominantly expressed in the root cortex and cotransported Na+ and K+ when expressed in Xenopus oocytes. Also, transgenic barley lines overexpressing HvHKT2;1 had a higher relative growth rate compared with wild plants under saline conditions (Mian et al. 2011). Furthermore, HKT2;1/2-like, HKT2;3/4-like, HKT1;1/2-like, HKT1;3-like, and HKT1;4-like genes have been mapped on respective regions of barley chromosomes 7HL, 7HL, 2HL, 6HS, and 2HL. These regions are in synteny with regions in rice that contain HKT genes (OsHKT2;1, OsHKT2;4, OsHKT1;1 & OsHKT1;2, OsHKT1;3, and OsHKT1;4, respectively); they could thus harbor rice HKT orthologs (Huang et al. 2008).
Little is known about the shoot ion-independent tolerance mechanism in plants (Roy et al. 2014). Rapid and long-distance sensing seems to require reactive oxygen species and calcium waves (Choi et al. 2014; Gilroy et al. 2014). The salt-overly sensitive (SOS) pathway is a putative signaling mechanism in salinity response (Munns and Tester 2008). In barley, the homolog of arabidopsis SOS3, HvCBL4, has been identified as a candidate gene underlying the HvNax4 locus (Rivandi et al. 2011). HvNax4 was initially identified using a mapping population of doubled-haploid lines derived from a cross between Clipper and Sahara 3771. It is located on chromosome 1HL (Lonergan et al. 2009). Rivandi et al. (2011) characterized HvNax4 using the same population. They found that HvNax4 governed the Na+-concentration in the shoots of plants grown in pots, but it had no significant effect on plants grown under hydroponic conditions. Moreover, the effect of HvNax4 on the Na+ content in shoots varied with soil type, suggesting that HvNax4 expression was strongly influenced by the environment. Furthermore, the mRNA expression of HvCBL4 did not vary between Clipper and Sahara 3771, but an amino acid residue change was proposed to alter the protein function between the two genotypes. Because HvNax4 expression was found to be highly dependent on the environment and because it did not significantly affect plant biomass, the agronomical importance of this locus has not yet been determined. Further progress in research on the shoot ion-independent tolerance mechanism in barley is expected due to the advances in high-throughput phenotyping methodologies (Saade et al., unpublished data). Such research employing a wide range of barley genotypes may clarify which genomic regions (and/or genes) are responsible for the shoot ion-independent component of salinity tolerance.
One should note that besides the QTLs presented here, a large body of genetic studies has identified QTLs associated with traits contributing to salinity tolerance in barley. For instance, Long et al. (2013) identified two QTLs on chromosome 3H (126.3 cM) and 6H (60.2 cM) that are associated with salinity tolerance, defined as the biomass production under saline relative to nonsaline conditions. Other study focused on plant survival and leaf chlorosis, and identified a QTL on chromosome 4H (145 cM) associated with these traits (Fan et al. 2016b). Liu et al. (2017) related stomatal traits and yield to salinity tolerance, and found QTLs on chromosomes 1H (11 cM) and 3H (58.9 cM) associated with grain yield, and QTLs on chromosome 1H (121.4 cM) and 2H (10.3 cM) associated with stomatal pore area and stomatal conductance, respectively. Moreover, QTLs associated with salinity tolerance were identified at different developmental stages such as salinity tolerance at germination (Mano and Takeda 1997), seedling (Ellis et al. 2002; Mano and Takeda 1997), and reproductive stage (Liu et al. 2017; Xue et al. 2009). Barati et al. (2017) identified several QTLs contributing to agronomic performance in the field under nonsaline and saline conditions using a doubled-haploid population derived from Clipper and Sahara. The barley high-quality reference genome (Mascher et al. 2017) will allow the identification of the candidate genes underlying many of these described QTLs. In addition, the publication of the high-quality barley genome sequence (Mascher et al. 2017) allows the identification of homologs of important players in salinity tolerance, such as the Na+/Ca2+ exchanger-like protein AtNCL (Wang et al. 2012a). Also, salt tolerance genes that are absent from the barley genome may be identified. For instance, AtCIPK16, a calcineurin B-like protein interacting protein kinase that reduces shoot Na+ in arabidopsis (Roy et al. 2013), does not have a homolog in barley (Amarasinghe et al. 2016). Moreover, the genetic position of some barley genes involved in salinity tolerance, such as HVP1 and other candidate genes underlying QTL regions associated with salinity tolerance (Long et al. 2013; Ma et al. 2015), can be further pinpointed using the barley high-quality reference genome (Mascher et al. 2017).
15.5 Drought
In a general sense, drought can be defined as a lack of precipitation over a long period of time affecting certain activities and sectors (National Drought Mitigation Center (NDMC), http://drought.unl.edu/DroughtBasics/WhatisDrought.aspx). However, drought is a complex phenomenon that can be divided into four categories: meteorological, hydrological, agricultural, and socioeconomic. In this section, we discuss agricultural drought (hereafter drought), which refers to a condition in which soil moisture does not meet the needs of a particular crop at a particular growth stage. The occurrence of drought has become more frequent due to climate change; this increasing frequency has severe consequences for agriculture and, consequently, for economic stability and food security. The impact of drought on crop yield is enormous. For example, severe drought in Kenya caused wheat yield to decrease by 45% in 2009 compared with the yield in 2010. The 2011 drought that affected the grain belt and large parts of the United States increased food prices across the globe (http://www.fao.org/docrep/017/aq191e/aq191e.pdf).
Symptoms of drought stress include slower plant growth and leaf expansion, deeper root systems, stomatal closure to reduce transpiration (which in turn increases leaf temperature and affects photosynthesis) and the accumulation of compatible solutes such as proline, sorbitol, and glycine betaine to adjust the cytoplasmic osmotic potential (Baerenfaller et al. 2012; Daszkowska-Golec and Szarejko 2013; Farooq et al. 2009; Singh et al. 2015; Tombesi et al. 2015; Uga et al. 2013). A pot experiment, using barley landraces from Iran, showed that drought can decrease the relative water content in leaves, the midday leaf water potential, and stomatal conductance (by 90%) compared with plants in well-watered pots, thus reducing photosynthetic activity (Robredo et al. 2007, 2010)
Another pot experiment, using 11-day-old barley seedlings showed that root mass aggregated towards the bottom of the pot and adventitious root formation was halted under drought conditions. In addition, a 10-fold increase in proline, involved in osmotic adjustment, was observed in barley roots in response to drought, as well as a severe reduction in the shoot-to-root ratio (Sicher et al. 2012).
Because of the importance of drought stress, several studies have focused on genetic mapping of drought-associated traits. Fan et al. (2015) examined a mapping population derived from a cross between TX9425 (a drought-tolerant Chinese landrace) and Franklin (a drought-sensitive genotype). They used wilting as the trait to evaluate drought response. This work led to the identification of a QTL on chromosome 5H that was independent of two other developmental traits—awn length and heading date. The gene underlying this QTL was suggested to be 9-cis-epoxycarotenoid dioxygenase 2 (HvNCED2), which is involved in the synthesis of abscisic acid (Fan et al. 2015). Mikolajczak et al. (2017) identified 103 QTLs associated with yield-related traits when 100 RILs from a cross between European and Syrian cultivars were grown under different water regimes.
Roots play a crucial role in acquiring water under drought stress. In fact, the smaller size of the root system in spring barley was associated with lower grain yield in dry environments (Chloupek et al. 2010). Reinert et al. (2016) used a barley diversity panel composed of modern cultivars, landraces, and wild accessions to identify a QTL on chromosome 5H that was induced by drought and associated with the dry weight of roots. HvCBF10B and HvCBF10A were suggested as candidate genes underlying this QTL because their conserved domains harbored a large deletion of 37 amino acids in wild barley compared with cultivated genotypes.
RILs from the mapping population between Tadmor (a drought-tolerant Syrian genotype) and Er/Apm (a moderately drought-tolerant genotype) were grown in pots in growth chambers under control and drought conditions. A QTL on chromosome 7H was associated with the relative water content and osmotic potential of leaves (Teulat et al. 1997, 1998, 2001a). It was cosegregated with the Acl3 locus (Hansen and von Wettstein-Knowles 1991). Acl3 encodes barley acyl carrier protein III, which is involved in the synthesis of the fatty acyl chain (Hansen and von Wettstein-Knowles 1991). Acl3 may play a role in membrane protection or fluidity under stress conditions and thus improve drought tolerance (Teulat et al. 1998). Another study associated this QTL with carbon isotope discrimination in the field, which is a measure of transpiration efficiency (Teulat et al. 2002). Under field conditions, Teulat et al. (2001b) and von Korff et al. (2008) evaluated the same RIL population for agronomic traits across different Mediterranean environments with varying drought severity. The genetic effects on flowering time, kernel weight, and plant height were stable across these environments, and QTLs associated with these traits were detected in both studies with comparable effects. However, yield seemed to be more influenced by the environment than other traits. In particular, the Tadmor allele at the pHva1 marker had a favorable effect on grain yield in the locations with more severe drought (von Korff et al. 2008). This marker colocates with HVA1 gene on chromosome 1H, which also plays a role in low-temperature tolerance (Tondelli et al. 2006; von Korff et al. 2008). The same locus also had a significant effect on the wilting score in a mapping population derived from a cross between Scarlett (a drought-sensitive German genotype) and ISR42-8 (a wild accession of H. spontaneum). The HVA1 QTL explained 12% of the phenotypic variation, with the wild allele contributing to a 17% decrease in the wilting score relative to the domesticated allele (Sayed et al. 2012). Transgenic studies showed that wheat, rice, and oat transformed with barley HVA1 had higher drought and salinity tolerance than control plants had (Babu et al. 2004; Bahieldin et al. 2005; Oraby et al. 2005; Xu et al. 1996).
Other studies, such as Suprunova et al. (2007), identified Hsdr4 (H. spontaneum dehydration-responsive 4), which codes for a Rho-GTPase-activating-like protein and is involved in drought tolerance in wild barley. The arabidopsis Rho GTPase homolog (AtRac1) is known to play a central role in abscisic-acid-mediated stomatal closure (Lemichez et al. 2001). Besides stomatal closure, the cuticle also plays a role in plants’ adaptation to drought because it limits water loss. In barley, several eceriferum (cer) genes involved in the deposition of epicuticular waxes were detected in a mutagenesis study (Lundqvist and Lundqvist 1988). Recently, Cer-cqu (Cer-c, Cer-q, and Cer-u) has been identified on chromosome 2HS as a cluster of three separate genes encoding: (1) a chalcone synthase-like polyketide synthase, (2) a lipase/carboxyl transferase, and (3) a P450 enzyme (Hen-Avivi et al. 2016; Schneider et al. 2016). Chen et al. (2004) identified a spontaneous wild barley mutant genotype, eibi1, which was found to be hypersensitive to drought with a low capacity to retain leaf water. The hypersensitivity of this genotype was due to a thin cuticle that resulted from low cutin deposition. The eibi1 gene was mapped to the pericentromeric region of chromosome 3H (Chen et al. 2009), and HvABCG31, an ATP-binding cassette subfamily G (ABCG) transporter, was identified as the gene underlying eibi1 (Chen et al. 2011).
Drought-induced root and leaf proteomic and metabolomic changes revealed that several proteins and metabolites, namely HSP70, proline, carbohydrates and ascorbic acid, accumulated in response to drought in drought-sensitive genotypes, but these proteins and metabolites were constitutively elevated in drought-tolerant genotypes. Hence, biochemical predisposition could confer increased tolerance to drought (Chmielewska et al. 2016). In fact, several barley genes have been successfully used to improve drought tolerance in barley and in other crops, e.g., overexpression of the isoform HvSNAC1 in barley (Al Abdallat et al. 2014) and overexpression of HvCBF4 in rice (Oh et al. 2007).
