Abstract
The functioning of electromyogram (EMG) driven prosthesis to control the performance of artificial prosthetic arms placed on people with missing limbs depends on the cumulative effect of multiple dynamic factors, some of which include electrode placement position, muscle contraction levels, forearm orientations, etc. However, the study of the combined influence of these dynamic factors has been limited and hence offered us scope to improve the accuracy of the previous studies. We used the data to extract multiple features through the Time Dependent Power Spectrum Descriptor (TD-PSD) algorithm, which has proven to be one of the best methods of feature extraction. Samples are classified using the Neural Pattern Recognition Toolbox with scaled conjugate gradient backpropagation as the training algorithm, which gives an improved accuracy over Support Vector Machine (SVM) classifier. Neural Network is trained using the EMG signals of 10 subjects performing multiple hand movements to achieve classification accuracy up to 94.7%. The results obtained are a testimony to the fact that the suggested method is competent to improve the operation of pattern recognition myoelectric signals.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In a country like India, with more than half the population working in the agricultural or labor sector where physical injuries are inevitable, a large number of people undergo limb losses. Apart from the limb loss due to physical injuries, India is also the third largest home to diabetes patients who eventually have to amputate their limbs to the disease [1]. The discovery of an artificial limb or prosthesis provides the much needed respite to the amputees in India and across the globe.
Prosthesis is defined as a man-made device attached to the subject body to replace a part which could have been lost through trauma, disease or other congenital conditions. The statistics of the National Limb Loss Information Centre reveal that the upper limb amputations account for a much greater part of the overall trauma related amputations [2] and hence in this entire paper, we focus on the arm amputations and the prosthetic arms.
Many different kinds of robotic arms have been developed since its inception [3], ranging from grippers capable of performing basic industrial applications like lifting, moving, etc., to extremely detailed ones capable of enacting every possible human hand function.
The functioning of prosthetic arms is indeed a complex mechanism and involves deciphering the intended movement with the help of the electrical activities of the muscles, which are known as electromyogram (EMG) signals. The recording of the EMG signals is done through non-invasive methods through the surface of the skin of the amputee.
An overall process of controlling a prosthetic arm usually involves an electrode placed directly on the surface of the amputee used to record the signals, which are amplified, filtered and sampled to get a refined data set to be considered for deciphering the movement to be performed. This data is used for EMG pattern classification which includes processing of EMG signals, extraction of features and classification [4].
Continuous research in this field has brought advancements in this field with around 90% accuracy [5]. A number of factors that influence pattern recognition have been studied, for example, strength exerted by muscle [6], limb orientation [7], and electrode placement [8]. Other forms of noise may also cause problems [9]. The effect of these factors has been studied individually but [10] recently studied the effects of a combination of muscle contraction levels and forearm orientation on classification of EMG signals.
Feature extraction methods are used to get useful information out of almost meaningless and random time series EMG signals. Feature extraction can be done using methods like Time Domain based features [11], Discrete Fourier Transform [12], or Time Domain-Power Spectral Descriptors (TD-PSD) [13], etc. TD-PSD method as it has been established to be the best among all methods by previous literature [10]. TD-PSD method quantifies the angle of the EMG pattern rather than the amplitude which gives more robust results in comparison to other feature extraction methods. Feature extraction is then followed by a classifier which ultimately differentiates between actions and force levels with which it needs to be performed by giving an input to the digital controllers which controls the prosthetic arm. Support Vector Machine classifier was used in [10]. It has been proven to be equally good if not better to other classifiers such as Linear Discriminant Analysis (LDA), kNN, Random Forest and Naive Bayes [10].
This paper has tried to study the combined influence of different orientations of the forearm and varied muscular contraction levels on EMG pattern recognition. It has tried to continue and improve on the work done in this domain previously [10] and used their data set which was recorded live in Iraq as well as Australia. The data consists of ten intact limbed amputee subjects on which multiple electrodes were placed. These electrodes captured the EMG signals generated by 6 different movement classes at 3 varied contraction levels, with 3 trials given to each recording performed in 3 different orientation of the forearm. This data was used to extract the features by using the TD-PSD feature extraction method. Here, we have utilised the Neural Network Pattern Recognition toolbox available on MATLAB 2014 and subsequently compared the classification accuracy of this classification method with the previously used support vector machine classification method.
