Abstract
Today we are witnessing an explosion in the size and the amount of the available RDF datasets. As such, conventional single node RDF management systems give their position to clustered ones. However most of the currently available clustered RDF database systems partition data using hash functions and/or vertical and horizontal partition algorithms with a significant impact on the number of nodes required for query answering, increasing the total cost of query evaluation. In this paper we present a novel semantic partitioning approach, exploiting both the structure and the semantics of an RDF Dataset, for producing vertical partitions that significantly reduce the number of nodes that should be visited for query answering. To construct these partitions, first we select the most important nodes in a dataset as centroids, using the notion of relevance. Then we use the notion of dependence to assign each remaining node to the appropriate centroid. We evaluate our approach using three real world datasets and demonstrate the nice properties that the constructed partitions possess showing that they significantly reduce the total number of nodes required for query answering while introducing minimal storage overhead.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
1 Introduction
The recent explosion of the Data Web and the associated Linked Open Data (LOD) initiative have led to an enormous amount of widely available RDF datasets. For example, data.gov comprises in more than 5 billion triples, the Linked Cancer Genome Atlas currently consists of more than 7 billion triples and is estimated to reach 30 billion [27] whereas the LOD cloud contained already 62 billion triples since January 2014 [25].
To store, manage and query these ever increasing RDF data, many systems were developed by the research community (e.g. Jena, Sesame etc.) and by many commercial vendors (e.g. Oracle and IBM) [10]. Although, these systems have demonstrated great performance on a single node, being able to manage millions, and, in some cases, billions of triples, as the amount of the available data continues to scale, it is no longer feasible to store the entire dataset on a single node. Consequently, under the light of the big data era, the requirement for clustered RDF database systems is becoming increasingly important [6].
In principle the majority of the available clustered RDF database systems, such as SHARD [23], YARS2 [6], and Virtuoso [20] partition triples across multiple nodes using hash functions. However, hash functions require in essence contacting all nodes for query answering and when the size of the intermediate results is large, the inter-node communication cost can be prohibitively high. To face this limitation, other systems try to partition RDF datasets into clusters such that the number of queries that hit partition boundaries is minimized. However most of these systems either treat RDF as simple graphs, exploiting graph partitioning algorithms, [7] or cluster triples based on locality measures with limited semantics [17].
Although RDF datasets can be interpreted as simple graphs, besides their structural information they have also attached rich semantics which could be exploited to improve the partition algorithms and dictate a different approach. As such, in this paper, we focus on effectively partitioning RDF datasets across multiple nodes exploiting all available information, both structural and semantic. More specifically our contributions are the following:
-
We present RDFCluster, a novel platform that accepts as input an RDF dataset and the number of the available computational nodes and generates the corresponding partitions, exploiting both the semantics of the dataset and the structure of the corresponding graph.
-
We view an RDF dataset as two distinct and interconnected graphs, i.e. the schema and the instance graph. Since query formulation is usually based on the schema, we generate vertical partitions based on schema clusters. To do so we select first the most important schema nodes as centroids and assign the rest of the schema nodes to their closest centroid similar to [11]. Then individuals are instantiated under the corresponding schema nodes producing the final partitions of the dataset.
-
To identify the most important nodes we reuse the notion of relevance based on the established measures of the relative cardinality and the in/out degree centrality of a node [30]. Then to assign the rest of the schema nodes to a centroid we define the notion of dependence assigning each schema node to the cluster with the maximum dependence between that node and the corresponding centroid.
-
We describe the aforementioned algorithm and we present the computational complexity for computing the corresponding partitions given a dataset and the available computational nodes.
-
Then, we experiment with three datasets, namely CRMdig, LUBM and eTMO, and the corresponding queries and we show the nice properties of the produced partitions with respect to query answering, i.e. the high quality of the constructed partitions and the low storage overhead it introduces.
Our partitioning scheme can be adopted for efficient storage of RDF data reducing communication costs and enabling efficient query answering. Our approach is unique in the way that constructs data partitions, based on schema clusters, constructed combining structural information with semantics. We have to note that in this paper we are not interested in benchmarking clustered RDF systems but only on the corresponding partition algorithm.
The rest of the paper is organized as follows. Section 2 introduces the formal framework of our solution and Sect. 3 describes the metrics used to determine how the cluster should be formulated and the corresponding algorithm. Then, Sect. 4 describes the evaluation conducted and Sect. 5 presents related work. Finally, Sect. 6 concludes the paper and presents directions for future work.
2 Preliminaries and Example
In this paper, we focus on datasets expressed in RDF, as RDF is the de-facto standard for publishing and representing data on the Web. The representation of knowledge in RDF is based on triples of the form (subject, predicate, object). RDF datasets have attached semantics through RDFSFootnote 1, a vocabulary description language. Here, we will follow an approach similar to [12], which imposes a convenient graph-theoretic view of RDF data that is closer to the way the users perceive their datasets.