Many traits, such as relative water content, have been shown to work as selection parameters for drought in barley (Matin et al. 1989). However, other traits have the potential to be used to map drought tolerance loci. Fv/Fm, the ratio of variable to maximal fluorescence in a dark-adapted state, evaluates the effect of stress on the reaction centers of Photosystem II and works as a selection criterion for drought tolerance (Guo et al. 2008). This trait has been used in a mapping population under greenhouse conditions (Guo et al. 2008). Two QTLs (116 and 135.7 cM) on the long arm of chromosome 2H respectively explained 15 and 9% of Fv/Fm phenotypic variation under drought conditions during the post-flowering stage. However, the use of Fv/Fm as a possible selection criterion should be validated in other populations.
Several studies have detected QTLs involved in drought tolerance under controlled and field conditions using different genetic material (Baum et al. 2003; Diab et al. 2004; Guo et al. 2008; Honsdorf et al. 2014; Lakew et al. 2013; Peighambari et al. 2005; Rollins et al. 2013; Talame et al. 2004; Teulat et al. 1997, 1998, 2001a, b, 2002, 2003; von Korff et al. 2008). However, these studies identify only the QTLs underlying the traits of interest; they do not identify genes underlying drought tolerance in barley. It is possible that many of these studies could be reanalyzed using new markers to validate discovered QTLs or to identify new QTLs using the improved barley genome sequence (Mascher et al. 2017). As such, the use of more markers and better physical maps of barley provided by the high-quality reference genome (Mascher et al. 2017) should lead to more accurate identification of QTLs and their underlying genes involved in drought tolerance. The publication of the high-quality barley genome sequence (Mascher et al. 2017) allows the identification of homologs of important players in drought tolerance. For example, the in silico and expression analyses of barley calcium-dependent protein kinases (CDPKs) demonstrated the involvement of CDPKs in barley signaling pathways in response to drought (Fedorowicz-Strońska et al. 2017).
15.6 Temperature Stress
Temperature stress has a strong effect on the growth and development of barley. Temperature can be divided into three ranges: (i) low-temperature range, which includes cold and freezing; (ii) optimum temperature range at which the plants grow and develop normally; and (iii) high-temperature range, which is associated with heat stress.
15.6.1 Low-Temperature Stress
Low-temperature stress, which includes cold (also known as chilling, above freezing point temperature) and freezing (below freezing point temperature), has a severe effect on small-grain crop plants in temperate climates (Kosova et al. 2011). In regions such as southern Australia, where winter temperatures are not low enough to cause freezing damage at the vegetative stage, radiation frost constitutes a problem at the reproductive stage. Radiation frost occurs during clear nights when the plant canopy is receiving less heat than what is radiated away, causing temperatures to drop below zero. Yield is severely affected by radiation frost as a result of floret and spike abortion as well as damage to developing grains (Reinheimer et al. 2004; Zheng et al. 2015). According to the 2006 and 2012 reports from the Grains Research & Development Corporation (https://grdc.com.au/uploads/documents/frost.pdf, https://grdc.com.au/uploads/documents/GRDC-FS-CrackingWheatsToughestNuts.pdf), frost causes yearly losses of more than $33 million in crop production in southern Australia and Victoria and about $100 million in barley and wheat production in northern New South Wales and Queensland.
Barley, like other temperate cereals, has an increase in freezing tolerance when exposed to cold at vegetative stages, a phenomenon known as cold acclimation or hardening. Under cold stress, plants have a short-term response including cold acclimation, and a long-term response, which involves developmental responses such as vernalization (Kosova et al. 2011; Pecchioni et al. 2014). Barley can be classified into spring, facultative, and winter types depending on its growth habitat, tolerance to low temperature, requirements for vernalization, and sensitivity to photoperiod (Tondelli et al. 2014). Photoperiod sensitivity and vernalization play roles in low-temperature tolerance by delaying the transition from the vegetative to the reproductive stage, thus delaying flowering, which allows plants to survive low-temperature stress (Fowler et al. 1996a, b, 2001; Mahfoozi et al. 2001a, b). In fact, genes that confer low-temperature-tolerance are expressed during the vegetative stage (Fowler et al. 1996b; Mahfoozi et al. 2001b), and processes that prolong this stage increase low-temperature tolerance. Some mechanisms that may contribute to freezing tolerance include the ability to prevent or reverse proteins’ denaturation and to reduce the physical damage of dehydration resulting from freezing-induced intercellular ice formation (Snyder and Melo-Abreu 2005; Thomashow 1998; Yadav 2010). Another important mechanism in freezing tolerance is the stabilization of membranes. In fact, the lipid composition of membranes and the accumulation of sucrose and other simple sugars seem to change in response to cold acclimation (Thomashow 1998). For instance, blt4, a barley gene involved in freezing tolerance, encodes a putative lipid transfer protein and plays a role in modifying the lipid composition of membranes (Thomashow 1998). Because blt4 expression is also induced under drought stress, this gene could be responsive to dehydration (Dunn et al. 1991). Hydrophilic polypeptides, encoded by cold-responsive (COR) genes, seem to contribute to membrane stabilization and increase stress tolerance during freezing (Thomashow 1998).
Genetic mapping of barley has identified loci associated with freezing tolerance on chromosomes 1H, 2H, 5H, and 6H. A doubled-haploid population resulting from a cross between a winter barley (Nure) and spring barley (Tremois) was assessed for low-temperature tolerance using three traits: winter survival in the field, frost resistance (controlled freeze test), and Fv/Fm. Two major QTLs were associated with the three traits and mapped on the long arm of chromosome 5H (Francia et al. 2004). The first QTL (FR-H1) was previously mapped by Hayes et al. (1993) in a barley population of Diktoo (winter) crossed with Morex (spring), which was assessed for field survival and the lethal temperature at which 50% of the plants die (LT50). Interestingly, FR-H1 was found to overlap with the VRN-H1 vernalization locus. Whether this association of both vernalization requirement and freezing tolerance is a result of having linked genes (Francia et al. 2004; Hayes et al. 1993) or having one gene with pleiotropic effects (Tondelli et al. 2014; von Zitzewitz et al. 2011) is still a debatable question (Fisk et al. 2013). If one considers the pleiotropy effect, then HvBM5A would be the candidate gene underlying VRN-H1 and FR-H1 (von Zitzewitz et al. 2005). HvBM5A, the barley ortholog of TmAP1, is a MADS-box transcription factor (von Zitzewitz et al. 2005; Yan et al. 2003), and spring accessions harbor a deletion in their first intron (Fu et al. 2005). Besides freezing tolerance at the vegetative stage (Francia et al. 2004; Hayes et al. 1993), the FR-H1/VRN-H1 locus is also associated with frost tolerance at the reproductive stage. In fact, FR-H1 has been associated with frost-induced floret sterility and grain damage in three different mapping populations (Reinheimer et al. 2004).
Another QTL specifically associated with frost-induced floret sterility was mapped on the long arm of chromosome 2H (Reinheimer et al. 2004). Approximately 30 cM away from FR-H1, the second QTL (FR-H2) detected by Francia et al. (2004) mapped to the same region where QTLs for the accumulation of two cold-responsive proteins, HvCOR14b (Crosatti et al. 1999) and TMC-Ap3 (Baldi et al. 1999; Mastrangelo et al. 2000), mapped. An HvCBF transcription factor was suggested as a candidate underlying FR-H2 because HvCBF4 was the peak marker and at least 12 HvCBF of the 20 barley CBFs mapped to FR-H2 (Francia et al. 2004, 2007; Pasquariello et al. 2014). Tondelli et al. (2006) suggested that HvCBF genes rather than their regulators, i.e., the inducer of CBF expression (ICE1) and fiery-1 (FRY1), were candidates for the FR-H2 locus. The higher copy number of CBF genes in winter genotypes compared with those in spring suggests the involvement of copy number/gene duplication in the degree of low-temperature tolerance in temperate climate cereals (Knox et al. 2010; Tondelli et al. 2011). In fact, transgenic barley overexpressing TaDREB2 and TaDREB3 exhibited increased drought and frost tolerance, as well as higher expression of stress-responsive genes involved in protecting cells from damage and desiccation (Morran et al. 2011).
The genetic mechanism of freezing tolerance exhibited dynamic control between FR-H1 and FR-H2. CBF genes at FR-H2 were more highly expressed in barley genotypes carrying the winter allele (vrn-h1) at the VRN-H1 locus compared with genotypes carrying the spring allele. In addition, the expression of CBF was reduced after vernalization (in vernalization-requiring plants), indicating that VRN-H1 attenuated the expression of FR-H2 (Stockinger et al. 2007). The involvement of FR-H1 and FR-H2 in cold acclimation was detected using a genome-wide association mapping approach, where the presence of specific alleles (winter alleles) at these loci conferred maximum low-temperature tolerance (von Zitzewitz et al. 2011). The allelic variation at FR-H1 and FR-H2 was also the main determinant of frost tolerance in a panel of a European two-row spring barley population (Tondelli et al. 2014). Besides FR-H1 and FR-H2, Fisk et al. (2013) mapped another QTL, FR-H3, which is associated with freezing tolerance on the short arm of chromosome 1H.
Research on low-temperature tolerance in barley is far from complete. Extensive high-quality research, including proteomics and metabolomics studies, is warranted to identify the genes involved in each of the two components of low-temperature tolerance, cold tolerance and freezing/frost tolerance. The barley high-quality reference genome (Mascher et al. 2017) can significantly contribute to a better understanding of the genetic mechanisms underlying low-temperature stress. For instance, the genes underlying the Fr-H3 QTL still need to be identified (Fisk et al. 2013). Also, whether or not freezing tolerance and vernalization requirements are due to a pleiotropic effect or linkage still needs to be clarified. Interestingly, copy number variation also plays a role in frost tolerance. Francia et al. (2016) found that within a collection of 41 barley genotypes, the genotypes with higher copy numbers at HvCBF2 and HvCBF4 showed increased frost tolerance (Francia et al. 2016). Therefore, sequencing different barley varieties could help in identifying variations in copy number and their role in enhancing stress tolerance.
15.6.2 High-Temperature Stress
The negative impact of climate change on crop yield has been established, with a projected increase in global temperature on the order of ~4 °C by the late twenty-first century (IPCC 2014). High temperature (hereafter heat stress) affects the duration of the growing season, geographic distribution (Dawson et al. 2015), and malting quality of barley (Savin et al. 1996). The effects of heat stress depend on several factors, such as stress intensity (temperature in degrees), duration and rate of temperature increase (Wahid et al. 2007). Symptoms of heat stress in barley include the reduction of yield and yield components, such as number of tillers per plant, spike length and thousand grain weight, and a lower final starch concentration (possibly caused by irreversible inactivation of sucrose synthase) (Abou-Elwafa and Amein 2016; Högy et al. 2013; MacLeod and Duffus 1988; Wallwork et al. 1998). Nevertheless, heat stress in barley is still an understudied topic, and further research is expected to take place due to climate change.
