Abstract
The current gold standard for interpreting patient tissue samples is the visual inspection of whole–slide histopathology images (WSIs) by pathologists. They generate a pathology report describing the main findings relevant for diagnosis and treatment planning. Searching for similar cases through repositories for differential diagnosis is often not done due to a lack of efficient strategies for medical case–based retrieval. A patch–based multimodal retrieval strategy that retrieves similar pathology cases from a large data set fusing both visual and text information is explained in this paper. By fine–tuning a deep convolutional neural network an automatic representation is obtained for the visual content of weakly annotated WSIs (using only a global cancer score and no manual annotations). The pathology text report is embedded into a category vector of the pathology terms also in a non–supervised approach. A publicly available data set of 267 prostate adenocarcinoma cases with their WSIs and corresponding pathology reports was used to train and evaluate each modality of the retrieval method. A MAP (Mean Average Precision) of 0.54 was obtained with the multimodal method in a previously unseen test set. The proposed retrieval system can help in differential diagnosis of tissue samples and during the training of pathologists, exploiting the large amount of pathology data already existing digital hospital repositories.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Health professionals often take decisions based on previously acquired textbook knowledge and their personal experience but rarely search for past cases to reinforce their medical assessment. Retrieval systems are developed to better exploit the large amount of digital medical data contained in hospital repositories for clinical decision support [1]. In retrieval systems, a query can be performed using text information, images or both (multimodal), resulting in a list of relevant cases ranked according to their similarity with the query case [2]. The integration of these systems into the clinical workflow remains a challenge [3].
In [4], a multimodal radiology case–based retrieval benchmark was reviewed. The cases included radiologic RadLex terms automatically extracted from radiology reports and 3D patient scans. Image retrieval systems have also been proposed in the growing field of digital pathology [5,6,7]. Nevertheless, multimodal case–based retrieval strategies for histopathology are rare, even though they could be a helpful tool for pathologists during training and to perform differential diagnosis. To our knowledge, only one multimodal retrieval system fusing histopathology image patches and semantics exists [5]. However, this method did not explore full pathology reports (since it was based on manual data annotations) and included only isolated image patches.
Whole Slide Image (WSI) scanning started to be applied at a large scale only recently, and a full digitization of pathology departments in hospitals will result in large scale digital WSI repositories [8]. Pathologists usually select candidate regions of interest (ROIs) in the WSIs at a low resolution and proceed to evaluate the selected regions in high–power fields. Currently available retrieval systems for histopathology are designed with either small tissue arrays, ROIs from WSIs or individual patches as visual input. To the best of our knowledge, there are no methods in literature proposed for WSI retrieval.
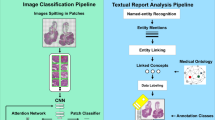
Hand–crafted visual features, such as texture and architecture features, are commonly used to represent images in retrieval systems [9]. In the past few years, deep learning (DL) methods have obtained a better performance for visual content description in comparison with traditional hand–crafted features in this regard [10,11,12]. In this paper, we propose a content–based retrieval system that uses the output features from a fine–tuned DL model, trained to classify cancer gradings in histopathology images, to represent the visual features from WSIs. An unsupervised analysis of the pathology report content was used to train the DL model and not time–consuming manual annotations from pathologists in the WSIs. This enables the reuse of already existing pathology cases for a more integral comparison of new cases to previously assessed ones. A search can in this case be a full case with WSIs and a report or only one of the two, giving several options for browsing.
2 Methods
2.1 Data Set
The Cancer Genome Atlas (TCGA) contains a large collection of digital pathology WSIs and their corresponding pathology reports [13]Footnote 1. Cases with prostate adenocarcinoma (PRAD), the second most common cancer in men, are available [14]. The Gleason grading is the standard evaluation of histopathological samples from prostate cancer patients [15]. 267 cases (WSIs and pathology reports) from prostatectomies of patients with prostate adenocarcinoma (PRAD) were included in this work, aiming at having balanced Gleason scores. The Gleason score (6–10) given to each WSI was manually obtained from the reports. The number of cases for each Gleason score were: G6 35, G7 87, G8 53, G9 83, G10 9. The cases were randomly divided as follows: 162 WSIs for training, 54 WSIs for validation and 51 WSIs for testing (approx. 60%–20%–20%). The pathology report length and content varied depending on the pathology center that generated them and the patient case. The hematoxylin and eosin stained WSIs do not contain any manual annotations (Fig. 1).