Representation of RDF data is based on three disjoint and infinite sets of resources, namely: URIs (U), literals (L) and blank nodes (B). We impose typing on resources, so we consider 3 disjoint sets of resources: classes (C \( \subseteq \) U \( \mathop \cup \nolimits \) B), properties (P \( \subseteq \) U), and individuals (I \( \subseteq \) U \( \mathop \cup \nolimits \) B). The set C includes all classes, including RDFS classes and XML datatypes (e.g., xsd:string, xsd:integer). The set P includes all properties, except rdf:type which connects individuals with the classes they are instantiated under. The set I includes all individuals (but not literals).
In this work, we separate between the schema and instances of an RDF dataset, represented in separate graphs (GS, GI respectively). The schema graph contains all classes and the properties they are associated with (via the properties’ domain/range specification); note that multiple domains/ranges per property are allowed, by having the property URI be a label on the edge (via a labelling function λ) rather than the edge itself. The instance graph contains all individuals, and the instantiations of schema properties; the labelling function λ applies here as well for the same reasons. Finally, the two graphs are related via the τc function, which determines which class(es) each individual is instantiated under. Formally:
Definition 1 (RDF Dataset).
An RDF dataset is a tuple \( V = \left\langle {G_{S} , G_{I} , \lambda , \tau_{c} } \right\rangle \) such that:
-
GS is a labelled directed graph GS = (VS, ES) such that VS, ES are the nodes and edges of GS, respectively, and VS \( \subseteq \) C \( \mathop \cup \nolimits \) L.
-
GI is a labelled directed graph GI = (VI, EI) such that VI, EI are the nodes and edges of GI respectively, and VI \( \subseteq \) I \( \mathop \cup \nolimits \) L.
-
A labelling function λ: ES \( \mathop \cup \nolimits \) EI \( \mapsto \) P that determines the property URI that each edge corresponds to (properties with multiple domains/ranges may appear in more than one edge).
-
A function τc: I \( \mapsto \) 2C associating each individual with the classes that it is instantiated under.
For simplicity, we forego extra requirements related to RDFS inference (subsumption, instantiation) and validity (e.g., that the source and target of property instances should be instantiated under the property’s domain/range respectively), because these are not relevant for our results below and would significantly complicate our definitions. In the following, we will write p(v1, v2) to denote an edge e in GS (where v1, v2 \( \in \) VS) or GI (where v1, v2 \( \in \) VI) from node v1 to node v2 such that λ(e) = p. In addition for brevity we will call schema node a node c \( \in \) VS, class node a node c \( \in \) C \( \mathop \cap \nolimits \) VS and instance node a node u \( \in \) I \( \mathop \cap \nolimits \) VI. In addition a path from v1 \( \in \) VS to v2 \( \in \) VS, i.e. path(v1, v2), is the finite sequence of edges, which connect a sequence of nodes, starting from the node v1 and ending in the node v2. In this paper we will focus on class and instance nodes due to lack of space, but our approach can be easily generalized to include literals as well.
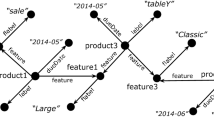
Now as an example consider the LUBM ontologyFootnote 2 part shown in Fig. 1 used to describe a university domain. This example contains 20 classes and many properties. Now assume that we would like to partition the corresponding RDF dataset into three partitions revealing discriminating features for each one of them. One way to do that for example would be to identify first the three most importance schema nodes of the dataset, allocate each one of those nodes to the corresponding cluster as a centroid and finally place into the same cluster the schema nodes that depend on those selected nodes. The clusters generated using our approach are shown in Fig. 1. The most important schema nodes, as identified by our algorithm, are the “Professor”, the “Publication” and the “Person” classes. These are used as centroids and the remaining schema nodes are assigned to the appropriate clusters by identifying the schema nodes that depend on those centroids. Finally the instance nodes are assigned to the class nodes that are instantiated under. In this paper we will use the term cluster to refer only to the schema graph and the term partition to refer to the entire dataset.
When data is partitioned across multiple machines the particular partitioning method, can have a significant impact on the amount of data that needs to be shipped over the network at query time. Ideally we would like the constructed partitions to increase the likelihood that query answers can be computed locally reducing the communication cost. In general, in distributed query processing, where multiple nodes are available, query answering proceeds by first breaking the query into pieces, all of which can be evaluated independently within individual partitions. The query pieces are then evaluated in the relevant partitions obtaining partial matches and they are joined to produce the final answer. Again, in this paper we are not interested on the technicalities of query answering but only on the aforementioned partitioning algorithm and how the careful placement of the nodes within partitions could optimize the overall number of nodes to be visited for query answering.