Barley, like other cereal crops, is particularly sensitive to heat stress during panicle development and meiosis, with high temperatures (over several days) causing abnormal pollen development and complete sterility (Sakata et al. 2000). Oshino et al. (2007) studied the effect of high-temperature injury on anther development in barley and observed a premature progression of early developmental stages and fate (e.g., progression to meiosis of pollen-mother cells). Transcriptional analysis suggested that the genetic control involved in the cessation of anther cell proliferation was related to the repression of cell proliferation factors, such as histones, replication licensing factors, and DNA polymerases (Oshino et al. 2007). Gene expression and differentiation of anthers were studied under normal (20 °C day/15 °C night) and heat stress (30 °C day/25 °C night) conditions, revealing that genes that are active in the anthers under normal temperature conditions, such as H3, H4, and glycine-rich RNA-binding protein genes, were transcriptionally inhibited under heat stress (Abiko et al. 2005). After anthesis, a field trial in Iran showed a 17% reduction in grain yield in barley under heat stress and significantly affected parameters, including the translocation of photosynthates to the grain, starch synthesis, and deposition in the developing grain (Modhej et al. 2015). Field trials have shown grain yield reduction caused by heat stress causing a higher penalty when temperature increased during stem elongation (Ugarte et al. 2007). In another field experiment using a panel of 138 spring barley genotypes, Ingvordsen et al. (2015b) corroborated the previously found decrease in grain yield (55.8%) caused by heat stress (Högy et al. 2013).
One field study in Egypt comprising 320 wild barley accessions and six local genotypes showed that several yield components, such as days to flowering, plant height and thousand kernel weight (among others), were affected by heat stress (Abou-Elwafa and Amein 2016). The screening of such a large number of individuals in the primary gene pool of barley quantified the effects of heat stress and grouped genotypes into two distinct groups based on heat stress tolerance. Cluster analysis revealed that group 1 consisted of 112 barley accessions mainly originating from cold regions while group 2 consisted of 224 barley genotypes with variable degrees of tolerance to heat stress (Abou-Elwafa and Amein 2016). These valuable data may provide the basis for future mapping populations and/or genome-wide association studies that could reveal the genetic mechanisms underlying heat stress in barley.
Also, Phillips et al. (2015) examined barley’s recombination landscape and observed that high temperature significantly changed the patterns of recombination only in male meiosis. Transcriptome changes in developing barley seeds in response to heat stress revealed the downregulation of genes related to storage compound biosynthesis and cell growth (Mangelsen et al. 2011). Metadata analysis also showed that seed embryo and endosperm were the primary locations of heat stress response genes and an overlap of heat- and drought-responsive genes in barley caryopses (Mangelsen et al. 2011).
The relative importance of day and night heat stress is still under debate. Garcia et al. (2015) observed approximately 7% reduction in grain yield per degree of night temperature increase during the critical period of grain filling. This yield reduction was caused by an accelerated development, which caused increased tillering, death and impacted biomass and grain production.
Despite the undeniable importance of heat stress, limited knowledge is available on the genetic mechanisms controlling barley’s response to high temperature. Heat shock proteins (HSP) are the obvious candidates underlying the plant’s heat stress response. Xia et al. (2013) investigated the allelic variation of HSP17.8, which encodes an HSP ubiquitously produced in response to heat stress, and its association with agronomic traits in barley. Nine haplotypes for HSP17.8 were identified, with wild accessions exhibiting greater allelic diversity than found in cultivated barley. These results suggest the role of HSP17.8 in response to heat stress as well as to drought. In addition to HSP, miRNAs are heat inducible and may play a role in barley thermotolerance through downregulation of their target genes (Kruszka et al. 2014).
To date, numerous studies have identified QTLs associated with general agronomic traits (e.g., yield and plant height) although little work has been done on traits specifically associated with heat stress. Gous et al. (2016) mapped a doubled-haploid population derived from a cross between ND24260 (stay-green genotype) and Flagship (high-quality malting genotype) to identify QTLs associated with the stay-green trait under abiotic stress conditions. Ten QTLs were identified, six of which were associated with heat stress and four with drought (Gous et al. 2016). In addition, Ingvordsen et al. (2015a) performed a genome-wide association study on spring barley genotypes under heat stress and elevated CO2 and identified a QTL associated with grain yield under heat stress on chromosome 2H. However, these loci should be further validated in field trials and the genes underlying these regions should be identified. Most strategies to avoid heat stress are related to early sowing and shorter season cultivars. This may be due to the complexity of heat stress and its strong interaction with the environment and to limited knowledge about heat-tolerant genotypes and the genetic regions underlying heat stress. The opportunity provided by the barley high-quality reference genome (Mascher et al. 2017) to identify new markers is expected to provide considerable advances in genetic mapping for heat stress. Studies with a design similar to the one adopted by Abou-Elwafa and Amein (2016) and Ingvordsen et al. (2015b) can be extremely valuable to future genetic mapping studies specifically targeting heat stress.
15.7 Nutrient Deficiency (Nitrogen and Phosphorus) Stress
Crop nutrition is a key determinant of yield potential. An adequate supply of essential macro- and micronutrients is required for crops to achieve sufficient vegetative growth to ensure the development of nutritious grain. Fertilizer is a major input cost for farmers but is nevertheless often applied in excess of a crop’s requirements. For example, estimates are that cereal crops take up only 40–60% of the applied nitrogen (N) fertilizer and that unused N may be leached from the soil or lost to volatilization (Raun and Johnson 1999; Sylvester-Bradley and Kindred 2009). The impact of this “lost” N is of critical concern as a source for major economic loss as an environmental pollutant that leads to release of potent greenhouse gases and eutrophication of aquatic environments (Zhang et al. 2015).
Insufficient availability of N and phosphorus (P) has the greatest impact on yield potential of crop plants (Hawkesford et al. 2012). Such deficiency directly limits yield potential as affected plants have a lower capacity for radiation capture and hence grain production, regardless of the application of fertilizer during the reproductive growth stage. The nutrient required in greatest supply is N because it is an integral component of major plant macromolecules. N-limited crops may yellow, particularly older leaves, due to loss of chlorophyll. P-deficient crops may, on the contrary, have older dark green leaves due to the accumulation of sugars and other compounds, and their root growth can be limited, reducing uptake capacity for other nutrients and water.
Nutrient use efficiency (NUE) is most commonly measured by relating crop yield to the amount, both applied and residual in the soil, of a particular nutrient available to the crop (Good et al. 2004). NUE may be improved through changing agronomic practices, such as the timing of fertilizer application and crop rotations (Fageria and Baligar 2005). The limited capacity for nutrient uptake and utilization in plants presents a real opportunity to improve NUE. In high input, high rainfall areas, what determines NUE is very different than in low input, dry areas (Cossani et al. 2010). In a similar way, increasing P use efficiency in P-fixing soils is very different than in P-available soils. Within these scenarios, the weather at a site influences NUE from season to season. However, there are a few underlying mechanisms which, if improved, would work across environments and management scenarios. First, in some environments, uptake efficiency may be improved by altering the architectural traits of roots such that the crop may access fertilizer deep in the soil profile or at shallow depths (Garnett et al. 2009). The families of transporters mediating both low- and high-affinity uptake of N and P have been described (Smith et al. 2003; Wang et al. 2012b), with the barley NRT2 and Pht1 transporters among the first N and P transporters identified and characterized in cereals (Rae et al. 2003; Vidmar et al. 2000a, b). Improving the efficiency of these transporters through the selection of superior alleles has improved uptake of target nutrients in rice (Fan et al. 2016a; Hu et al. 2015). Another important aspect for P uptake is the plant’s interaction with mycorrhizae. Selecting varieties of barley and mycorrhizae that maximize P uptake in an agricultural environment is a possible strategy to improve NUE (Grace et al. 2009).
Similarly, there is scope to improve the utilization efficiency of nutrients once they have been taken up from the soil. Much is known about the biochemical pathways in plants that are responsible for the conversion of inorganic nutrients into the organic building blocks for protein synthesis (Xu et al. 2012). The role of glutamine synthetase (GS) in N assimilation in barley has been well documented (Avila-Ospina et al. 2015; Goodall et al. 2013) and GS is regarded as a candidate gene in both reverse and forward genetic approaches to improve NUE (Hirel et al. 2007; Thomsen et al. 2014). However, the gene encoding alanine aminotransferase (AlaAT) in barley has exhibited the most promise to increase NUE via overexpression because it has improved NUE in canola and rice (Good et al. 2007; Shrawat et al. 2008). Importantly, uptake and utilization systems are often intricately intertwined such that the status of one system provides important feedback signaling, which alters the capacity of the other. In general, the remobilization of nutrients, particularly N, stored in vacuoles, proteins and other molecules in older leaves is another determinant of NUE for which underlying genetic loci have been identified in barley (Havé et al. 2016; Hollmann et al. 2014; Mickelson et al. 2003; Yang et al. 2004). However, for malting barley production, it is important the grain has 9–12% protein content, as excess protein interferes with the malting process (Bertholdsson 1999). Grain N has been the focus of most efforts to identify genetic regions regulating NUE in barley and a locus has been identified on chromosome 6 that regulates leaf senescence and N reallocation (Bezant et al. 1997; Cai et al. 2013; Heidlebaugh et al. 2008; Jukanti and Fischer 2008; Jukanti et al. 2008). The newly published high-quality barley genomic sequence (Mascher et al. 2017) will aid in efforts to identify the causal gene(s).
Genetic improvements in NUE have been made “inadvertently” during yield-based selection in barley breeding over the last 75 years, even though breeding trials were conducted with extensive application of N fertilizer (Bingham et al. 2012). However, these gains are not sufficient to alleviate concerns over rising fertilizer costs and increasing environmental regulations of fertilizer application. Variation in NUE has been identified in adapted barley varieties (Anbessa et al. 2009, 2010; Beatty et al. 2010); however, variation in the transcriptomic responses of contrasting Tibetan wild barley lines to low N or K barley lines suggests that this collection may be a source of greater variation in NUE and component traits (Quan et al. 2016; Wei et al. 2016; Zeng et al. 2014). To assess the potential of exotic germplasm to be a source of superior NUE more fully will require its incorporation into more advanced populations such as the nested association mapping (NAM) or multi-parent advanced generation intercross (MAGIC) populations (Nice et al. 2016; Sannemann et al. 2015) to reduce the “wildness” of the material, which can create artifacts in data due to issues such as phenology (Karley et al. 2011). A field-based analysis of a barley association mapping panel revealed how large the environmental influence is when measuring traits like P uptake and assimilation efficiency, requiring multiple site-years of data to produce meaningful genetic associations (George et al. 2011; George et al. 2014). The studies also showed the potential for identifying surrogate traits for NUE, such as rhizosheath production, as a way to screen more quickly for NUE potential.
There has been very little success in improving barley NUE through genetic modification approaches (McAllister et al. 2012). It is clear that improved genetic modification tools will be necessary and the identification of N- and P-responsive promoter elements will play an important role in fine-tuning transgene expression to change nutrient supply in the field (Schünmann et al. 2004a, b). The arrival of the barley high-quality reference genome (Mascher et al. 2017) will help identify further regulatory motifs and targets for application of genome editing technologies, such as CRISPR/CAS (Teotia et al. 2016).
15.8 Other Stresses
Other stresses, mainly waterlogging and manganese (Mn) and cadmium (Cd) toxicity, affect barley to a lesser extent.
Cultivated barley shows a genetic diversity in waterlogging tolerance, and this tolerance varies according to life stage, with barley being more susceptible to waterlogging at preemergence, seedling growth, and reproductive stages (Setter and Waters 2003). Breeding for waterlogging tolerance in barley is still in its early stages with few studies examining waterlogging tolerance in barley. Li et al. (2008) mapped waterlogging tolerance using two doubled-haploid populations and targeting traits such as leaf chlorosis, plant survival, and plant biomass reduction. Some QTLs (on chromosomes 1H, 3H, and 7H) were significant in both populations and some QTLs associated with more than one trait, such as the 4H QTL, which associated with leaf chlorosis and plant biomass. In addition to these mapping studies, Mendiondo et al. (2016) showed that HvPRT6, an N-end rule pathway N-recognin E3 ligase PROTEOLYSIS6, played a role in waterlogging tolerance in barley. In fact, the role of the N-end rule pathway in controlling plant response to hypoxia, which results from waterlogging, has already been established in arabidopsis. Transgenic barley plants with decreased HvPRT6 expression exhibited increased waterlogging tolerance as manifested by biomass maintenance, increased yield, and decreased chlorophyll degradation under stress (Mendiondo et al. 2016).
