Abstract
Liver diseases have produced a big data such as metabolomics analyses, electronic health records, and report including patient medical information, and disorders. However, these data must be analyzed and integrated if they are to produce models about physiological mechanisms of pathogenesis. We use machine learning based on classifier for big datasets in the fields of liver to Predict and therapeutic discovery. A dataset was developed with twenty three attributes that include the records of 7000 patients in which 5295 patients were male and rests were female. Support Vector Machine (SVM), Boosted C5.0, and Naive Bayes (NB), data mining techniques are used with the proposed model for the prediction of liver diseases. The performance of these classifier techniques are evaluated with accuracy, sensitivity, specificity.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
According to the WHO in Egypt the chronic diseases are responsible for 78% (382,000/495,000) of total deaths and in next 10 years twenty lakhs of people will die due to chronic diseases [1]. Liver diseases also come in the category of chronic diseases. A large number of infections affect the liver which led to the various liver diseases. The deaths due to the liver diseases have reached 208185 or 7.34% of total death in Egypt [2]. Liver is one of the strongest organs in our body that sits on the right side of the belly. Color of the liver is reddish-brown. The liver has two sections in our body, one is called right section and the other is left section. The place of gallbladder is under the liver, alongside parts of the pancreas and guts. The liver and these organs cooperate to assimilate and process sustenance. Liver sanitizes the blood originating from the digestive tract. Likewise, cleans chemicals and metabolizes drugs. The liver shrouds bile that winds up back in the inner parts. The liver additionally makes proteins imperative for blood coagulating. The liver is in charge of the evacuation of pathogens and exogenous antigens from the systemic flow. Liver infection is alluded to as hepatic ailment. Liver infection is a general term that covers all the potential issues that bring about the liver to neglect to perform its assigned capacities. This study mainly discusses about five types of liver diseases such as alcoholic liver damage (ALD), liver cirrhosis (LC), primary hepatoma (PM), cholelithiasis (C) [3] and HCC [4].
One of the main causes of increased liver diseases in Egypt is obesity, inhale of harmful gases, intake of contaminated food, excessive consumption pickles and drugs, alcohol [2]. The objective of this paper is to propose a Machine learning techniques based on Classification of Liver disorders for reduce burden on doctors.
The organization of this paper is as follows. In Sect. 2, some related work on data mining, liver disease, classification algorithms, machine learning, and related works are provided. Section 3 describes our method in the implementation of Support Vector Machine (SVM), Boosted C5.0, and Naive Bayes (NB) classification algorithms for the early detection of liver diseases. Finally, in Sect. 4, Conclusion and future works.
2 Related Work
2.1 Machine Learning and Knowledge Discovery
Biomedical science is one of the important areas where data mining is used. Since this branch of science deals with human life, it is highly sensitivities. In recent years, a lot of researches have been done on a variety of diseases using data mining.
Looking more closely at the research done in recent years in this field, specifically, in the biomedical field, we can see many works that use data mining for forecasting, prevention and treatment of patients [5].
In biomedical science, accuracy and speed are two important factors that should be considered chiefly in dealing with any disease. In this regard, data mining techniques can be of great help to physicians.
With advances, several machines have entered in our lives. One of the most famous areas where computers as the mostly used machines can be helpful is knowledge extraction with the help of a machine (machine learning).
This approach that can be of great help to all scientific fields is called data mining orKnowledge Discovery of the Datasets as shown in Fig 1. Supervised and unsupervised learning are two main methods for machine learning [6]. The purpose of these methods is to learn by use of data mining approaches and to use part of data for training and the other part for test.
The Data mining or Knowledge Discovery in Datasets (KDD) process [7].
2.2 Liver Diseases
Liver is largest internal organ of the body. It plays a significant role in transfer of blood throughout our body. The levels of most chemicals in our blood are regulated by the liver. It helps in metabolism of the alcohol, drugs and destroys toxic substances. Liver can be infected by parasites, viruses which cause inflammation and diminish its function [8]. It has the potential to maintain the customary function, even when a part of it is damaged. However, it is important to diagnose liver disease early which can increase the patient’s survival rate. Expert physicians are required for various examination tests to diagnose the liver disease, but it cannot assure the correct diagnosis [9]. Accordingly, the mortality and morbidity impact of chronic liver disease is greatest among the population of Egypt.
2.3 Big Data
Big data turns into segments due to multidisciplinary combined effort of machine learning, datasets and statistics. Today, in biomedical sciences disease diagnostic test is a serious task [10]. In biomedical sciences disease diagnostic test is a serious task.