Assume for example the following SPARQL query involving 3 classes and 2 user-defined properties, requesting all publications of the persons belonging to an organization:

If data was partitioned using a simple hash partitioning algorithm, then obviously all nodes would have to be examined. If however, the data was partitioned as shown in Fig. 1 then only two nodes would have to be contacted, as instances of the “Organization” and “Publication” classes can be found in the second partition and the instances of “Person” can be located at the third partition. We therefore, instead of using simple hash or graph partitioning algorithm are looking for a more advanced method, partitioning the schema into appropriate clusters, considering the semantics of the nodes and the structural information of the corresponding graph.
3 Metrics
Our clustering framework follows the K-Medoids clustering method [11]; we select the most centrally located point in a cluster as a centroid, and assign the rest of points to their closest centroids. To identify the most centrally located point in a cluster we use the notion of relevance. Then dependence is used for extracting nodes, highly relevant to the specific important nodes (centroids) connecting other nodes to the most important ones.
3.1 Identifying Centroids
Initially, the notion of centrality [30] is used to quantify how central is a class node in a specific RDF dataset. To identify the centrality of a class node c in a dataset V, we initially consider the instances it contains by calculating its relative cardinality. The relative cardinality RC(p(c, ci)) of an edge p(c, ci), which connects the class nodes c and ci in the schema graph, is defined as the number of the specific instance connections between the corresponding two classes divided by the total number of the connections of the instances that these two classes have. Then, we combine the data distribution with the number of the incoming/outgoing edges, aka properties, of this class. As such, the in/out-centrality (Cin/Cout) is defined as the sum of the weighted relative cardinalities of the incoming/outgoing edges:
Definition 2 (Centrality).
Assume a node c \( \in \) C \( \mathop \cap \nolimits \) VS in a dataset V = \( \left\langle {G_{S} , G_{I} , \lambda , \tau_{c} } \right\rangle \). The in-centrality Cin(c) (respectively, the out-centrality Cout(c)) of c is defined as the sum of the weighted relative cardinality of the incoming p(ci, c) \( \in \) Es (respectively, outgoing p(c, ci) \( \in \) ES) edges:
The weights in the above formula have been experimentally defined [30] and vary depending on whether edges that correspond to properties are user-defined or RDF/S, giving higher importance to user-defined ones (in our experiments we used wp = 0.8 for user-defined properties and wp = 0.2 for RDF/S ones). This is partly because user-defined properties correlate classes, each exposing the connectivity of the entire schema, in contrast to hierarchical or other kinds (e.g., rdfs:label) of RDF/S properties. Consider now the “Article” class shown in Fig. 1. Assume also that there are not any instances in the corresponding dataset. Then the relative cardinality of all nodes is initialized to a constant a = 0.03. As such Cin(University) = 0 since there are no incoming edges and Cout(University) = RC(rdf:type) * wrdf:type = 0.03 * 0.2 = 0.06.
Now that centrality is defined we are going to define relevance. The notion of relevance [30] has been proposed as adequate for quantifying the importance of a class in an RDF dataset. In particular, relevance is based on the idea that the importance of a class should describe how well the class could represent its neighborhood. Intuitively, classes with many connections with other classes in a dataset should have a higher importance than classes with fewer connections. Thus, the relevance of a class is affected by the centrality of the class itself, as well as by the centrality of its neighboring classes. Moreover, since the version might contain huge amounts of data, the actual data instances of the class should also be considered when trying to estimate its importance, namely relevance. Formally, relevance is defined as follows:
Definition 3 (Relevance).
Assume a node c \( \in \) C \( \mathop \cap \nolimits \) VS in a dataset V = \( \left\langle {G_{S} , G_{I} , \lambda , \tau_{c} } \right\rangle \). Assume also that c1, …, cn \( \in \) ES are the incoming edges of c (p(ci, c) \( \in \) ES) and \( c_{1}^{\prime } , \cdots c_{k}^{\prime } \) \( \in \) ES are the outgoing edges of \( c\left( {p\left( {c,\,c_{i}^{\prime } ,} \right) \in E_{s} } \right) \). Then the relevance of c, i.e. Relevance(c), is the following:
The aforementioned metric identifies class nodes being able to represent an entire area and as a consequence those nodes can be used as the centroids of the corresponding graph. In our example, shown in Fig. 1, Relevance(University) = Cin(University) + Cout(University)/Cout(Organization) + 0 = 0 + 0.06/0.048 = 1.25.
3.2 Assigning Nodes to Centroids
Having a method to identify the most important nodes (centroids) in an RDF dataset we are now interested on identifying to which cluster the remaining nodes should be assigned to. Our first idea to this direction comes from the classical information theory; that infrequent words are more informative that frequent ones. The idea is also widely used in the field of instance matching [24]. The basic hypothesis here is that the greater the influence of a property on identifying a corresponding instance the less times it is repeated. According to this idea, we try to initially identify the dependence between two classes based on their data instances.