Although barley is sensitive to Mn toxicity, a large genetic variation in Mn tolerance exists among cultivated genotypes (Hebbern et al. 2005). This variation could be harnessed to identify chromosomal regions not yet identified that are involved in Mn tolerance. Huang et al. (2015) proposed that breeding for Mn tolerance could improve waterlogging tolerance in barley, which could be due to the decrease in soil redox potential under waterlogging, which in turn increases the soil Mn2+ concentration and causes Mn toxicity effects in plants. On the other hand, Leplat et al. (2016) examined 248 barley varieties that were cultivated in six distinct environments prone to Mn deficiency, and identified several putative QTLs. Then, the authors were able to explore putative candidate genes in the flanking region of the SNP associations, using the barley genome sequence.
Cadmium is a heavy metal usually associated with polluted soils. It can affect human health, and barley grains have the lowest threshold value for Cd concentration for food safety among cereal crops (Wu et al. 2015). Barley genotypes have differences in tolerance to Cd toxicity (Chen et al. 2008; Persson et al. 2006). A genome-wide association study demonstrated a large genotypic variation in the Cd concentration in different organs and detected several QTLs associated with Cd accumulation, some of which were organ-specific (Wu et al. 2015). Furthermore, Cao et al. (2014) used microarray expression profiling on two genotypes contrasting in their Cd tolerance to identify novel genes that might play a role in various mechanisms of Cd tolerance.
15.9 Conclusions
The responses of plants to abiotic stresses are complex. While certain tolerance mechanisms are specific to a particular stress (e.g., Na+ exclusion from the leaf blades under salinity), plants have to cope with the co-occurrence of multiple stresses under field conditions. Co-occurrence of stresses can trigger common responses, with some molecular pathways likely to play roles in more than one stress, e.g., pathways involving reactive oxygen species. In barley, three out of the 13 dehydrin genes were induced by low temperature and drought (Tommasini et al. 2008). The role of dehydrins in barley in response to several stresses including cold, drought, and salinity was reviewed by Kosova et al. (2014). In addition, as previously mentioned, transcription factor-encoding genes such as C-repeat binding factors (CBFs) are involved in low-temperature and drought stress (see Hasegawa et al. 2000). Another example is HVA1, which encodes a stress-induced class III late embryogenesis abundant (LEA III) protein (Hong et al. 1992) and is associated with salinity, drought, and low-temperature tolerance (Hong et al. 1992; Xu et al. 1996). In fact, due to the resilient nature of barley to abiotic stresses, numerous barley genes have been used to improve stress tolerance as reviewed by Gürel et al. (2016).
In this chapter, we have described the broader impact of each stress and the physiological and genetic components of barley stress responses. Many of the responses to stresses, such as to heat stress and Cd toxicity, remain understudied. The previously released barley genome sequence (Mayer et al. 2012) enabled the identification of candidate genes involved in stress tolerance. The completion of the barley high-quality reference genome (Mascher et al. 2017) will undoubtedly contribute to an improved and more comprehensive picture of barley genetics and genomics in response to abiotic stress (Fig. 15.1).
References
Abiko M et al (2005) High-temperature induction of male sterility during barley (Hordeum vulgare L.) anther development is mediated by transcriptional inhibition. Sex Plant Reprod 18:91–100. https://doi.org/10.1007/s00497-005-0004-2
Abou-Elwafa SF, Amein KA (2016) Genetic diversity and potential high temperature tolerance in barley (Hordeum vulgare). WJAS 4:1–8. Doi:https://doi.org/10.12691/wjar-4-1-1
Al Abdallat AM, Ayad JY, Abu Elenein JM, Al Ajlouni Z, Harwood WA (2014) Overexpression of the transcription factor HvSNAC1 improves drought tolerance in barley (Hordeum vulgare L.) Mol Breed 33:401–414. https://doi.org/10.1007/s11032-013-9958-1
Allaby RG (2015) Barley domestication: the end of a central dogma? Genome Biol 16:176. https://doi.org/10.1186/s13059-015-0743-9
Amarasinghe S, Watson-Haigh NS, Gilliham M, Roy S, Baumann U (2016) The evolutionary origin of CIPK16: a gene involved in enhanced salt tolerance. Mol Phylogenet Evol 100:135–147. https://doi.org/10.1016/j.ympev.2016.03.031
Anbessa Y, Juskiw P, Good A, Nyachiro J, Helm J (2009) Genetic variability in nitrogen use efficiency of spring barley. Crop Sci 49:1259–1269. https://doi.org/10.2135/cropsci2008.09.0566
Anbessa Y, Juskiw P, Good A, Nyachiro J, Helm J (2010) Selection efficiency across environments in improvement of barley yield for moderately low nitrogen environments. Crop Sci 50:451–457. https://doi.org/10.2135/cropsci2009.02.0058
Avila-Ospina L, Marmagne A, Talbotec J, Krupinska K, Masclaux-Daubresse C (2015) The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J Exp Bot 66:2013–2026. https://doi.org/10.1093/jxb/erv003
Babu RC, Zhang JX, Blum A, Ho THD, Wu R, Nguyen HT (2004) HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci 166:855–862. https://doi.org/10.1016/j.plantsci.2003.11.023
Badr A et al (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17:499–510. https://doi.org/10.1093/oxfordjournals.molbev.a026330
Baerenfaller K et al (2012) Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol Syst Biol 8:606. https://doi.org/10.1038/msb.2012.39
Bahieldin A et al (2005) Field evaluation of transgenic wheat plants stably expressing the HVA1 gene for drought tolerance. Physiol Plant 123:421–427. https://doi.org/10.1111/j.1399-3054.2005.00470.x
Baldi P, Grossi M, Pecchioni N, Vale G, Cattivelli L (1999) High expression level of a gene coding for a chloroplastic amino acid selective channel protein is correlated to cold acclimation in cereals. Plant Mol Biol 41:233–243. https://doi.org/10.1023/A:1006375332677
Barati A, Moghaddam M, Mohammadi SA, Ghazvini HA, Sadeghzadeh B (2017) Identification of QTLs associated with agronomic and physiological traits under salinity stress in barley. J Agric Sci Technol-Iran 19:185–200
Barragan V et al (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24:1127–1142. https://doi.org/10.1105/tpc.111.095273
Bassil E et al (2011) The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23:3482–3497. https://doi.org/10.1105/tpc.111.089581
Baum M, Grando S, Backes G, Jahoor A, Sabbagh A, Ceccarelli S (2003) QTLs for agronomic traits in the Mediterranean environment identified in recombinant inbred lines of the cross ‘Arta’ × H. spontaneum 41-1. Theor Appl Genet 107:1215–1225. https://doi.org/10.1007/s00122-003-1357-2
Bayat F et al (2010) Potato plants bearing a vacuolar Na+/H+ antiporter HvNHX2 from barley are characterized by improved salt tolerance. Russ J Plant Physiol 57:696–706. https://doi.org/10.1134/s1021443710050134
Bayat F, Shiran B, Belyaev DV (2011) Overexpression of HvNHX2, a vacuolar Na+/H+ antiporter gene from barley, improves salt tolerance in Arabidopsis thaliana. Aust J Crop Sci 5:428–432
Beatty PH, Anbessa Y, Juskiw P, Carroll RT, Wang J, Good AG (2010) Nitrogen use efficiencies of spring barley grown under varying nitrogen conditions in the field and growth chamber. Ann Bot 105:1171–1182. https://doi.org/10.1093/aob/mcq025
Bertholdsson NO (1999) Characterization of malting barley cultivars with more or less stable grain protein content under varying environmental conditions. Eur J Agron 10:1–8. https://doi.org/10.1016/S1161-0301(98)00043-4
Bezant H, Laurie DA, Pratchett N, Chojecki J, Kearsey MJ (1997) Mapping of QTL controlling NIR predicted hot water extract and grain nitrogen content in a spring barley cross using marker-regression. Plant Breed 116:141–145. https://doi.org/10.1111/j.1439-0523.1997.tb02168.x
Bingham IJ, Karley AJ, White PJ, Thomas WTB, Russell JR (2012) Analysis of improvements in nitrogen use efficiency associated with 75 years of spring barley breeding. Eur J Agron 42:49–58. https://doi.org/10.1016/j.eja.2011.10.003
Byrt CS et al (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 143:1918–1928. https://doi.org/10.1104/pp.106.093476
Cai S, Yu G, Chen X, Huang Y, Jiang X, Zhang G, Jin X (2013) Grain protein content variation and its association analysis in barley. BMC Plant Biol 13:1–11. https://doi.org/10.1186/1471-2229-13-35
Cao FB, Chen F, Sun HY, Zhang GP, Chen ZH, Wu FB (2014) Genome-wide transcriptome and functional analysis of two contrasting genotypes reveals key genes for cadmium tolerance in barley. BMC Genomics 15. https://doi.org/10.1186/1471-2164-15-611
Cartwright B, Zarcinas BA, Spouncer LR (1986) Boron toxicity in South Australian barley crops. Aust J Agric Res 37:351–359. https://doi.org/10.1071/Ar9860351
Chen GX et al (2009) Mapping of the eibi1 gene responsible for the drought hypersensitive cuticle in wild barley (Hordeum spontaneum). Breed Sci 59:21–26. https://doi.org/10.1270/jsbbs.59.21
Chen G, Sagi M, Weining S, Krugman T, Fahima T, Korol AB, Nevo E (2004) Wild barley eibi1 mutation identifies a gene essential for leaf water conservation. Planta 219:684–693. https://doi.org/10.1007/s00425-004-1277-7
Chen F, Wang F, Zhang G, Wu F (2008) Identification of barley varieties tolerant to cadmium toxicity. Biol Trace Elem Res 121:171–179. https://doi.org/10.1007/s12011-007-8042-2
Chen G et al (2011) An ATP-binding cassette subfamily G full transporter is essential for the retention of leaf water in both wild barley and rice. Proc Natl Acad Sci USA 108:12354–12359. https://doi.org/10.1073/pnas.1108444108
Chloupek O, Dostal V, Streda T, Psota V, Dvorackova O (2010) Drought tolerance of barley varieties in relation to their root system size. Plant Breed 129:630–636. https://doi.org/10.1111/j.1439-0523.2010.01801.x
Chmielewska K et al (2016) Analysis of drought-induced proteomic and metabolomic changes in barley (Hordeum vulgare L.) leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.01108
Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S (2014) Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA 111:6497–6502. https://doi.org/10.1073/pnas.1319955111
Cossani CM, Slafer GA, Savin R (2010) Co-limitation of nitrogen and water, and yield and resource-use efficiencies of wheat and barley. Crop Pasture Sci 61:844–851. https://doi.org/10.1071/cp10018
Crosatti C, de Laureto PP, Bassi R, Cattivelli L (1999) The interaction between cold and light controls the expression of the cold-regulated barley gene cor14b and the accumulation of the corresponding protein. Plant Physiol 119:671–680. https://doi.org/10.1104/pp.119.2.671