It is important to understand the exact diagnosis of patients by assessment and clinical examination. Medical field generates big data about report regarding patient, clinical assessment, cure, follow-ups, and medication. Enhancement in big data needs some proper means to extract and process data effectively and efficiently [11]. One of the many machine-learning is employed to build such classifier that can divide the data on the basis of their attributes. Dataset is divided into two or more than two classes. Such classifiers are used for medical big data analysis and disease detection.
2.4 Classification Algorithms
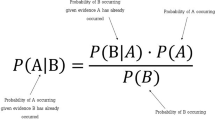
Naive Bayes classifier
Naive Bayes is a statistical classifier based on Bayes theorem for Thomas Bayes who worked in decision theory and probability [12]. Some literation mentioned that the Naive Bayes has simplicity, traceability and fast learner. On the other hand, many authors concentrated on class conditional independence assumption’s advantages and disadvantages due to its influence on Naive Bayes performance whereas the Naive Bayes assumes that the attributes are independent among each other on a given class.
To compute the posterior probability that tuple X = (x1, x2, x3,..xn) belongs to the class Ci, we use the Eq. 1 below where xi is the value of attribute Ai and xn is the value of attribute An.
Where
-
P (c/x) is the posterior probability of class (target) given predictor (attribute).
-
P(c) is the prior probability of class.
-
P (x/c) is the likelihood which is the probability of predictor given class.
-
P(x) is the prior probability of predictor
C5.0 classifier
The decision tree such as C5.0, Id3, or CART can handle the real world datasets efficiently [13]. C5.0 is a Decision Tree was designed ID3 which is based on information gain. Because of the ID3 biases of multivalued attributes. C5.0 was designed to solve that problem by computing the information gain ration for each attribute then select the attribute has the maximal Information Gain ration value to be a root node of the training dataset. The attribute of the maximum gain ratio is picked up for splitting to reduce the needed information to predict a given instance in the resulting attribute’s partition, the Gain Ration for attribute A is computed as follows:
Where D is the training dataset.
\( P\left( {Ci} \right) = |C_{\text{i,D}} | / | {\text{D|}} \) Where \( \left| { C_{\text{i,D }} } \right| \) is the number of the tuples of the class Ci in the training dataset and |D| is the number of the tuples of the training dataset, and n is the number of the class’s values.
Where |ai,D| is the number of the tuples of the value aJ of the attribute A in the training dataset and |D| is the tuples of the training dataset and n is the number of the values of attribute A. |CJ,D| is the number of the tuples of class CJ related with the value ai of the attribute A and m is the number of the classes of class C.
As mentioned above that Information Gain is biased for multivalued attributes for example, serial number attribute will get the maximal value in Eq. 3 but will be useless in classification stage. To avoid this bias, the gain value of attribute A is dived by a measure gives the potential information after splitting the training dataset into v datasets according to the values of attribute A. This measure is called split information which is used in information gain ratio in Eq. 2. The split information is defined in Eq. 6:
Where |DJ| is the number of the tuples of the value of attribute A and |D| is the number of the tuples of the training dataset. For continuous attributes, firstly, sort the values in ascending order, secondly, set a split point for each pair of adjacent values \( \frac{{{\text{a}}_{\text{i}} {\text{ + a}}_{\text{i + 1}} }}{2} \) then compute the \( Info_{\text{A}} ( {\text{D)}} \) for each a1 < Split point and a2 > split point where a1 represents all values before the split point and a2 represents all values after the split point of the continuous attribute A in Eq. 6. Finally, the best split point is selected based on the minimal value of \( Info_{\text{A}} \left( {\text{D}} \right) \) in Eq. 5.
Support Vector Machine
SVM is a supervised learning method used for both classification and regression. It has very high generalization performance. There is no requirement to add a prior knowledge, even when it has very high input space dimension. This makes it a very good quality classifier. The main intend of the SVM classifier is to discriminate between members of two classes in the training data by finding best classification function. SVM is a generalized linear classification method. It simultaneously maximizes the geometric margin and minimizes the classification error [14].
Viewing input data as two sets of vectors in n dimensional space, a separating hyper-plane will be constructed by SVM which maximizes the margin between two data sets. Two parallel hyperplanes are constructed in order to calculate the margin, one on each side of separating hyperplane. The largest distance to the neighboring data points of both the classes helps to achieve good separation. If the margin is large then the generalization error will become less. Thus, the support vectors and margins help to find the hyper-planes. Here the data points are considered in the form of:
Here zi = 1/−1 is a constant which donates the class to which zi belongs where i is the number of samples. zi represents m-dimensional real vector. With the help of separating hyperplane the training data can be easily viewed, which is
Where u is m-dimensional vector and c is scalar.