In our running example, the node “Person” has a high relevance in the graph and as a consequence a great probability to be used as a centroid. Assume also two nodes “SSN” and “Work” directly connected it. Although an instance of “Person” can have only one social security number, many persons can be employed by the same employer and as such a person cannot be characterized by his work. As a consequence, the dependence between “Person” and “SSN” is higher than the dependence between “Person” and “Telephone”. Based on this observation, we define the measurement of cardinality closeness of two adjacent schema nodes.
Definition 4 (Cardinality Closeness).
Let ck, cs be two adjacent schema nodes and ui, uj \( \in G_{I} \) such that τc(ui) = ck and τc(uj) = cs. The cardinality closeness of p(ck, cs), namely the CC(p(ck, cs)), is the following:
where |c|, c \( \in \) C \( \mathop \cap \nolimits \) VS the number of nodes in the schema graph, DistinctValues(p(ui, uj)) the number of distinct p(ui, uj) and Instances(p(ui, uj)) the number of p(ui, uj). When there are no instances Instances(p(ui, uj)) = 1 and DistinctValues(p(ui, uj)) = 0.
The constant 1/|c| is added in order to have a minimum value for the CC in case of no available instances. Having defined the cardinality closeness of two adjacent schema nodes we proceed further to identify their dependence. As such we identify the dependence between two classes as a combination of their cardinality closeness, the relevance of the classes and the number of edges between these two classes:
Definition 5 (Dependence of two schema nodes).
The dependence of two schema nodes cs and ce, i.e. the Dependence(cs, ce), is given by the following formula
Obviously as we move away from a node, the dependence becomes smaller by calculating the differences of relevance across a selected path in the graph. We penalize additionally dependence dividing by the distance of the two nodes. The highest the dependence of a path, the more appropriate is the first node to describe the final node of the path. Also note that the Dependence(cs, ce) is different than Dependence(ce, cs). For example, Dependence(Publication, Book) ≥ Dependence(Book, Publication). This is happening, since the dependence of a more relevant node toward a less relevant node is higher than the other way around, although, they share the same cardinality closeness.
3.3 The Clustering Algorithm
Having defined both the relevance for identifying the most important nodes and the dependence of two schema nodes we are now ready to define the semantic partitioning problem:
Definition 6 (Semantic Partitioning Problem).
Given an RDF Dataset V = \( \left\langle {G_{S} , G_{I} , \lambda , \tau_{c} } \right\rangle \), partition V into k subsets V1, V2, …, Vk such that:
-
1.
V = \( \bigcup\nolimits_{i = 1}^{k} {V_{i} } \)
-
2.
Let topk = {c1, …, ck} be the k schema nodes with the highest relevance in V. Then c1 \( \in \) V1, …, ck \( \in \) Vk
-
3.
Let dj be a schema node and dj \( \notin \) topk. Then
Dependence(dj, cp) = \( \max_{0 \le x \le k} Dependence({\text{d}}_{\text{j}} , {\text{c}}_{\text{x}} ) \to \exists \;d_{j} \) in Vp, (1 ≤ p ≤ k)
-
4.
\( \forall u \in G_{I} , \) such that τc(u) \( \in G_{S} \) and τc(u) \( \in \) \( V_{j} \to \exists \) u in Vj
The first requirement says that we should be able to recreate V by taking the union of all Vι (1 ≤ i ≤ k). The second one that each cluster should be based on one of the nodes with the top k relevance (the topk set) as a centroid, and the third that each node that does not belong to the topk should appear at least in the cluster with the maximum dependence between the specific node and the corresponding centroid. Note that a node can appear in multiple clusters. The idea originates from social networks where an individual can simultaneously belong to several communities (family, work etc.), similarly an RDF resource might belong to more than one clusters. As such, in order to include a schema node in the Vp cluster (1 ≤ p ≤ k) we are looking for the path maximizing the Dependence. In the selected path however there might exist nodes not directly assigned to Vp. We include those nodes in the cluster as well since they have also high dependence to the centroid. Finally all instances are replicated under the corresponding schema nodes.
The corresponding algorithm is shown in Fig. 2. The algorithm gets as input an RDF dataset and the number of computational nodes (k) and partitions the dataset into k partitions. Bellow we explain in more detail each of the steps of the algorithm.
The algorithm starts by calculating the relevance of all schema nodes (lines 2–3). More specifically for each node in \( G_{S} \) we calculate the corresponding relevance according to Definition 3. Having calculated the relevance of each node we would like to get the k most important ones to be used as centroids in our clusters. Those are selected (line 4) and then assigned to the corresponding cluster (lines 5–6).
Then the algorithm examines the remaining schema nodes to determine to which cluster they should be placed at. For each node we calculate the dependence between the selected node and all centroids (line 7). We select to place the node in the cluster with the maximum dependence between the aforementioned node and the k centroids (line 8). However we are not only interested in placing the selected node to the identified cluster but we place the whole path and specifically the nodes contained in the path, which connects the selected node with the appropriate centroid (path_with_max_depedence), maximizing the dependence of the selected node in that cluster (line 9) as well. Next, we add to each cluster the corresponding instance nodes to the schema nodes they are instantiated under. Finally, we return the partitions to the user. The correctness of the algorithm is immediately proved by construction.