Section 2 discusses the data acquisition as well as the major methodologies and concepts used in feature extraction as well as classification involved in the study. Section 3 discusses the results obtained followed by the concluding remarks in Sect. 4.
2 Methods
2.1 Experimental Protocol
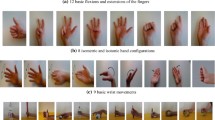
A normal computer display is placed in front of the subjects. The subjects were made to perform six actions or movements, which are, closed fist, opened fist, extension of wrist, flexion of wrist, wrist ulnar deviation, and wrist radial deviation. Three forearm orientations were considered: wrist fully supinated, at rest, and fully pronated, marked as 1st, 2nd, and 3rd orientations. Movements are performed at varied contraction levels: low, average and high in each orientation of all the six movements. Overall, each subject gave 162 trials: 6 classes of moves × 3 orientations of the arm × 3 levels of muscular contraction × 3 trials per movement.
2.2 Feature Extraction Method: Time Domain Power Spectrum Descriptors
Recent studies in Electromyogram (EMG) pattern recognition show the error when the implementation of myoelectric control system is carried out [14]. When tests are carried out on EMG patterns for the same movement at different position, the controller shows limited performance. The feature extraction method is known as TD-PSD [13] is utilized to minimize the effect of limb position on classification.
The feature vector is obtained in two steps. In the first step, using the sampled time series EMG signal and transforming them through Fourier transform and Parseval’s relations, a set of power spectrum features is extracted. Then the sampled time domain EMG signal is logarithmically scaled and the power spectrum moments are obtained from it. This is also called cepstral feature extraction method.
In the final step, the total six features are extracted which are nothing but the orientation between the power spectrum moments for original electromyogram signal and its cepstral version using a cosine similarity rule. The next section explains the feature extraction method in detail.
In Fig. 1 x[j] with j = 1, 2,… N, of length N denotes the sampled set of EMG signal. EMG trace within a certain epoch can be expressed as a function of frequency by means of Discrete Fourier transform (DFT). The feature extraction process begins by observing Parseval’s theorem which states that the sum of the square of the function is equal to the sum of the square of its transform.
Frequency index is denoted by k and P[k] is the power spectrum without phase [15]. This method will deal with the whole spectrum because the full frequency description is symmetric in nature which we obtained from Fourier Transform and we cannot obtain the power spectral density directly from the time-domain. So, all odd moments will be considered as zero. So, m is denoted as moment and n as order of the moment of the power spectrum P[k].
In the equation shown above, if the value of n is nonzero then the Fourier transform’s time-differentiation property is used and when n = 0 then Parseval’s theorem is used. This kind of property states that Δn is denoted as discrete time signals, which can also be written as multiplying the X[K] by k to the n th power.
The moments which will help in extraction of feature sets as shown in Fig. 1 are:
Root squared zero order moment: This feature mainly denotes the strength of muscle contraction, or the frequency-domain’s total power.
Root squared second order moments: Power spectrum is denoted as the second order moments.
Root squared fourth order moments: For the fourth order moment we raise the power of frequency index by 2 in second order moment.
So, the second and the fourth moment derivatives of the signals are used to minimize the signal’s full energy; hence, we normalize (λ = 0.1) to limit the influence of noise on all moments.
Now, the first three features using the above moments are:
The other three features are:
Sparseness: Sparseness measures the amount of energy is packed in only minor components of a vector. This feature can be expressed as:
Such a feature describes a vector with all elements equal with a sparseness measure of zero that is m 2 and m 4 equal to zero because of differentiation and so f 4 = 0. For all other sparseness levels, value should be greater than 0 [14].