Daszkowska-Golec A, Szarejko I (2013) Open or close the gate—stomata action under the control of phytohormones in drought stress conditions. Front Plant Sci 4:138. https://doi.org/10.3389/fpls.2013.00138
Dawson IK, Russell J, Powell W, Steffenson B, Thomas WTB, Waugh R (2015) Barley: a translational model for adaptation to climate change. New Phytol 206:913–931. https://doi.org/10.1111/nph.13266
Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA 101:15249–15254. https://doi.org/10.1073/pnas.0406258101
Diab AA, Teulat-Merah B, This D, Ozturk NZ, Benscher D, Sorrells ME (2004) Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor Appl Genet 109:1417–1425. https://doi.org/10.1007/s00122-004-1755-0
Dunn MA, Hughes MA, Zhang L, Pearce RS, Quigley AS, Jack PL (1991) Nucleotide sequence and molecular analysis of the low temperature induced cereal gene, BLT4. Mol Gen Genet 229:389–394
Ellis RP et al (2002) Phenotype/genotype associations for yield and salt tolerance in a barley mapping population segregating for two dwarfing genes. J Exp Bot 53:1163–1176. https://doi.org/10.1093/jexbot/53.371.1163
Emebiri L, Michael P, Moody D (2009) Enhanced tolerance to boron toxicity in two-rowed barley by marker-assisted introgression of favourable alleles derived from Sahara 3771. Plant Soil 314:77–85. https://doi.org/10.1007/s11104-008-9707-0
Fageria NK, Baligar VC (2005) Enhancing nitrogen use efficiency in crop plants. In: Advances in agronomy, vol 88. Elsevier, pp 97–185. https://doi.org/10.1016/s0065-2113(05)88004-6
Fan Y, Shabala S, Ma YL, Xu RG, Zhou MX (2015) Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genomics 16. https://doi.org/10.1186/s12864-015-1243-8
Fan X et al (2016a) Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci USA 113:7118–7123. https://doi.org/10.1073/pnas.1525184113
Fan Y, Zhou G, Shabala S, Chen ZH, Cai S, Li C, Zhou M (2016b) Genome-wide association study reveals a new QTL for salinity tolerance in barley (Hordeum vulgare L.). Front Plant Sci 7:946. https://doi.org/10.3389/fpls.2016.00946
FAO (2015) Global soil status, processes and trends. In: Status of the world’s soil resources (SWSR)—main report
Farooq M, Wahid A, Lee DJ, Ito O, Siddique KHM (2009) Advances in drought resistance of rice. Crit Rev Plant Sci 28:199–217. https://doi.org/10.1080/07352680902952173
Fedorowicz-Strońska O, Koczyk G, Kaczmarek M, Krajewski P, Sadowski J (2017) Genome-wide identification, characterisation and expression profiles of calcium-dependent protein kinase genes in barley (Hordeum vulgare L.). J Appl Genet 58:11–22. https://doi.org/10.1007/s13353-016-0357-2
Fisk SP et al (2013) FR-H3: a new QTL to assist in the development of fall-sown barley with superior low temperature tolerance. Theor Appl Genet 126:335–347. https://doi.org/10.1007/s00122-012-1982-8
Fowler DB, Chauvin LP, Limin AE, Sarhan F (1996a) The regulatory role of vernalization in the expression of low-temperature-induced genes in wheat and rye. Theor Appl Genet 93:554–559. https://doi.org/10.1007/Bf00417947
Fowler DB, Limin AE, Wang SY, Ward RW (1996b) Relationship between low-temperature tolerance and vernalization response in wheat and rye. Can J Plant Sci 76:37–42. https://doi.org/10.4141/cjps96-007
Fowler DB, Breton G, Limin AE, Mahfoozi S, Sarhan F (2001) Photoperiod and temperature interactions regulate low-temperature-induced gene expression in barley. Plant Physiol 127:1676–1681. https://doi.org/10.1104/pp.010483
Foy CD, Chaney RL, White MC (1978) The physiology of metal toxicity in plants. Annu Rev Plant Physiol 29:511–566. https://doi.org/10.1146/annurev.pp.29.060178.002455
Francia E et al (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ’Tremois’ (spring) barley map. Theor Appl Genet 108:670–680. https://doi.org/10.1007/s00122-003-1468-9
Francia E et al (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115:1083–1091. https://doi.org/10.1007/s00122-007-0634-x
Francia E et al (2016) Copy number variation at the HvCBF4-HvCBF2 genomic segment is a major component of frost resistance in barley. Plant Mol Biol 92:161–175. https://doi.org/10.1007/s11103-016-0505-4
Fu DL et al (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65. https://doi.org/10.1007/s00438-004-1095-4
Fujii M et al (2012) Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 3:713. https://doi.org/10.1038/ncomms1726
Fukuda A, Chiba K, Maeda M, Nakamura A, Maeshima M, Tanaka Y (2004) Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+-pyrophosphatase, H+-ATPase subunit A, and Na+/H+ antiporter from barley. J Exp Bot 55:585–594. https://doi.org/10.1093/jxb/erh070
Furukawa J et al (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48:1081–1091. https://doi.org/10.1093/pcp/pcm091
Garcia GA, Dreccer MF, Miralles DJ, Serrago RA (2015) High night temperatures during grain number determination reduce wheat and barley grain yield: a field study. Glob Change Biol 21:4153–4164. https://doi.org/10.1111/gcb.13009
Garnett T, Conn V, Kaiser BN (2009) Root based approaches to improving nitrogen use efficiency in plants. Plant, Cell Environ 32:1272–1283. https://doi.org/10.1111/j.1365-3040.2009.02011.x
Garthwaite AJ, von Bothmer R, Colmer TD (2005) Salt tolerance in wild Hordeum species is associated with restricted entry of Na+ and Cl− into the shoots. J Exp Bot 56:2365–2378. https://doi.org/10.1093/jxb/eri229
George TS, Brown LK, Newton AC, Hallett PD, Sun BH, Thomas WTB, White PJ (2011) Impact of soil tillage on the robustness of the genetic component of variation in phosphorus (P) use efficiency in barley (Hordeum vulgare L.). Plant Soil 339:113–123. https://doi.org/10.1007/s11104-009-0209-5
George TS et al (2014) Understanding the genetic control and physiological traits associated with rhizosheath production by barley (Hordeum vulgare). New Phytol 203:195–205. https://doi.org/10.1111/nph.12786
Gilroy S, Suzuki N, Miller G, Choi WG, Toyota M, Devireddy AR, Mittler R (2014) A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci 19:623–630. https://doi.org/10.1016/j.tplants.2014.06.013
Goldberg S (1997) Reactions of boron with soils. Plant Soil 193:35–48. https://doi.org/10.1023/A:1004203723343
Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9:597–605. https://doi.org/10.1016/j.tplants.2004.10.008
Good AG et al (2007) Engineering nitrogen use efficiency with alanine aminotransferase. Can J Bot 85:252–262. https://doi.org/10.1139/B07-019
Goodall AJ, Kumar P, Tobin AK (2013) Identification and expression analyses of cytosolic glutamine synthetase genes in barley (Hordeum vulgare L.). Plant Cell Physiol 54:492–505. https://doi.org/10.1093/pcp/pct006
Gous PW, Hickey L, Christopher JT, Franckowiak J, Fox GP (2016) Discovery of QTL for stay-green and heat-stress in barley (Hordeum vulgare) grown under simulated abiotic stress conditions. Euphytica 207:305–317. https://doi.org/10.1007/s10681-015-1542-9
Grace EJ, Cotsaftis O, Tester M, Smith FA, Smith SE (2009) Arbuscular mycorrhizal inhibition of growth in barley cannot be attributed to extent of colonization, fungal phosphorus uptake or effects on expression of plant phosphate transporter genes. New Phytol 181:938–949. https://doi.org/10.1111/j.1469-8137.2008.02720.x
Gruber BD et al (2010) HvALMT1 from barley is involved in the transport of organic anions. J Exp Bot 61:1455–1467. https://doi.org/10.1093/jxb/erq023
Guo PG, Baum M, Varshney RK, Graner A, Grando S, Ceccarelli S (2008) QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica 163:203–214. https://doi.org/10.1007/s10681-007-9629-6
Guo W, Chen T, Hussain N, Zhang G, Jiang L (2016) Characterization of salinity tolerance of transgenic rice lines harboring HsCBL8 of wild barley (Hordeum spontaneum) line from Qinghai-Tibet Plateau. Front Plant Sci 7:1678. https://doi.org/10.3389/fpls.2016.01678
Gürel F, Öztürk ZN, Uçarlı C, Rosellini D (2016) Barley genes as tools to confer abiotic stress tolerance in crops. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.01137
Hansen L, von Wettstein-Knowles P (1991) The barley genes Acl1 and Acl3 encoding acyl carrier proteins I and III are located on different chromosomes. Mol Gen Genet 229:467–478
Haro R, Banuelos MA, Senn ME, Barrero-Gil J, Rodriguez-Navarro A (2005) HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol 139:1495–1506. https://doi.org/10.1104/pp.105.067553
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499. https://doi.org/10.1146/annurev.arplant.51.1.463
Hauser F, Horie T (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant, Cell Environ 33:552–565. https://doi.org/10.1111/j.1365-3040.2009.02056.x
Havé M, Marmagne A, Chardon F, Masclaux-Daubresse C (2016) Nitrogen remobilisation during leaf senescence: lessons from Arabidopsis to crops. J Exp Bot. https://doi.org/10.1093/jxb/erw365
Hawkesford M, Horst W, Kichey T, Lambers H, Schjoerring J, Møller IS, White P (2012) Functions of macronutrients. In: Marschner’s mineral nutrition of higher plants, 3 edn. Elsevier, San Diego, pp 135–189. https://doi.org/10.1016/b978-0-12-384905-2.00006-6
Hayes JE, Reid RJ (2004) Boron tolerance in barley is mediated by efflux of boron from the roots. Plant Physiol 136:3376–3382. https://doi.org/10.1104/pp.103.037028
Hayes PM, Blake T, Chen TH, Tragoonrung S, Chen F, Pan A, Liu B (1993) Quantitative trait loci on barley (Hordeum vulgare L.) chromosome 7 associated with components of winterhardiness. Genome 36:66–71. https://doi.org/10.1139/g93-009
Hayes JE, Pallotta M, Garcia M, Oz MT, Rongala J, Sutton T (2015) Diversity in boron toxicity tolerance of Australian barley (Hordeum vulgare L.) genotypes. BMC Plant Biol 15. https://doi.org/10.1186/s12870-015-0607-1
Hebbern CA, Pedas P, Schjoerring JK, Knudsen L, Husted S (2005) Genotypic differences in manganese efficiency: field experiments with winter barley (Hordeum vulgare L.). Plant Soil 272:233–244. https://doi.org/10.1007/s11104-004-5048-9
Heidlebaugh NM, Trethewey BR, Jukanti AK, Parrott DL, Martin JM, Fischer AM (2008) Effects of a barley (Hordeum vulgare) chromosome 6 grain protein content locus on whole-plant nitrogen reallocation under two different fertilisation regimes. Funct Plant Biol 35:619–632. https://doi.org/10.1071/FP07247
Hen-Avivi S et al (2016) A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell 28:1440–1460. https://doi.org/10.1105/tpc.16.00197
Hirel B, Le Gouis J, Ney B, Gallais A (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58:2369–2387. https://doi.org/10.1093/jxb/erm097
Högy P, Poll C, Marhan S, Kandeler E, Fangmeier A (2013) Impacts of temperature increase and change in precipitation pattern on crop yield and yield quality of barley. Food Chem 136:1470–1477. https://doi.org/10.1016/j.foodchem.2012.09.056
Hollmann J, Gregersen PL, Krupinska K (2014) Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of field grown barley. J Exp Bot. https://doi.org/10.1093/jxb/eru094