To identify the complexity of the algorithm we should first identify the complexity of its various components. Assume |V| the number of nodes, |E| the number of edges and |I| the number of instances. For identifying the relative cardinality of the edges we should visit all instances and edges once. Then for calculating the schema node centralities we should visit each node once whereas for calculating the relevance of each node we should visit twice all nodes Ο(|Ι| + |Ε| + 2|V|). Then we have to sort all nodes according to their relevance and select the top k ones O(|V|log|V|). To calculate the dependence of each node we should visit each node once per selected node O(k|V|), whereas to identify the path maximizing the dependence we use the weighted Dijkstra algorithm with cost O(|V|2). Finally we should check once all instances for identifying the clusters to be assigned O(|I|). As such the time complexity of the algorithm is polynomial Ο(|Ι| + |Ε| + 2|V|) + O(|V|log|V|) + O(k|V|) + O(|V|2) ≤ O(|V|2).
4 Evaluation
To evaluate our approach and the corresponding algorithm we used three RDF datasets:
CRMdigFootnote 3. CRMdig is an ontology to encode metadata about the steps and methods of production (“provenance”) of digitization products and synthetic digital representations created by various technologies. The ontology contains 126 classes and 435 properties. For our experiments we used 900 real instances from the 3D-SYSTEKFootnote 4 project. In addition we used 9 template queries published in [28] with an average size of 6 triple patterns.
LUBM. The Lehigh University Benchmark (LUBM) is a widely used benchmark for evaluating semantic web repositories. It contains 43 classes, and 32 properties modeling information about universities and is accompanied by a synthetic data generator. For our tests we used the default 1555 instances coming from a real dataset. The benchmark provides 14 test queries that we used in our experiments with an average size of 4 triple patterns.
eTMO. This ontology has been defined in the context of MyHealthAvatarFootnote 5 EU project [16] and is used to model various information within the e-health domain. It is consisted of 335 classes and 67 properties and it is published with 7848 real instances coming from the MyHealthAvatar EU project. For querying we used 8 template queries specified within the project for retrieving relevant information, with an average size of 17 triple patterns per query.
Each dataset was split into 2, 5 and 10 partitions and we used all queries available for query answering. For a fixed dataset, increasing the number of partitions is likely to increase the number of nodes required for answering queries as the data becomes more fragmented. However, it increases the number of queries that can be answered independently in parallel reducing the computation task for a single node. As we have already mentioned, our task is not to measure end-to-end query answering times involving multiple systems but to evaluate the quality of the constructed partitions with respect to the query answering
As such, for each V1, …, Vk (k = 2, 5, 10) we measure the following characteristics: (i) The quality of constructed partitioning algorithms, i.e. the percentage of the test queries that can be answered only by a single partition, (ii) the number of partitions that are needed in order to answer each query and (iii) the space overhead that our algorithm introduces in both schema nodes and the dataset.
We compare our approach with (a) subject-based hash partitioning similar to YARS2 [6] and Trinity.RDF [34] called Hashing, and (b) METIS used by [7, 17] for clustering RDF Datasets. Hashing is distributing triples in partitions by applying a hash function to the subject of the triple in order to guarantee that star queries can be evaluated locally. METIS [13] on the other hand calculates n disjoint sets of nodes such that all sets are of similar sizes and the number of edges in connecting nodes in distinct sets is minimized. In this work we focus only on the partitioning schemes of the aforementioned works. All datasets and queries used in our experiments along with the detailed results can be found onlineFootnote 6.
4.1 Quality
We perceive the quality of a partitioning algorithm with respect to query answering as the percentage of queries that can be answered by a single computational node without requiring to visit additional nodes to provide answers to the user. The results for all queries in our three datasets for the three algorithms in 2, 5 and 10 partitions are shown in Table 1.
We can easily identify that RDFCluster is better in almost all the cases showing the high quality of the produced partitions with respect to query answering. The only case that METIS is better than RDFCluster is in LUBM when we have 5 partitions where one more query can be answered by a single one. However, for LUBM, even in 5 partitions as we shall see in the sequel (Sect. 4.2) our algorithm requires less nodes to be visited on average for answering the benchmark queries. In addition we expect that as the number of partitions increases the average number of queries that can be answered by an individual partition decreases as the data are distributed to more nodes. Our expectations are confirmed by our results.
In addition as expected, smaller queries (LUBM with an average of 4 triple patterns per query and CRMdig with an average of 6 triple patterns per query) show a greater likelihood to be answered by a single node than queries with more triple patterns such as eTMO with an average of 17 triple patterns per query.