Irregularity Factor (IF): It denotes the measure of ratio of the count of upward zero crossings to the number of peaks. This feature can be expressed as in terms of spectral moments:
Waveform Length Ratio (WL): The summation of absolute value of the first and second derivative of EMG signal over its entire length is calculated to find the waveform length (WL) feature using the formula:
Now on the basis of Fig. 1, we form a matrix a = [a 1, a 2, a 3, a 4, a 5, a 6] by using the six extracted features. We also add another feature vector, expressed as b = [b 1, b 2, b 3, b 4, b 5, b 6], which is extracted logarithmically scaled version log (\( x^{{ 2^{{x^{2} }} }} \)). For each EMG channel the final 6 features are extracted which are nothing but the orientation between the vectors obtained earlier. A cosine similarity rule is used to find these features given as
2.3 Artificial Neural Network
It makes a network of artificial neurons that map the input to the output, both of which are known to us with certainty. A backpropagation network consists of at least three layer which are one input layer, one output layer, and one or more hidden layers in between the input and output layers. The hidden layer has a number of hidden neurons. The network is trained, that is, the various connection weights and bias values are adjusted so as to generate the desired outputs for the given inputs. The error generated at the output is the difference in our desired output and the present output at the output nodes. This error is backpropagated from the output layer to the input layer through the hidden layer(s). This changes the connection weights to reduce the error. This process is called backpropagation.
For the classification process, Neural Pattern Recognition toolbox has been used, available in MATLAB 2014, where the extracted TD-PSD features from EMG signals obtained from 10 subjects are treated as input, that is, the data to be classified. Networks of pattern recognition are feed-forward networks which can be used to train and classify inputs according to their target classes.
To train the network, we use the scaled conjugate gradient backpropagation method of classification. This training function comprises of three layers in total. There is just one hidden layer. The number of neurons is selected according to our network size.
3 Experimental Results
To report our results, we combine all the data and classification results from our 10 subjects. There are a total of 1620 samples from 162 trials of our 10 subjects. We first divided our samples randomly to feed into the pattern recognition tool using scaled conjugate gradient backpropagation algorithm. We used 70% of those samples for training, 15% of those samples for validation and 15% of those samples for testing. Figure 2 shows the confusion matrix for training with TD-PSD features:
We now conducted a systematic study where, at each forearm orientation, we selected the data from one contraction level to train our classifier and then used the data from all contraction levels for testing our model.
For the case when we use the data of low contraction level in orientation 1 for training, Fig. 3 shows the confusion matrix and receiver operating characteristics (ROC), and Fig. 4 shows the performance curve. These plots help in the gauging the efficiency of a supervised learning algorithms (Table 1).
The classification accuracy was the highest when training data from medium contraction level was used, at each forearm orientation.
Next, the classifier was trained and tested with data from different orientations. Specifically, for training, each orientation was selected one by one and the TD-PSD features extracted from data of all three muscle contraction levels was used. The TD-PSD features exacted from all the contraction levels of the other two orientations was the testing data.
There was a decline in classification performance from the earlier scenario, predictably so. For the case when the network was trained using data from orientation 1, and tested using data from orientations 2 and 3, Fig. 5 shows the confusion matrix and ROC. Figure 6 shows the performance curve.
Training the classifier with data from orientation 2 (as opposed to orientations 1 or 3) and testing the network with the other orientations resulted in the highest classification accuracy, which is 69.1% on average.
These results can be compared to those obtained by using support vector machine (SVM) classifier with SVM parameters as C = 32 and γ = 0.0625 (Table 2).
4 Concluding Remarks
Using TD-PSD as the feature extraction method, a comprehensive study of how varying muscle contraction levels and different forearm orientations together affect the EMG pattern recognition was conducted. It has been well established that the TD-PSD is a superior feature extraction method in comparison to other feature extraction methods. This paper has employed a new classifier, namely, Neural Network Classifier with scaled conjugate gradient back propagation training algorithm on MATLAB to improve the classification accuracy. Training this new classifier with data of six movement classes, each carried out with multiple muscular contraction levels and at various forearm orientations, by using TD-PSD features offers satisfactory classification accuracy and an improvement over Support Vector Machine classifier.