There is a vector u which is perpendicular to the dividing hyperplane and the scalar parameter c helps to increase the margin. In case of absence of c the hyperplane is passed through the origin by force. In order to maximize the margin, parallel hyper-planes are required. These parallel hyperplanes are described by the equation.
If training data is separated linearly, the parallel hyperplane are chosen as there are no points among them. Here by geometrically, the distance between the hyper planes can be find which is 2/|u|. This is the reason why there is need to lessen the |u|. The equation is:
To define formally a hyper plane the following notation can be used:
Where \( \alpha \) weight is vector and \( \alpha_{o} \) is the bias.
The optimal hyperplane can be represented by scaling of \( \alpha \) and \( \alpha_{o} \). There is one representation has been chosen from different possible representations of hyperplane which is as follows:
z represents the training examples that are close to the hyperplane and these are known as support vectors. This representation is also known as canonical hyperplane.
The following equation gives the distance between the point z and a hyperplane
But for canonical hyperplane the numerator is equal to one and distance to the support vector is:
Here, margin which is used in the above, is denoted by M is twice the distance to the closest examples \( M = \frac{2}{\alpha } \).
Here, the problem of maximizing margin M is identical to the problem of minimizing a function L(α) subject to some constraints. To classify all the training examples zj correctly, there is a constraint model which is the requirement for the hyperplane. Formally the equation is,
Where xj represents the labels of training examples.
This is actually a lagrangian optimization problem which can be solved by using the lagrange multipliers in order to obtain the weight vector α and the \( \alpha_{o} \) bias of optimal hyperplane.
Propose positive lagrange multipliers, one for each of the inequality constraints. This offers lagrangian:
Minimize Lp with relevance to u, c. This is a convex quadratic programming problem. In the solution, those points for which β J > 0 are called “support vectors”. Model selection of SVM is also a difficult approach. SVM has shown a good performance in data classification recently. Tuning of several parameters is an effective approach which affects the generalization error and this acts as the model selection procedure. In case of linear SVM there is a need to tune the cost parameter C. However, linear SVM is generally applied to linearly separable problems. In cross validation, grid search method can be used to find the paramount parameter set. Then we obtain the classifier after applying this parameter set to the training dataset and this classifier is used to classify the testing dataset to obtain the generalization accuracy [15].
3 Experimental Results
3.1 Dataset
The dataset is collected from the Egyptian Liver Research Institute and the Mansoura Central Hospital, Dakahlia Governorate, Egypt. Till data, there is no availability of standard big dataset that is used for diagnosing of liver diseases in Egypt using conventional factors. Therefore, these databases are collected by efforts of individual research groups. Each collected data contains of the lesions of the liver includes alcoholic liver damage (ALD), primary hepatoma (PH), liver cirrhosis (LC), cholelithiasis (C), and HCC. We have collected 7000 patient’s data, in which the patient’s age in the dataset ranges from 4 to 90 years. Table 1 shows the details of the collecting data. A dataset was developed with twenty three attributes as shown in Table 1 that include the records of 7000 patients in which 5295 patients were male and rests were female.
3.2 Classifications Performance
Our study focuses on integrating machine learning with individual and hybrid classifiers (NB, C5.0, and SVM). In this proposed technique, the machine learning is employed as a feature selection technique and C5.0, SVM, and NB are employed as an ensemble model. The proposed machine learning showing in Fig. 2.
The dataset is collected from the Egyptian Liver Research Institute and the Mansoura Central Hospital, Dakahlia Governorate, Egypt.
The dataset should be prepared to eliminate the redundancy, and check for the missing values. The data preparation process is often the mainly time-consuming and computational stage. In this step, data sets have been split into the training set, which is used to build the machine learning and testing set that is used to evaluate the proposed machine learning technique.
The dataset is first divided as (90% of them are used for classifier training and 10% for classifier testing). The accuracies from each of the dataset are averaged to given an overall accuracy. It avoids the problem of overlapping test dataset and makes optimal employment of the obtainable data. We used accuracy, sensitivity, and specificity to test the classification performance of the proposed technique.
We used as a feature selection technique to generate a subset of features from the original features that make machine learning easier and less time-consuming. After generating a subset, NB, C5.0, and SVM are separately used as classification techniques. Then, an ensemble classifier (NB + C5.0 + SVM) is proposed to classify the data.
-
Accuracy is the percent of correct classifications and can be defined as:
-
Sensitivity is the rate of true positive and can be defined as:
-
Specificity is the true negative rate and can be defined as:
Where:
-
TP = the number of positive examples correctly classified.