Holloway RE, Alston AM (1992) The effects of salt and boron on growth of wheat. Aust J Agric Res 43:987–1001. https://doi.org/10.1071/Ar9920987
Hong B, Barg R, Ho TH (1992) Developmental and organ-specific expression of an ABA- and stress-induced protein in barley. Plant Mol Biol 18:663–674. https://doi.org/10.1007/BF00020009
Honsdorf N, March TJ, Berger B, Tester M, Pillen K (2014) High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLOS ONE 9:e97047. https://doi.org/10.1371/journal.pone.0097047
Hu B et al (2015) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47:834–838. https://doi.org/10.1038/ng.3337
Huang S, Spielmeyer W, Lagudah ES, Munns R (2008) Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J Exp Bot 59:927–937. https://doi.org/10.1093/jxb/ern033
Huang X, Shabala S, Shabala L, Rengel Z, Wu X, Zhang G, Zhou M (2015) Linking waterlogging tolerance with Mn2+ toxicity: a case study for barley. Plant Biol 17:26–33. https://doi.org/10.1111/plb.12188
Ingvordsen CH, Backes G, Lyngkjaer MF, Peltonen-Sainio P, Jahoor A, Mikkelsen TN, Jorgensen RB (2015a) Genome-wide association study of production and stability traits in barley cultivated under future climate scenarios. Mol Breed 35. https://doi.org/10.1007/s11032-015-0283-8
Ingvordsen CH et al (2015b) Significant decrease in yield under future climate conditions: stability and production of 138 spring barley accessions. Eur J Agron 63:105–113. https://doi.org/10.1016/j.eja.2014.12.003
IPCC (2014) Climate change 2014: synthesis report. IPCC, Geneva, Switzerland
Islam S, Malik AI, Islam AK, Colmer TD (2007) Salt tolerance in a Hordeum marinum-Triticum aestivum amphiploid, and its parents. J Exp Bot 58:1219–1229. https://doi.org/10.1093/jxb/erl293
James RA, Munns R, Von Caemmerer S, Trejo C, Miller C, Condon T (2006) Photosynthetic capacity is related to the cellular and subcellular partitioning of Na+, K+ and Cl− in salt-affected barley and durum wheat. Plant, Cell Environ 29:2185–2197. https://doi.org/10.1111/j.1365-3040.2006.01592.x
Jefferies SP et al (1999) Mapping of chromosome regions conferring boron toxicity tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 98:1293–1303. https://doi.org/10.1007/s001220051195
Jukanti AK, Fischer AM (2008) A high-grain protein content locus on barley (Hordeum vulgare) chromosome 6 is associated with increased flag leaf proteolysis and nitrogen remobilization. Physiol Plant 132:426–439. https://doi.org/10.1111/j.1399-3054.2007.01044.x
Jukanti AK, Heidlebaugh NM, Parrott DL, Fischer IA, McInnerney K, Fischer AM (2008) Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol 177:333–349. https://doi.org/10.1111/j.1469-8137.2007.02270.x
Karley AJ, Valentine TA, Squire GR (2011) Dwarf alleles differentially affect barley root traits influencing nitrogen acquisition under low nutrient supply. J Exp Bot 62:3917–3927. https://doi.org/10.1093/jxb/err089
Knox AK, Dhillon T, Cheng HM, Tondelli A, Pecchioni N, Stockinger EJ (2010) CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121:21–35. https://doi.org/10.1007/s00122-010-1288-7
Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260. https://doi.org/10.1146/annurev.arplant.46.1.237
Kosova K, Vitamvas P, Prasil IT (2011) Expression of dehydrins in wheat and barley under different temperatures. Plant Sci 180:46–52. https://doi.org/10.1016/j.plantsci.2010.07.003
Kosova K, Vitamvas P, Prasil IT (2014) Wheat and barley dehydrins under cold, drought, and salinity—what can LEA-II proteins tell us about plant stress response? Front Plant Sci 5:343. https://doi.org/10.3389/fpls.2014.00343
Kruszka K et al (2014) Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J Exp Bot 65:6123–6135. https://doi.org/10.1093/jxb/eru353
Lakew B, Henry RJ, Ceccarelli S, Grando S, Eglinton J, Baum M (2013) Genetic analysis and phenotypic associations for drought tolerance in Hordeum spontaneum introgression lines using SSR and SNP markers. Euphytica 189:9–29. https://doi.org/10.1007/s10681-012-0674-4
Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH (2001) Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Gene Dev 15:1808–1816. https://doi.org/10.1101/gad.900401
Leplat F, Pedas PR, Rasmussen SK, Husted S (2016) Identification of manganese efficiency candidate genes in winter barley (Hordeum vulgare) using genome wide association mapping. BMC Genom 17:775. https://doi.org/10.1186/s12864-016-3129-9
Li HB, Vaillancourt R, Mendham N, Zhou MX (2008) Comparative mapping of quantitative trait loci associated with waterlogging tolerance in barley (Hordeum vulgare L.). BMC Genomics 9. https://doi.org/10.1186/1471-2164-9-401
Ligaba A, Katsuhara M (2010) Insights into the salt tolerance mechanism in barley (Hordeum vulgare) from comparisons of cultivars that differ in salt sensitivity. J Plant Res 123:105–118. https://doi.org/10.1007/s10265-009-0272-2
Liu X et al (2017) QTLs for stomatal and photosynthetic traits related to salinity tolerance in barley. BMC Genomics 18:9. https://doi.org/10.1186/s12864-016-3380-0
Lonergan PF, Pallotta MA, Lorimer M, Paull JG, Barker SJ, Graham RD (2009) Multiple genetic loci for zinc uptake and distribution in barley (Hordeum vulgare). New Phytol 184:168–179. https://doi.org/10.1111/j.1469-8137.2009.02956.x
Long NV, Dolstra O, Malosetti M, Kilian B, Graner A, Visser RGF, van der Linden CG (2013) Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 126:2335–2351. https://doi.org/10.1007/s00122-013-2139-0
Lundqvist U, Lundqvist A (1988) Mutagen specificity in barley for 1580 eceriferum mutants localized to 79 loci. Hereditas 108:1–12. https://doi.org/10.1111/j.1601-5223.1988.tb00676.x
Ma JF, Ryan PR, Delhaize E (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6:273–278. https://doi.org/10.1016/S1360-1385(01)01961-6
Ma JF, Nagao S, Sato K, Ito H, Furukawa J, Takeda K (2004) Molecular mapping of a gene responsible for Al-activated secretion of citrate in barley. J Exp Bot 55:1335–1341. https://doi.org/10.1093/jxb/erh152
Ma Y, Shabala S, Li C, Liu C, Zhang W, Zhou M (2015) Quantitative trait loci for salinity tolerance identified under drained and waterlogged conditions and their association with flowering time in barley (Hordeum vulgare. L). PLoS ONE 10:e0134822. https://doi.org/10.1371/journal.pone.0134822
Ma Y et al (2016) A new allele for aluminium tolerance gene in barley (Hordeum vulgare L.). BMC Genom 17:186. https://doi.org/10.1186/s12864-016-2551-3
MacLeod L, Duffus C (1988) Reduced starch content and sucrose synthase activity in developing endosperm of barley plants grown at elevated temperatures. Funct Plant Biol 15:367–375
Mahfoozi S, Limin AE, Fowler DB (2001a) Developmental regulation of low-temperature tolerance in winter wheat. Ann Bot 87:751–757. https://doi.org/10.1006/anbo.2001.1403
Mahfoozi S, Limin AE, Fowler DB (2001b) Influence of vernalization and photoperiod responses on cold hardiness in winter cereals. Crop Sci 41:1006–1011. https://doi.org/10.2135/cropsci2001.4141006x
Mangelsen E, Kilian J, Harter K, Jansson C, Wanke D, Sundberg E (2011) Transcriptome analysis of high-temperature stress in developing barley caryopses: early stress responses and effects on storage compound biosynthesis. Mol Plant 4:97–115. https://doi.org/10.1093/mp/ssq058
Mano Y, Takeda K (1997) Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L). Euphytica 94:263–272. https://doi.org/10.1023/A:1002968207362
Maršálová L, Vítámvás P, Hynek R, Prášil IT, Kosová K (2016) Proteomic response of Hordeum vulgare cv. Tadmor and Hordeum marinum to salinity stress: similarities and differences between a glycophyte and a halophyte. Front Plant Sci 7:1154. https://doi.org/10.3389/fpls.2016.01154
Mascher M et al (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544:427–433. https://doi.org/10.1038/nature22043
Maser P et al (2002) Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA 99:6428–6433. https://doi.org/10.1073/pnas.082123799
Mastrangelo AM, Baldi P, Mare C, Terzi V, Galiba G, Cattivelli L, Di Fonzo N (2000) The cold dependent accumulation of COR TMC-AP3 in cereals with contrasting, frost tolerance is regulated by different mRNA expression and protein turnover. Plant Sci 156:47–54. https://doi.org/10.1016/S0168-9452(00)00228-4
Matin MA, Brown JH, Ferguson H (1989) Leaf water potential, relative water content, and diffusive resistance as screening techniques for drought resistance in barley. Agron J 81:100–105. https://doi.org/10.2134/agronj1989.00021962008100010018x
Mayer KFX et al (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716. https://doi.org/10.1038/nature11543
McAllister CH, Beatty PH, Good AG (2012) Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol J 10:1011–1025. https://doi.org/10.1111/j.1467-7652.2012.00700.x
McDonald GK (2006) Effects of soil properties on variation in growth, grain yield and nutrient concentration of wheat and barley. Aust J Exp Agr 46:93–105. https://doi.org/10.1071/ea04015
McDonald GK, Eglinton JK, Barr AR (2010) Assessment of the agronomic value of QTL on chromosomes 2H and 4H linked to tolerance to boron toxicity in barley (Hordeum vulgare L.). Plant Soil 326:275–290. https://doi.org/10.1007/s11104-009-0006-1
Mendiondo GM et al (2016) Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnol J 14:40–50. https://doi.org/10.1111/pbi.12334
Mian A, Oomen RJ, Isayenkov S, Sentenac H, Maathuis FJ, Véry AA (2011) Over-expression of an Na+-and K+-permeable HKT transporter in barley improves salt tolerance. Plant J 68:468–479. https://doi.org/10.1111/j.1365-313X.2011.04701.x
Mickelson S, See D, Meyer FD, Garner JP, Foster CR, Blake TK, Fischer AM (2003) Mapping of QTL associated with nitrogen storage and remobilization in barley (Hordeum vulgare L.) leaves. J Exp Bot 54:801–812. https://doi.org/10.1093/jxb/erg084
Mikolajczak K et al (2017) Quantitative trait loci for plant height in Maresi × CamB barley population and their associations with yield-related traits under different water regimes. J Appl Genet 58:23–35. https://doi.org/10.1007/s13353-016-0358-1
Minella E, Sorrells ME (1992) Aluminum tolerance in barley: Genetic relationships among genotypes of diverse origin. Crop Sci 32:593–598
Minella E, Sorrells ME (1997) Inheritance and chromosome location of Alp, a gene controlling aluminum tolerance in ‘Dayton’ barley. Plant Breed 116:465–469. https://doi.org/10.1111/j.1439-0523.1997.tb01032.x
Modhej A, Farhoudi R, Afrous A (2015) Effect of post-anthesis heat stress on grain yield of barley, durum and bread wheat genotypes. JSRAD 2:127–131
Morran S et al (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9:230–249. https://doi.org/10.1111/j.1467-7652.2010.00547.x
Munns R (2002) Comparative physiology of salt and water stress. Plant, Cell Environ 25:239–250. https://doi.org/10.1046/j.0016-8025.2001.00808.x
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Munns R et al (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30:360–364. https://doi.org/10.1038/nbt.2120