2.2 Whole–Slide Image Representation
A Convolutional Neural Network (CNN) is a specialized type of neural network that can learn abstract and complex representations of visual data using a large number of training samples [16]. Manually annotating WSIs in order to obtain exclusively tumor patches from the WSIs, is a time–consuming and challenging task. In [17], it was shown that a CNN can be successfully trained to classify WSIs for prostate cancer grading with a fully automatic sampling of weakly labeled patches i.e. only using the global Gleason score. A subset of 5000 random patches were initially sampled per WSI. The number of cells in tissue increases in the presence of tumors, which results in dark blue areas in the WSI due to the eosin staining of cell nuclei. The sampled patches were subsequently ranked according to the energy of the blue–ratio (BR), which is a feature that can be closely related to cell density. Only the top 2000 patches from each WSI are considered for characterizing the tissue samples.
To encode the visual features of every WSI, the CNN features generated from the fine–tuned deep CNN network for prostate Gleason grading classification were extracted from the patches. The network architecture for pathology images presented in [17] was fine–tuned to classify five Gleason scores (6–10) instead of high vs. low cancer grading. In the end, 1024 dimensional feature vectors from the layer previous to the class probability output were extracted from each patch with the trained network.
Let \(S_{a},S_{b}\) be the sets of selected RGB patches from two WSI at 40\(\times \) resolution, in our setup \(|S_a|=|S_b|=2000\). Let \(f(p)\in \mathbb {R}^{1024}\) be the function that takes a patch as input and computes the forward pass through the deep learning network up to the previous–to–last layer. For two patches \(p_{k} \in S_{a}, p_{l} \in S_{b}\) we have \(v^{k}=f(p_k)\) and \(v^{l}=f(p_l)\), as the two CNN patch codes (or embeddings). The similarity between two patches is computed using the cosine similarity:
The visual similarity between the two slides is calculated adding all the similarities of the individual patches from each WSI:
The cosine similarity is then computed between the vectors of a test query image and the vectors corresponding to each of the training WSIs. This results in a \(2000\,\times \,2000\) similarity matrix for each pair of cases, which is added up to obtain a final visual similarity score.
2.3 Pathology Report Representation
Pathology reports contain information not only from the tissue samples but also from the surgical procedure performed to remove the tissue. This means that information not present in the histopathology images, such as tumor invasion to other body parts, is reported as well. Five criteria of diagnostic relevance for PRAD cases selected by a pathologist in addition to the Gleason score were extracted manually from the pathology reports. The selected criteria include the TNM classification of malignant tumors, with T (0–4) corresponding to the size of the tumor and invasion to nearby tissue and N (0–1) was marked as positive if lymph nodes were involved. Additionally, if the case showed angiolymphatic invasion of the tumor, perineural invasion or if the seminal vesicles were involved then each of these criteria was represented as 1, or 0 if absent. In cases with missing data, the lowest score was given to the corresponding criteria as their absence from the report could have signaled that it was not present during the interpretation. In the experiments, the Gleason score was excluded from the input criteria for the retrieval system.
Extracting the data from the reports automatically is not straightforward, as many regular expressions need to be formulated. For example, it is common to encounter the same grading written as Gleason Score: 9, Primary pattern: 5, Secondary pattern: 4, score = 5 + 4, ... This restricts the use of bag of words models because the pathology reports are not standardized. A more general approach was implemented to make use of unsupervised distributed models for embedding text content. We propose the representation of the text content from each report, embeddeding an n–dimensional space using an unsupervised distribution to a paragraph vector model of doc2vec [18]. Doc2vec is a suitable model for variable–length documents, as is the case of the pathology reports in the data set that were embedded into a 100 dimensional space. The text similarity was computed with the cosine similarity between the case embeddings and the similarity to the query cases was ranked according to this score. The proposed strategy can also be used for different types of tissue and different pathologies.
2.4 Multimodal Fusion
Let \(R_{v},R_{t}\), be the ranking for each query case, sorting the visual and text similarities. The generated late multimodal fusion rank R ranks the most relevant cases for the query by weighting the visual and textual similarities:
In Fig. 2 a flowchart of the full approach is shown.
3 Experimental Results
The four retrieval methods for pathology cases presented in this paper were tested and compared. A retrieved case was considered relevant if the Gleason score from the case matched the query. To evalute the results, retrieval metrics from the NIST (US National Institute of Standards and Technology) evaluation procedures used in the Text Retrieval Conference (TREC) [19] were considered. The following five evaluation metrics were selected: mean average precision (MAP), geometric mean average precision (GM-MAP), binary preference (bpref), precision after 10 cases retrieved (P10) and precision after 30 cases retrieved (P30). The performance of each method is shown in Fig. 3.