-
FP = the number of positive examples misclassified as negative
-
FN = the number of negative examples misclassified as positive
-
TN = the number of negative examples correctly classified [5].
Table 2 shows the accuracy, sensitivity, and specificity of our proposed machine learning before and after the fine-tuning. From the table, it can be noticed that the accuracy of the technique after fine-tuning the overall is higher than the accuracy before the fine-tuning. Thus, the fine-tuning step is very important for improving the accuracy of classification.
Figure 3 shows a diagram that represents the overall performance evaluation of the proposed technique SVM, C5.0, and Naïve Bayes. It is shown that the accuracy, sensitivity, and specificity of our proposed technique are better than the three state-of-the-art techniques.
Figure 4 shows ten relevant factors in the prediction of liver disease according to our technique. The rules generated by our technique is shown in Table 3. According to the Table 1, it can be seen that 9 rules have been produced by our technique.
4 Conclusion and Feature Work
In this paper, we proposed and built a machine learning based on a hybrid classifier to be used as a classification model for liver diseases diagnosis to improve performance and experts to identify the chances of disease and conscious prescription of further treatment healthcare and examinations.
In future work, the use of fast datasets technique like Apache Hadoop or Spark can be incorporated with this technique. In addition to this, we can use distributed refined algorithms like Forest Tree implemented in Apache Hadoop to increase scalability and efficiency.
References
Kumar, Y., Sahoo, G.: Prediction of different types of liver diseases using rule based classification model. Technol. Health Care 21(5), 417–432 (2013)
Roy, S., Singh, A., Shadev, S.K.: Machine learning method for classification of liver disorders. Far East J. Electron. Commun. 16(4), 789 (2016)
Zhang, Y., et al.: A systems biology-based classifier for hepatocellular carcinoma diagnosis. PLoS One 6(7), e22426 (2011)
Kavakiotis, I., et al.: Machine learning and data mining methods in diabetes research. Comput. Struct. Biotech. J. 15, 104–116 (2017)
Ayodele, T.O.: Types of machine learning algorithms. In: New Advances in Machine Learning. InTech (2010)
Gullo, F.: From patterns in data to knowledge discovery: what data mining can do. Phys. Procedia 62, 18–22 (2015)
Pandey, B., Singh, A.: Intelligent techniques and applications in liver disorders. Survey, January 2014
Takkar, S., Singh, A., Pandey, B.: Application of machine learning algorithms to a well defined clinical problem: liver disease. Int. J. E-Health Med. Commun. (IJEHMC) 8(4), 38–60 (2017)
Siuly, S., Zhang, Y.: Medical big data: neurological diseases diagnosis through medical data analysis. Data Sci. Eng. 1(2), 54–64 (2016)
Luo, J., et al.: Big data application in biomedical research and health care: a literature review. Biomed. Inform. Insights 8, 1 (2016)
Ramana, B.V., Babu, M.S.P., Venkateswarlu, N.B.: A critical study of selected classification algorithms for liver disease diagnosis. Int. J. Database Manag. Syst. (IJDMS) 3(2), 1–14 (2011)
Bujlow, T., Riaz, T., Pedersen, J.M.: A method for classification of network traffic based on C5.0 machine learning algorithm. In: 2012 International Conference on Computing, Networking and Communications (ICNC). IEEE (2012)
Ozcift, A., Gulten, A.: Classifier ensemble construction with rotation forest to improve medical diagnosis performance of machine learning algorithms. Comput. Methods Programs Biomed. 104(3), 443–451 (2011)
Fatima, M., Pasha, M.: Survey of machine learning algorithms for disease diagnostic. J. Intell. Learn. Syst. Appl. 9(01), 1 (2017)
Singh, A., Pandey, B.: Intelligent techniques and applications in liver disorders: a survey. Int. J. Biomed. Eng. Technol. 16(1), 27–70 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this paper
Cite this paper
El-Shafeiy, E.A., El-Desouky, A.I., Elghamrawy, S.M. (2018). Prediction of Liver Diseases Based on Machine Learning Technique for Big Data. In: Hassanien, A., Tolba, M., Elhoseny, M., Mostafa, M. (eds) The International Conference on Advanced Machine Learning Technologies and Applications (AMLTA2018). AMLTA 2018. Advances in Intelligent Systems and Computing, vol 723. Springer, Cham. https://doi.org/10.1007/978-3-319-74690-6_36
Download citation
DOI: https://doi.org/10.1007/978-3-319-74690-6_36
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-74689-0
Online ISBN: 978-3-319-74690-6
eBook Packages: EngineeringEngineering (R0)