Newton AC et al (2011) Crops that feed the world 4. Barley: a resilient crop? Strengths and weaknesses in the context of food security. Food Secur 3:141–178. https://doi.org/10.1007/s12571-011-0126-3
Nice LM et al (2016) Development and genetic characterization of an advanced backcross-nested association mapping (AB-NAM) population of wild × cultivated barley. Genetics 203:1453–1467. https://doi.org/10.1534/genetics.116.190736
Oh SJ, Kwon CW, Choi DW, Song SI, Kim JK (2007) Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol J 5:646–656. https://doi.org/10.1111/j.1467-7652.2007.00272.x
Oraby HF, Ransom CB, Kravchenko AN, Sticklen MB (2005) Barley HVA1 gene confers salt tolerance in R3 transgenic oat. Crop Sci 45:2218–2227. https://doi.org/10.2135/cropsci2004-0605
Oshino T, Abiko M, Saito R, Ichiishi E, Endo M, Kawagishi-Kobayashi M, Higashitani A (2007) Premature progression of anther early developmental programs accompanied by comprehensive alterations in transcription during high-temperature injury in barley plants. Mol Genet Genomics 278:31–42. https://doi.org/10.1007/s00438-007-0229-x
Pasquariello M et al (2014) The barley frost resistance-H2 locus. Funct Integr Genomics 14:85–100. https://doi.org/10.1007/s10142-014-0360-9
Pecchioni N et al (2014) Genomics of low-temperature tolerance for an increased sustainability of wheat and barley production. In: Genomics of plant genetic resources. Springer, Netherlands, pp 149–183. https://doi.org/10.1007/978-94-007-7575-6_6
Peighambari SA, Samadi BY, Nabipour A, Charmet G, Sarrafi A (2005) QTL analysis for agronomic traits in a barley doubled haploids population grown in Iran. Plant Sci 169:1008–1013. https://doi.org/10.1016/j.plantsci.2005.05.018
Persson DP et al (2006) Multi-elemental speciation analysis of barley genotypes differing in tolerance to cadmium toxicity using SEC-ICP-MS and ESI-TOF-MS. J Anal At Spectrom 21:996–1005. https://doi.org/10.1039/b608701a
Phillips D et al (2015) The effect of temperature on the male and female recombination landscape of barley. New Phytol 208:421–429. https://doi.org/10.1111/nph.13548
Platten JD et al (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11:372–374. https://doi.org/10.1016/j.tplants.2006.06.001
Poets AM, Fang Z, Clegg MT, Morrell PL (2015) Barley landraces are characterized by geographically heterogeneous genomic origins. Genome Biol 16:173. https://doi.org/10.1186/s13059-015-0712-3
Qadir M et al (2014) Economics of salt-induced land degradation and restoration. Nat Resour Forum 38:282–295. https://doi.org/10.1111/1477-8947.12054
Quan X, Zeng J, Ye L, Chen G, Han Z, Shah JM, Zhang G (2016) Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol 16:1–16. https://doi.org/10.1186/s12870-016-0721-8
Rae AL, Cybinski DH, Jarmey JM, Smith FW (2003) Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53:27–36. https://doi.org/10.1023/b:plan.0000009259.75314.15
Raman H, Moroni S, Sato K, Read J, Scott J (2002) Identification of AFLP and microsatellite markers linked with an aluminium tolerance gene in barley (Hordeum vulgare L.). Theor Appl Genet 105:458–464. https://doi.org/10.1007/s00122-002-0934-0
Raman H et al (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48:781–791. https://doi.org/10.1139/g05-054
Raun WR, Johnson GV (1999) Improving nitrogen use efficiency for cereal production. Agron J 91:357–363. https://doi.org/10.2134/agronj1999.00021962009100030001x
Rea PA, Poole RJ (1993) Vacuolar H+-translocating pyrophosphatase. Annu Rev Plant Physiol Plant Mol Biol 44:157–180. https://doi.org/10.1146/annurev.arplant.44.1.157
Reid DA (1971) Genetic control of reaction to aluminum in winter barley. In: Second international barley genetics symposium, Washington State University, pp 409–413
Reid R (2010) Can we really increase yields by making crop plants tolerant to boron toxicity? Plant Sci 178:9–11. https://doi.org/10.1016/j.plantsci.2009.10.006
Reid DA, Jones GD, Armiger WH, Foy CD, Koch EJ, Starling TM (1969) Differential aluminum tolerance of winter barley varieties and selections in associated greenhouse and field experiments. Agron J 61:218–222. https://doi.org/10.2134/agronj1969.00021962006100020014x
Reinert S, Kortz A, Leon J, Naz AA (2016) Genome-wide association mapping in the global diversity set reveals new QTL controlling root system and related shoot variation in barley. Front Plant Sci 7:1061. https://doi.org/10.3389/fpls.2016.01061
Reinheimer JL, Barr AR, Eglinton JK (2004) QTL mapping of chromosomal regions conferring reproductive frost tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 109:1267–1274. https://doi.org/10.1007/s00122-004-1736-3
Rivandi J, Miyazaki J, Hrmova M, Pallotta M, Tester M, Collins NC (2011) A SOS3 homologue maps to HvNax4, a barley locus controlling an environmentally sensitive Na+ exclusion trait. J Exp Bot 62:1201–1216. https://doi.org/10.1093/jxb/erq346
Robredo A, Perez-Lopez U, de la Maza HS, Gonzalez-Moro B, Lacuesta M, Mena-Petite A, Munoz-Rueda A (2007) Elevated CO2 alleviates the impact of drought on barley improving water status by lowering stomatal conductance and delaying its effects on photosynthesis. Environ Exp Bot 59:252–263. https://doi.org/10.1016/j.envexpbot.2006.01.001
Robredo A, Perez-Lopez U, Lacuesta M, Mena-Petite A, Munoz-Rueda A (2010) Influence of water stress on photosynthetic characteristics in barley plants under ambient and elevated CO2 concentrations. Biol Plant 54:285–292. https://doi.org/10.1007/s10535-010-0050-y
Rollins JA et al (2013) Variation at the vernalisation genes Vrn-H1 and Vrn-H2 determines growth and yield stability in barley (Hordeum vulgare) grown under dryland conditions in Syria. Theor Appl Genet 126:2803–2824. https://doi.org/10.1007/s00122-013-2173-y
Roy SJ, Huang W, Wang XJ, Evrard A, Schmockel SM, Zafar ZU, Tester M (2013) A novel protein kinase involved in Na+ exclusion revealed from positional cloning. Plant, Cell Environ 36:553–568. https://doi.org/10.1111/j.1365-3040.2012.02595.x
Roy SJ, Negrão S, Tester M (2014) Salt resistant crop plants. Curr Opin Biotechnol 26:115–124. https://doi.org/10.1016/j.copbio.2013.12.004
Ryan J, Singh M, Yau SK (1998) Spatial variability of soluble boron in Syrian soils. Soil Tillage Res 45:407–417. https://doi.org/10.1016/S0933-3630(97)00037-8
Ryan P, Delhaize E, Jones D (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52:527–560. https://doi.org/10.1146/annurev.arplant.52.1.527
Saade S et al. (2016) Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Sci Rep 6:32586. https://doi.org/10.1038/srep32586
Sakata T, Takahashi H, Nishiyama I, Higashitani A (2000) Effects of high temperature on the development of pollen mother cells and microspores in barley Hordeum vulgare L. J Plant Res 113:395–402. https://doi.org/10.1007/Pl00013947
Sannemann W, Huang BE, Mathew B, Léon J (2015) Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol Breed 35:86. https://doi.org/10.1007/s11032-015-0284-7
Santa-Maria GE, Rubio F, Dubcovsky J, Rodriguez-Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9:2281–2289. https://doi.org/10.1105/tpc.9.12.2281
Sasaki T et al (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37:645–653. https://doi.org/10.1111/j.1365-313X.2003.01991.x
Savin R, Stone P, Nicolas M (1996) Responses of grain growth and malting quality of barley to short periods of high temperature in field studies using portable chambers. Aust J Agric Res 47:465–477. https://doi.org/10.1071/AR9960465
Sayed MA, Schumann H, Pillen K, Naz AA, Leon J (2012) AB-QTL analysis reveals new alleles associated to proline accumulation and leaf wilting under drought stress conditions in barley (Hordeum vulgare L.). BMC Genet 13. https://doi.org/10.1186/1471-2156-13-61
Schilling RK, Marschner P, Shavrukov Y, Berger B, Tester M, Roy SJ, Plett DC (2014) Expression of the Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) improves the shoot biomass of transgenic barley and increases grain yield in a saline field. Plant Biotechnol J 12:378–386. https://doi.org/10.1111/pbi.12145
Schneider LM et al (2016) The Cer-cqu gene cluster determines three key players in a β-diketone synthase polyketide pathway synthesizing aliphatics in epicuticular waxes. J Exp Bot 67:2715–2730. https://doi.org/10.1093/jxb/erw105
Schnurbusch T et al (2010) Boron toxicity tolerance in barley through reduced expression of the multifunctional aquaporin HvNIP2;1. Plant Physiol 153:1706–1715. https://doi.org/10.1104/pp.110.158832
Schünmann PHD, Richardson AE, Smith FW, Delhaize E (2004a) Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J Exp Bot 55:855–865. https://doi.org/10.1093/jxb/erh103
Schünmann PHD, Richardson AE, Vickers CE, Delhaize E (2004b) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136:4205–4214. https://doi.org/10.1104/pp.104.045823
Setter TL, Waters I (2003) Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil 253:1–34. https://doi.org/10.1023/A:1024573305997
Shavrukov Y et al (2010) HvNax3—a locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum). Funct Integr Genomics 10:277–291. https://doi.org/10.1007/s10142-009-0153-8
Shavrukov Y, Bovill J, Afzal I, Hayes JE, Roy SJ, Tester M, Collins NC (2013) HVP10 encoding V-PPase is a prime candidate for the barley HvNax3 sodium exclusion gene: evidence from fine mapping and expression analysis. Planta 237:1111–1122. https://doi.org/10.1007/s00425-012-1827-3
Shrawat AK, Carroll RT, DePauw M, Taylor GJ, Good AG (2008) Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol J 6:722–732. https://doi.org/10.1111/j.1467-7652.2008.00351.x
Sicher RC, Timlin D, Bailey B (2012) Responses of growth and primary metabolism of water-stressed barley roots to rehydration. J Plant Physiol 169:686–695. https://doi.org/10.1016/j.jplph.2012.01.002
Singh M, Kumar J, Singh S, Singh VP, Prasad SM (2015) Roles of osmoprotectants in improving salinity and drought tolerance in plants: a review. Rev Environ Sci Biotechnol 14:407–426. https://doi.org/10.1007/s11157-015-9372-8
Smith FW, Mudge SR, Rae AL, Glassop D (2003) Phosphate transport in plants. Plant Soil 248:71–83. https://doi.org/10.1023/a:1022376332180
Snyder RL, Melo-Abreu JPd (2005) Frost protection: fundamentals, practice and economics. Environment and natural resources series. Food and Agriculture Organization of the United Nations, Rome
Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N (2007) Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J 51:308–321. https://doi.org/10.1111/j.1365-313X.2007.03141.x
Stolen O, Andersen S (1978) Inheritance of tolerance to low soil pH in barley. Hereditas 88:101–105. https://doi.org/10.1111/j.1601-5223.1978.tb01608.x
Suprunova T, Krugman T, Distelfeld A, Fahima T, Nevo E, Korol A (2007) Identification of a novel gene (Hsdr4) involved in water-stress tolerance in wild barley. Plant Mol Biol 64:17–34. https://doi.org/10.1007/s11103-006-9131-x