Maximum classification accuracy is obtained when the training data includes all forearm orientations. It is also observed that using training data from medium muscle contraction level provides the best accuracy when testing in comparison to low or high force levels.
References
Nazarpour, K., Sharafat, A.R., Firoozabadi, S.M.P.: Application of higher order statistics to surface electromyogram signal classification. IEEE Trans. Biomed. Eng. 54(10), 1762–1769 (2007)
Owings, M.F., Kozak, L.J.: Ambulatory and inpatient procedures in the United States, 1996. Vital Health Stat. 139, 1–119 (1998)
Belter, J.T.: Mechanical design and performance specifications of anthropomorphic prosthetic hands: a review. J. Rehabil. Res. Dev. 50(5), 599 (2013)
Boostani, R., Moradi, M.H.: Evaluation of the forearm EMG signal features for the control of a prosthetic hand. Physiol. Meas. 24(2), 309 (2003)
Rasool, G., et al.: Real-time task discrimination for myoelectric control employing task-specific muscle synergies. IEEE Trans. Neural Syst. Rehabil. Eng. 24(1), 98–108 (2016)
Al-Timemy, A.H., et al.: A preliminary investigation of the effect of force variation for myoelectric control of hand prosthesis. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE (2013)
Peng, L., et al.: Combined use of semg and accelerometer in hand motion classification considering forearm rotation. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE (2013)
Hargrove, L., Englehart, K., Hudgins, B.: A training strategy to reduce classification degradation due to electrode displacements in pattern recognition based myoelectric control. Biomed. Signal Process. Control 3(2), 175–180 (2008)
Spanias, J.A., Perreault, E.J., Hargrove, L.J.: Detection of and compensation for EMG disturbances for powered lower limb prosthesis control. IEEE Trans. Neural Syst. Rehabil. Eng. 24(2), 226–234 (2016)
Khushaba, R.N., et al.: Combined influence of forearm orientation and muscular contraction on EMG pattern recognition. Expert Syst. Appl. 61, 154–161 (2016)
Hakonen, M., Piitulainen, H., Visala, A.: Current state of digital signal processing in myoelectric interfaces and related applications. Biomed. Signal Process. Control 18, 334–359 (2015)
He, J., et al.: Invariant surface EMG feature against varying contraction level for myoelectric control based on muscle coordination. IEEE J. Biomed. Health Inf. 19(3), 874–882 (2015)
Al-Timemy, A.H., et al.: Improving the performance against force variation of EMG controlled multifunctional upper-limb prostheses for transradial amputees. IEEE Trans. Neural Syst. Rehabil. Eng. 24(6), 650–661 (2016)
Khushaba, R.N., et al.: Towards limb position invariant myoelectric pattern recognition using time-dependent spectral features. Neural Netw. 55, 42–58 (2014)
Khushaba, N., et al.: A fusion of time-domain descriptors for improved myoelectric hand control. In: 2016 IEEE Symposium Series on Computational Intelligence (SSCI). IEEE (2016)
Bhardwaj, N., et al.: Extraction of EMG signal in a software compatible format from an online database using WFDB package. Persp Sci. 8, 767–769 (2016)
Agarwal, S., et al.: EEG signal enhancement using cascaded S-Golay filter. Biomed. Signal Process. Cont. 36, 194–204 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this paper
Cite this paper
Gupta, T., Yadav, J., Chaudhary, S., Agarwal, U. (2018). EMG Pattern Classification Using Neural Networks. In: Thampi, S., Mitra, S., Mukhopadhyay, J., Li, KC., James, A., Berretti, S. (eds) Intelligent Systems Technologies and Applications. ISTA 2017. Advances in Intelligent Systems and Computing, vol 683. Springer, Cham. https://doi.org/10.1007/978-3-319-68385-0_20
Download citation
DOI: https://doi.org/10.1007/978-3-319-68385-0_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-68384-3
Online ISBN: 978-3-319-68385-0
eBook Packages: EngineeringEngineering (R0)