4.2 Number of Clusters Required for Answering a Query
Besides evaluating the quality of our algorithm, another interesting dimension is to evaluate how much work is required for answering the queries in each case in terms of the nodes required to be visited. The nodes to be visited give us an intuition about how many joins will be required to construct the final answer that will be returned to the user. This is critical because, in order to ensure the completeness of query answers, all partial matches in all partition elements must be computed and joined together.
The results are shown in Fig. 3 where we can see that in all cases RDFCluster requires on average less nodes to be visited for query answering, showing again the nice properties of our algorithm. Note that even for large queries (eTMO with an average of 17 triple patterns) our algorithm requires only three partitions to be visited on average for query answering and this applies even in the case of 10 partitions.
4.3 Storage Overhead
The storage overhead provides us with an indication of how much space is needed for our approach compared to the space required for storing all datasets in a single node. Since Hashing and METIS algorithms construct non-overlapping clusters they have no storage overhead. However for simple variations of hash allowing duplication the overhead can be really high (e.g. 2-hop duplication can lead to an overhead up to 430% [7]). In our case, since we allow a class node and the corresponding instances to be replicated in multiple nodes we expect as the number of clusters increases to increase the storage overhead as well.
To identify and understand the overhead introduced by our algorithm first we focus only on the schema graph and identify the overhead introduced there. This is shown in Table 2 calculating the percentage \( \left| {G_{SV1} } \right| + \ldots + \left| {G_{SVK} } \right| - \left| {G_{S} } \right|/\left| {G_{S} } \right| \). As shown the overhead is minimal introducing at most 15.00% additional schema nodes for LUBM whereas for eTMO and CRMdig is only 7.19% and 6.20% respectively.
The impact of these additional schema nodes to the overhead of the entire dataset is shown in Table 3. The table shows the total storage overhead introduced by our algorithm, i.e. the percentage |V1| + … + |Vk | − |V|/|V|. As shown, the total storage overhead introduced from our algorithm is at most 16.73% for CRMdig and for the majority of the cases less than 1%. Another interesting observation is that in almost all the cases the schema nodes overhead is greater than the corresponding total storage overhead showing that our algorithm succeeds in replicating only nodes with small additional overhead that however significantly improve query answering as shown in previous sections.
Overall, as the experiments show although our algorithm chooses to sacrifice equal data distribution on the nodes to achieve a better performance with respect to query answering the imposed overhead is really low reaching at most 16.73% overhead on our test cases.
5 Related Work
Graph clustering has received much attention over the latest years [35], aiming to partition large graphs into several densely connected components, with many application such as community detection in social networks, identification of interactions in protein interaction networks etc. The problem proved to be an NP-complete problem [5]. Typical algorithms of this class include local search based solutions (such as KL [15] and FM [4]), which swap heuristically selected pairs of nodes, simulated annealing [8], and genetic algorithms [3]. Algorithms in this category focus on the topological structure of a graph so that each partition achieves a cohesive internal structure and there are approaches based on normalized-cut [26], modularity [20], structural density [33], attribute similarity [29] or combinations between those [35]. To scale up to graphs with millions of nodes, multi-level partitioning solutions, such as Metis [13], Chaco [9], and Scotch [22], and variations over these have been proposed.
To this direction, several approaches try to represent RDF datasets as graphs and exploit variations of the aforementioned data for data partitioning. For example, Wang et al. [31] focus on providing semantic-aware highly parallelized graph partitioning algorithms for generic-purpose distributed memory systems whereas Huang et al. [7] apply graph partitioning over the Hadoop MapReduce framework trying to reduce as much as possible the communication costs. Our approach however, does not focus only on the structural part of the graph for partitioning the RDF datasets but considers in addition semantic information (such as the number of instances, the distinct instance values, assigns different weights according to the type of the properties) with the same target however, i.e. to reduce as much as possible the communication costs among partitions when these partitions are used for query answering.
Other clustered RDF database systems, such as SHARD [23], YARS2 [6], and Virtuoso [20] partition triples across multiple nodes using hash functions. However, portioning data using hashing requires a lot of communication overhead for query answering since in essence all nodes have to be contacted. The same problem appear in other works that adopt vertical [2] or horizontal partitioning schemes based on triples [18] ignoring however the correlation between triples, leading to a large number of join operators among the compute nodes. Other algorithms, but with the same problem use hybrid hierarchical clustering [19] combining an affinity propagation clustering algorithm and the k-Means clustering algorithms. To overcome that problem Lee et al. [17] proposed to by use locality sensitive hashing schemes. Although this approach moves to the same direction with ours, trying to exploit semantics, the adopted solution is limited to only the fact that triples are anchored at the same subject or object node. In addition according to our experiments our solution outperforms similar approaches.
Finally there are approaches that try to monitor the execution of SPARQL queries [1] or assume that query patterns are already available [32] and keep track of records that are co-accessed and physically cluster them using locality sensitive hashing schemes. Our approach uses a similar “profiling” mechanism but instead of focusing on queries, we focus on profiling “data” identifying and combining the knowledge of the instance distribution with structure and semantics. A more thorough overview of the different partition schemes for RDF datasets can be found on [10].