The method WS_CNN_Codes used only visual features obtained from a fine–tuned CNN for Gleason grading classification, with 2000 selected patches at a 40\(\times \) resolution per WSI. The model was trained using the Caffe framework and took 15 h to train with 2 NVIDIA Tesla K80 GPUs.
For text representation we tested two approaches, the first (RepCateg) computed a ranking based on the similarity of the report categories manually extracted from the pathologist reports, without including the Gleason score. The second text approach (Rep2Vec) was based on the unsupervised distributed representation of doc2vec [18]. Including or exculding information from the report regarding the Gleason grading was unsupervised. The gensim library was used with the default parameters and a total vocabulary of 3730 words, obtaining 100 dimensional vectors for each report. The ranking was generated using the cosine similarity between the doc2vec report representation.
The proposed multimodal approach (Multimodal) retrieved similar histopathology cases fusing the ranking generated by the deep CNN representation of the WSIs and the ranking from the embedded pathology report text using doc2vec. 10 values of \(\alpha \) were explored in the range of [0, 1]. The best scores were obtained by the multimodal approach with \(\alpha =0.3\).
4 Discussions
A multimodal case–based retrieval approach for histopathology cases based on visual features obtained with deep learning is presented in this paper with an automatic description of pathology reports. The main contributions are:
-
This is the first multimodal histopathology strategy fusing visual features from WSIs and text embeddings of pathology reports, resulting in a novel case–based retrieval system.
-
The method uses visual deep learning features for retrieval, representing WSIs, generated with a CNN trained to classify cancer gradings.
-
The visual CNN model was trained with weakly annotated data (global Gleason scores from WSIs, without any manual annotations), and the free–form text embeddings obtained with an unsupervised approach.
The retrieval methods were trained, evaluated and compared on a publicly available test set. The visual–only approach (WS CNN codes) had better scores than both of the two text–only approaches: RepCateg, using 5 report criteria manually extracted and Rep2Vec, an unsupervised report to vector representation. This could be the result of training the visual representation of the cases with the 5 Gleason scores classes used to evaluate the relevance of the retrieved cases. Moreover, there is an intensive similarity computation among the CNN featires of the query case versus the remaining cases in the data set. When comparing both text–only approaches, embedding full–text reports to a vector, Rep2Vec, resulted in higher retrieval scores than RepCateg. Rep2Vec was able to better mimic the defined relevance of the retrieved cases, mainly because the selected criteria by a pathologist in the reports are only indirectly linked to the Gleason score. These criteria are focused on the surrounding organs and metastasic events which can be considered for another relevance measure of the cases.
The methods were trained and tested with images from several scanners and with no staining normalization. Adding such a normalization can improve performance. The TCGA data and the manual categories extracted from the reports are available for a fully reproducible setup of the proposed strategy. The multimodal fusion tested in this paper is simple as this is the very first example of retrieval fusing real medical reports and WSIs. More advanced fusion techniques can be implemented in a straightforward manner.
Most of the computations can be performed offline and a full case query can be performed in less than 8 s once the patches are extracted. The unsupervised retrieval system strategy was successful in obtaining cases with the same cancer grading even if these scores were not explicitly used in the text representations. The proposed retrieval system could be implemented, with minor modifications, for other organs and diseases. The task of assigning cancer gradings is strongly subjective. The cases retrieved could be better exploited to harmonize pathology case assessment and as a valuable resource for pathologists in training without depending on expensive and time consuming manual annotations.
Notes
- 1.
http://cancergenome.nih.gov/, as of 11 June 2017.