Sutton T et al (2007) Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318:1446–1449. https://doi.org/10.1126/science.1146853
Sylvester-Bradley R, Kindred DR (2009) Analysing nitrogen responses of cereals to prioritize routes to the improvement of nitrogen use efficiency. J Exp Bot 60:1939–1951. https://doi.org/10.1093/jxb/erp116
Sze H, Ward JM, Lai SP (1992) Vacuolar H+-translocating ATPases from plants: Structure, function, and isoforms. J Bioenerg Biomembr 24:371–381. https://doi.org/10.1007/Bf00762530
Takano J, Wada M, Ludewig U, Schaaf G, von Wiren N, Fujiwara T (2006) The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18:1498–1509. https://doi.org/10.1105/tpc.106.041640
Talame V et al (2004) Identification of Hordeum spontaneum QTL alleles improving field performance of barley grown under rainfed conditions. Ann Appl Biol 144:309–319. https://doi.org/10.1111/j.1744-7348.2004.tb00346.x
Teotia S, Singh D, Tang X, Tang G (2016) Essential RNA-based technologies and their applications in plant functional genomics. Trends Biotechnol 34:106–123. https://doi.org/10.1016/j.tibtech.2015.12.001
Teulat B, Monneveux P, Wery J, Borries C, Souyris I, Charrier A, This D (1997) Relationships between relative water content and growth parameters under water stress in barley: a QTL study. New Phytol 137:99–107. https://doi.org/10.1046/j.1469-8137.1997.00815.x
Teulat B et al (1998) Several QTLs involved in osmotic adjustment trait variation in barley (Hordeum vulgare L.). Theor Appl Genet 96:688–698. https://doi.org/10.1007/s001220050790
Teulat B, Borries C, This D (2001a) New QTLs identified for plant water status, water-soluble carbohydrate and osmotic adjustment in a barley population grown in a growth-chamber under two water regimes. Theor Appl Genet 103:161–170. https://doi.org/10.1007/s001220000503
Teulat B, Merah O, Souyris I, This D (2001b) QTLs for agronomic traits from a Mediterranean barley progeny grown in several environments. Theor Appl Genet 103:774–787. https://doi.org/10.1007/s001220100619
Teulat B, Merah O, Sirault X, Borries C, Waugh R, This D (2002) QTLs for grain carbon isotope discrimination in field-grown barley. Theor Appl Genet 106:118–126. https://doi.org/10.1007/s00122-002-1028-8
Teulat B, Zoumarou-Wallis N, Rotter B, Ben Salem M, Bahri H, This D (2003) QTL for relative water content in field-grown barley and their stability across Mediterranean environments. Theor Appl Genet 108:181–188. https://doi.org/10.1007/s00122-003-1417-7
Thomashow MF (1998) Role of cold-responsive genes in plant freezing tolerance. Plant Physiol 118:1–7. https://doi.org/10.1104/pp.118.1.1
Thomsen HC, Eriksson D, Møller IS, Schjoerring JK (2014) Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency? Trends Plant Sci 19:656–663. https://doi.org/10.1016/j.tplants.2014.06.002
Tombesi S et al (2015) Stomatal closure is induced by hydraulic signals and maintained by ABA in drought-stressed grapevine. Sci Rep 5. https://doi.org/10.1038/srep12449
Tommasini L et al (2008) Dehydrin gene expression provides an indicator of low temperature and drought stress: transcriptome-based analysis of Barley (Hordeum vulgare L.). Funct Integr Genomics 8:387–405. https://doi.org/10.1007/s10142-008-0081-z
Tondelli A et al (2006) Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet 112:445–454. https://doi.org/10.1007/s00122-005-0144-7
Tondelli A, Francia E, Barabaschi D, Pasquariello M, Pecchioni N (2011) Inside the CBF locus in Poaceae. Plant Sci 180:39–45. https://doi.org/10.1016/j.plantsci.2010.08.012
Tondelli A et al (2014) Allelic variation at Fr-H1/Vrn-H1 and Fr-H2 loci is the main determinant of frost tolerance in spring barley. Environ Exp Bot 106:148–155. https://doi.org/10.1016/j.envexpbot.2014.02.014
Uga Y et al (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45:1097–1102. https://doi.org/10.1038/ng.2725
Ugarte C, Calderini DF, Slafer GA (2007) Grain weight and grain number responsiveness to pre-anthesis temperature in wheat, barley and triticale. Field Crops Res 100:240–248. https://doi.org/10.1016/j.fcr.2006.07.010
Vasekina AV, Yershov PV, Reshetova OS, Tikhonova TV, Lunin VG, Trofimova MS, Babakov AV (2005) Vacuolar Na+/H+ antiporter from barley: identification and response to salt stress. Biokhim Biochem (Mosc) 70:100–107. https://doi.org/10.1007/pl00021768
Venema K, Quintero FJ, Pardo JM, Donaire JP (2002) The Arabidopsis Na+/H+exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J Biol Chem 277:2413–2418. https://doi.org/10.1074/jbc.M105043200
Vidmar JJ, Zhuo D, Siddiqi MY, Glass ADM (2000a) Isolation and characterization of HvNRT2.3 and HvNRT2.4, cDNAs encoding high-affinity nitrate transporters from roots of barley. Plant Physiol 122:783–792. https://doi.org/10.1104/pp.122.3.783
Vidmar JJ, Zhuo D, Siddiqi MY, Schjoerring JK, Touraine B, Glass ADM (2000b) Regulation of high-affinity nitrate transporter genes and high-affinity nitrate influx by nitrogen pools in roots of barley. Plant Physiol 123:307–318. https://doi.org/10.1104/pp.123.1.307
von Korff M, Grando S, Del Greco A, This D, Baum M, Ceccarelli S (2008) Quantitative trait loci associated with adaptation to Mediterranean dryland conditions in barley. Theor Appl Genet 117:653–669. https://doi.org/10.1007/s00122-008-0787-2
von Zitzewitz J et al (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59:449–467. https://doi.org/10.1007/s11103-005-0351-2
von Zitzewitz J et al (2011) The genetics of winterhardiness in barley: perspectives from genome-wide association mapping. Plant Genome 4:76–91. https://doi.org/10.3835/plantgenome2010.12.0030
Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223. https://doi.org/10.1016/j.envexpbot.2007.05.011
Wallwork M, Logue S, MacLeod L, Jenner C (1998) Effects of a period of high temperature during grain filling on the grain growth characteristics and malting quality of three Australian malting barleys. Aust J Agric Res 49:1287–1296. https://doi.org/10.1071/A98004
Wang JP et al (2007) High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 115:265–276. https://doi.org/10.1007/s00122-007-0562-9
Wang P, Li ZW, Wei JS, Zhao ZL, Sun DY, Cui SJ (2012a) A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J Biol Chem 287:44062–44070. https://doi.org/10.1074/jbc.M112.351643
Wang Y-Y, Hsu P-K, Tsay Y-F (2012b) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17:458–467. https://doi.org/10.1016/j.tplants.2012.04.006
Wei Z et al (2016) Comparative transcriptome analysis revealed genes commonly responsive to varied nitrate stress in leaves of Tibetan hulless barley. Front Plant Sci 7:1067. https://doi.org/10.3389/fpls.2016.01067
Wicke B, Smeets E, Dornburg V, Vashev B, Gaiser T, Turkenburg W, Faaij A (2011) The global technical and economic potential of bioenergy from salt-affected soils. Energ Environ Sci 4:2669–2681. https://doi.org/10.1039/c1ee01029h
Wu H, Shabala L, Barry K, Zhou M, Shabala S (2013) Ability of leaf mesophyll to retain potassium correlates with salinity tolerance in wheat and barley. Physiol Plant. https://doi.org/10.1111/ppl.12056
Wu DZ, Sato K, Ma JF (2015) Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol 208:817–829. https://doi.org/10.1111/nph.13512
Xia YS et al (2013) Single nucleotide polymorphisms in HSP17.8 and their association with agronomic traits in barley. PLOS ONE 8. https://doi.org/10.1371/journal.pone.0056816
Xu D, Duan X, Wang B, Hong B, Ho T, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110:249–257. https://doi.org/10.1104/pp.110.1.249
Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182. https://doi.org/10.1146/annurev-arplant-042811-105532
Xue DW et al (2009) Identification of QTLs associated with salinity tolerance at late growth stage in barley. Euphytica 169:187–196. https://doi.org/10.1007/s10681-009-9919-2
Yadav SK (2010) Cold stress tolerance mechanisms in plants. Agron Sustain Dev 30(3):515–527. https://doi.org/10.1051/agro/2009050
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268. https://doi.org/10.1073/pnas.0937399100
Yang L, Mickelson S, See D, Blake TK, Fischer AM (2004) Genetic analysis of the function of major leaf proteases in barley (Hordeum vulgare L.) nitrogen remobilization. J Exp Bot 55:2607–2616. https://doi.org/10.1093/jxb/erh267
Yau SK, Ryan J (2008) Boron toxicity tolerance in crops: A viable alternative to soil amelioration. Crop Sci 48:854–865. https://doi.org/10.2135/cropsci2007.10.0539
Zeng J et al (2014) Comparative transcriptome profiling of two Tibetan wild barley genotypes in responses to low potassium. PLOS ONE 9:e100567. https://doi.org/10.1371/journal.pone.0100567
Zhang G, Li C (2016) Exploration and utilization of aluminum-tolerant barley germplasm. In: Exploration, identification and utilization of barley germplasm, pp 181–208
Zhang X, Davidson EA, Mauzerall DL, Searchinger TD, Dumas P, Shen Y (2015) Managing nitrogen for sustainable development. Nature 528:51–59. https://doi.org/10.1038/nature15743
Zheng BY, Chapman SC, Christopher JT, Frederiks TM, Chenu K (2015) Frost trends and their estimated impact on yield in the Australian wheatbelt. J Exp Bot 66:3611–3623. https://doi.org/10.1093/jxb/erv163
Zhou GF, Delhaize E, Zhou MX, Ryan PR (2013) The barley MATE gene, HvAACT1, increases citrate efflux and Al3+ tolerance when expressed in wheat and barley. Ann Bot 112:603–612. https://doi.org/10.1093/aob/mct135
Zhou GF, Pereira JF, Delhaize E, Zhou MX, Magalhaes JV, Ryan PR (2014) Enhancing the aluminium tolerance of barley by expressing the citrate transporter genes SbMATE and FRD3. J Exp Bot 65:2381–2390. https://doi.org/10.1093/jxb/eru121
Zhou G, Broughton S, Zhang XQ, Ma Y, Zhou M, Li C (2016) Genome-wide association mapping of acid soil resistance in barley (Hordeum vulgare L.). Front Plant Sci 7:406. https://doi.org/10.3389/fpls.2016.00406
Zohary D (1999) Monophyletic versus polyphyletic origin of the crops on which agriculture was founded in the near east. Genet Resour Crop Evol 46:133–142. https://doi.org/10.1023/a:1008692912820
Acknowledgements
We thank Ivan Gromicho, scientific illustrator from King Abdullah University of Science and Technology, for the scientific illustration in this book chapter. Financial support from King Abdullah University of Science and Technology (KAUST) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Saade, S., Negrão, S., Plett, D., Garnett, T., Tester, M. (2018). Genomic and Genetic Studies of Abiotic Stress Tolerance in Barley. In: Stein, N., Muehlbauer, G. (eds) The Barley Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-319-92528-8_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-92528-8_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-92527-1
Online ISBN: 978-3-319-92528-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)