6 Conclusions and Future Work
In this paper we present a novel method that gets as input and RDF dataset and the number of available computational nodes and returns a set of partitions to be stored on the aforementioned nodes. To select the centroids for the each cluster initially our algorithm selects the most important nodes based on the notion of relevance. Then to assign the remaining nodes to a cluster we use the notion of dependence eventually assigning the remaining schema nodes to the cluster maximizing the dependence with the corresponding centroid. Having constructed the appropriate “schema clusters” we place next the instances on the corresponding classes they belong to. Our algorithm exploits both structural and semantic information in order to both select the most important nodes and then to assign the remaining nodes to the proper clusters. In addition, since both our constructed clusters and user queries are based on schema information we argue that this partitioning scheme will have a beneficial impact on query evaluation limiting significantly the nodes that should be visited to answer frequent queries.
The quality of our partitioning scheme is verified by our experiments. We use three RDF Datasets, namely CRMdig, LUBM and eTMO with their corresponding template queries and we show that the clusters produced significantly limit the number of clusters to be contacted for query answering. Obviously, as the number of clusters increases, eventually the number of nodes required for query answering increases as well, leading to trade-offs among load-balancing and the number of nodes to be used. However, as shown, our algorithm achieves better performance than existing systems with respect to query answering, requiring at most 3 nodes for our template queries even when the dataset is partitioned in 10 nodes. In addition, although in order to these results we allow replication, we show that the impact is minimal imposing at most at most 16.73% total storage overhead.
As future work we intend to explore how our algorithm shall be adapted when no schema is available in an RDF dataset; it is true that RDF datasets do not have always a predefined schema which limits their use to express queries or to understand their content. To this direction approaches are starting to emerge discovering the types of the data using clustering algorithms [14]. Furthermore, we plan to deploy our clustering algorithm in a real clustered environment and to measure the actual improvement on query execution times, comparing our solution with other competitive approaches. In addition our clustering method does not considers limiting the number of nodes that are included in each cluster. However, an idea would be to try to limit the nodes assigned to each cluster trying in parallel to maximize the total dependence of the selected nodes. The problem is well-known to be NP-complete, requires complex variation algorithms over Steiner-Tree problem and we have already started to explore interesting approximations [21]. Obviously as the size and complexity of data increases, partitioning schemes are becoming more and more important and several challenges remain to be investigated in the near future.
References
Aluç, G., Özsu, M.T., Daudjee, K.: Clustering RDF databases using tunable-LSH. CoRR abs/1504.02523 (2015)
Álvarez-García, S., Brisaboa, N.R., Fernández, J.D., Martínez-Prieto, M.A., Navarro, G.: Compressed vertical partitioning for efficient RDF management. Knowl. Inf. Syst. 44(2), 439–474 (2015)
Bui, T.N., Moon, B.R.: Genetic algorithm and graph partitioning. IEEE Trans. Comput. 45(7), 841–855 (1996)
Fiduccia, C.M., et al.: A linear-time heuristic for improving network partitions. In: DAC (1982)
Garey, M.R., Johnson, D.S., Stockmeyer, L.J.: Some simplified NP-complete problems. In: STOC (1974)
Harth, A., Umbrich, J., Hogan, A., Decker, S.: YARS2: a federated repository for querying graph structured data from the web. In: Aberer, K., et al. (eds.) ASWC/ISWC -2007. LNCS, vol. 4825, pp. 211–224. Springer, Heidelberg (2007). doi:10.1007/978-3-540-76298-0_16
Huang, J., Abadi, D.J., Ren, K.: Scalable SPARQL querying of large RDF graphs. PVLDB 4(11), 1123–1134 (2011)
Johnson, D.S., Aragon, C.R., McGeoch, L.A., et al.: Optimization by simulated annealing: an experimental evaluation. part i, graph partitioning. Oper. Res. 37, 865–892 (1989)
Hendrickson, B., Leland, R.: The chaco user’s guid, version 2.0. Technical report SAND94–2692, Sandia National Laboratories (1995)
Kaoudi, Z., Manolescu, I.: RDF in the clouds: a survey. VLDB J. 24(1), 67–91 (2015)
Kaufman, L., Rousseeuw, P.J.: Clustering by means of medoids. Statistical Data Analysis based on the L1 Norm (1987)