References
Müller, H., Michoux, N., Bandon, D., Geissbuhler, A.: A review of content-based image retrieval systems in medicine-clinical benefits and future directions. Int. J. Med. Inform. 73(1), 1–23 (2004)
Begum, S., Ahmed, M.U., Funk, P., Xiong, N., Folke, M.: Case-based reasoning systems in the health sciences: a survey of recent trends and developments. IEEE Trans. Syst. Man Cybern. 41(4), 421–434 (2011)
Welter, P., Deserno, T.M., Fischer, B., Günther, R.W., Spreckelsen, C.: Towards case-based medical learning in radiological decision making using content-based image retrieval. BMC Med. Inform. Decis. Making 11, 68 (2011)
Jiménez-del-Toro, O.A., Hanbury, A., Langs, G., Foncubierta-Rodríguez, A., Müller, H.: Overview of the VISCERAL retrieval benchmark 2015. In: Müller, H., Jimenez del Toro, O.A., Hanbury, A., Langs, G., Foncubierta Rodriguez, A. (eds.) MRMD 2015. LNCS, vol. 9059, pp. 115–123. Springer, Cham (2015). doi:10.1007/978-3-319-24471-6_10
Caicedo, J.C., Vanegas, J.A., Páez, F., González, F.A.: Histology image search using multimodal fusion. J. Biomed. Inform. 51, 114–128 (2014)
Zhang, X., Liu, W., Dundar, M., Badve, S., Zhang, S.: Towards large-scale histopathological image analysis: hashing-based image retrieval. IEEE Trans. Med. Imaging 34(2), 496–506 (2015)
Kwak, J.T., Hewitt, S.M., Kajdacsy-Balla, A.A., Sinha, S., Bhargava, R.: Automated prostate tissue referencing for cancer detection and diagnosis. BMC Bioinform. 17(1), 227 (2016)
Weinstein, R.S., Graham, A.R., Richter, L.C., Barker, G.P., Krupinski, E.A., Lopez, A.M., Erps, K.A., Bhattacharyya, A.K., Yagi, Y., Gilbertson, J.R.: Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future. Hum. Pathol. 40(8), 1057–1069 (2009)
Doyle, S., Hwang, M., Naik, S., Feldman, M., Tomaszeweski, J., Madabhushi, A.: Using manifold learning for content-based image retrieval of prostate histopathology. In: MICCAI 2007 Workshop on Content-based Image Retrieval for Biomedical Image Archives: Achievements, Problems, and Prospects, pp. 53–62. Citeseer (2007)
Krizhevsky, A., Hinton, G.E.: Using very deep autoencoders for content-based image retrieval. In: ESANN (2011)
Wu, P., Hoi, S.C., Xia, H., Zhao, P., Wang, D., Miao, C.: Online multimodal deep similarity learning with application to image retrieval. In: Proceedings of the 21st ACM International Conference on Multimedia, pp. 153–162. ACM (2013)
Wan, J., Wang, D., Hoi, S.C.H., Wu, P., Zhu, J., Zhang, Y., Li, J.: Deep learning for content-based image retrieval: a comprehensive study. In: Proceedings of the 22nd ACM International Conference on Multimedia, pp. 157–166. ACM (2014)
Gutman, D.A., Cobb, J., Somanna, D., Park, Y., Wang, F., Kurc, T., Saltz, J.H., Brat, D.J., Cooper, L.A.D., Kong, J.: Cancer digital slide archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J. Am. Med. Inform. Assoc. 20(6), 1091–1098 (2013)
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., Parkin, D.M., Forman, D., Bray, F.: Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136(5), E359–E386 (2015)
Humphrey, P.A.: Gleason grading and prognostic factors in carcinoma of the prostate. Mod. Pathol. 17(3), 292–306 (2004)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521(7553), 436–444 (2015)
Jimenez-del-Toro, O., Atzori, M., Otálora, S., Andersson, M., Eurén, K., Hedlund, M., Rönnquist, P., Müller, H.: Convolutional neural networks for an automatic classification of prostate tissue slides with high-grade gleason score. In: SPIE Medical Imaging. International Society for Optics and Photonics (2017)
Le, Q.V., Mikolov, T.: Distributed representations of sentences and documents. In: ICML, vol. 14, pp. 1188–1196 (2014)
Voorhees, E.M., Ellis, A. (eds.): Proceedings of The Twenty-Fourth Text REtrieval Conference, TREC 2015, Gaithersburg, Maryland, USA, 17–20 November 2015, vol. Special Publication 500–319. National Institute of Standards and Technology (NIST) (2015)
Acknowledgments
This work was partially supported by the Eurostars project E! 9653 SLDESUTO-BOX. The authors would like to thank pathologist Lis Vázquez for her counsel regarding the handling of the pathology reports.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Jimenez-del-Toro, O., Otálora, S., Atzori, M., Müller, H. (2017). Deep Multimodal Case–Based Retrieval for Large Histopathology Datasets. In: Wu, G., Munsell, B., Zhan, Y., Bai, W., Sanroma, G., Coupé, P. (eds) Patch-Based Techniques in Medical Imaging. Patch-MI 2017. Lecture Notes in Computer Science(), vol 10530. Springer, Cham. https://doi.org/10.1007/978-3-319-67434-6_17
Download citation
DOI: https://doi.org/10.1007/978-3-319-67434-6_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67433-9
Online ISBN: 978-3-319-67434-6
eBook Packages: Computer ScienceComputer Science (R0)