Karvounarakis, G., Alexaki, S., Christophides, V., Plexousakis, D., Scholl, M.: RQL: a declarative query language for RDF. In: WWW (2002)
Karypis, G., Kumar, V.: A fast and high quality multilevel scheme for partitioning irregular graphs. SIAM J. Sci. Comput. 20(1), 359 (1999)
Kellou-Menouer, K., Kedad, Z.: A clustering based approach for type discovery in RDF data sources. In: EGC (2015)
Kernighan, B., Lin, S.: An efficient heuristic procedure for partitioning graphs. Bell Syst. J. 49, 291–307 (2013)
Kondylakis, H., Spanakis, M., Sfakianakis, S., et al.: Digital patient: personalized and translational data management through the MyHealthAvatar EU project. In: EMBC (2015)
Lee, K., Liu, L.: Scaling queries over big RDF graphs with semantic hash partitioning. PVLDB 6(14), 1894–1905 (2013)
Lee, K., Liu, L., Tang, Y., Zhang, Q., Zhou, Y.: Efficient and customizable data partitioning framework for distributed big RDF data processing in the cloud. In: CLOUD (2013)
Leng, Y., Chen, Z., Zhong, F., Zhong, H.: BRDPHHC: A Balance RDF data partitioning algorithm based on hybrid hierarchical clustering. In: HPCC/CSS/ICESS (2015)
Newman, M.E.J., Girvan, M.: Finding and evaluating community structure in networks. Phys. Rev. E 69, 026113 (2004)
Pappas, A., Troullinou, G., Roussakis, G., Kondylakis, H., Plexousakis, D.: Exploring importance measures for summarizing RDF/S KBs. In: Blomqvist, E., Maynard, D., Gangemi, A., Hoekstra, R., Hitzler, P., Hartig, O. (eds.) ESWC 2017. LNCS, vol. 10249, pp. 387–403. Springer, Cham (2017). doi:10.1007/978-3-319-58068-5_24
Pellegrini, F., Roman, J.: Scotch: a software package for static mapping by dual recursive bipartitioning of process and architecture graphs. In: Liddell, H., Colbrook, A., Hertzberger, B., Sloot, P. (eds.) HPCN-Europe 1996. LNCS, vol. 1067, pp. 493–498. Springer, Heidelberg (1996). doi:10.1007/3-540-61142-8_588
Rohloff, K., Schantz, R.E.: High-performance, massively scalable distributed systems using the MapReduce software framework: the SHARD triple-store. In: PSI EtA, 4 (2010)
Seddiqui, H., Nath, R.P.D., Aono, M.: An efficient metric of automatic weight generation for properties in instance matching technique. JWS 6(1), 1–17 (2015)
Schmachtenberg, M., Bizer, C., Paulheim, H.: State of the LOD Cloud. http://linkeddatacatalog.dws.informatik.uni-mannheim.de/state/. Accessed 30 Apr 2016
Shi, J., Malik, J.: Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 22(8), 888–905 (2000)
The Cancer Genome Atlas project. http://cancergenome.nih.gov/. Accessed 30 Apr 2016
Theodoridou, M., Tzitzikas, Y., Doerr, M., et al.: Modeling and querying provenance by extending CIDOC CRM. Distrib. Parallel Databases 27(2), 169–210 (2010)
Tian, Y., Hankins, R.A., Patel, J.M.: Efficient aggregation for graph summarization. In: SIGMOD (2008)
Troullinou, G., Kondylakis, H., Daskalaki, E., Plexousakis, D.: RDF digest: efficient summarization of RDF/S KBs. In: Gandon, F., Sabou, M., Sack, H., d’Amato, C., Cudré-Mauroux, P., Zimmermann, A. (eds.) ESWC 2015. LNCS, vol. 9088, pp. 119–134. Springer, Cham (2015). doi:10.1007/978-3-319-18818-8_8
Wang, L., Xiao, Y., Shao, B., Wang, H.: How to partition a billion-node graph. In: ICDE (2014)
Wang, X., Yang, T., Chen, J., He, L., Du, X.: RDF partitioning for scalable SPARQL query processing. Front. Comput. Sci. 9(6), 919–933 (2015)
Xu, X., Yuruk, N., Feng, Z., Schweiger, T.A.J.: Scan: a structural clustering algorithm for networks. In: KDD (2007)
Zeng, K., Yang, J., Wang, H., Shao, B., Wang, Z.: A distributed graph engine for web scale RDF data. PVLDB 6(4), 265–276 (2013)
Zhou, Y., Cheng, H., Yu, J.X.: Graph clustering based on structural/attribute similarities. PVLDB 2(1), 718–729 (2009)
Acknowledgements
This research is implemented through IKY scholarships programme and co-financed by the European Union and Greek national funds through the action entitled “Reinforcement of Postdoctoral Researchers”, in the framework of the Operational Programme “Human Resources Development Program, Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) 2014−2020.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Troullinou, G., Kondylakis, H., Plexousakis, D. (2017). Semantic Partitioning for RDF Datasets. In: Kotzinos, D., Laurent, D., Petit, JM., Spyratos, N., Tanaka, Y. (eds) Information Search, Integration, and Personlization. ISIP 2016. Communications in Computer and Information Science, vol 760. Springer, Cham. https://doi.org/10.1007/978-3-319-68282-2_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-68282-2_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-68281-5
Online ISBN: 978-3-319-68282-2
eBook Packages: Computer ScienceComputer Science (R0)