Abstract
Coronary artery segmentation in 3D images is a fundamental step in evaluating the degree of Coronary Artery Disease (CAD) in cardiac clinical diagnosis and surgical planning. In this paper, we study the effect of vessel filtering and enhancement on coronary artery segmentation from Computed Tomography Angiography (CTA) datasets. The method mainly consists of two steps: (1) CTA datasets enhancement using Hessian-based analysis; and (2) coronary vessels segmentation in enhanced images using Otsu thresholding. The experiments are carried on 18 different CTA datasets and segmentation results of enhanced and non-enhanced datasets are quantitatively measured and compared using three different evaluation metrics. Experimental results show that segmenting coronary vessels in enhanced CTA images gives more accurate extraction of coronary arteries than non-enhanced images.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Vessel enhancement
- Vessel segmentation
- Coronary arteries
- Three-dimensional segmentation
- Medical imaging
- CTA
- Computed tomography angiography
- Thresholding
- Otsu method
- Region growing
1 Introduction
Based on the reports of the world health organization (WHO) [1], CAD is one of the main causes of death worldwide. It’s caused by some narrowed places along coronaries that slow down the blood flow. As a result, oxygen-rich blood can’t reach the heart muscle causing angina or heart attack. There are many cardiac tests that are performed for the assessment of CAD. The reference standard test is the quantitative coronary angiography (QCA) or catheterization. Although, it provides accurate estimation of CAD due to the perfect visualization of vessels, it is invasive, high-cost and may cause some complications [2].
Recent advances in cardiac medical imaging provide high resolution 3D images of heart like Computed Tomography Angiography (CTA) and Magnetic Resonance Angiography (MRA). Such image modalities have recently become an alternative way for coronary disease assessment [3]. It is better than QCA as it is non-invasive but its interpretation is difficult, time consuming and requires special training [4]. These drawbacks motivate the development of automatic accurate coronary segmentation techniques.
The main problems that disturb the coronary segmentation process are: structures attached to coronaries and have similar intensities which mostly cause over segmentation and the variations in geometry and size along the vessels that may cause under-segmentation at low resolution regions. Many segmentation techniques have been developed for segmenting coronary tree and other vessel trees within human body [5].
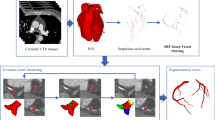
The coronary segmentation framework usually consists of three main steps: (1) initialization, (2) enhancement and (3) segmentation. Figure 1 shows the main components of that framework. The initialization step guides the segmentation algorithm to the regions of interest which can be done by manual initialization or automatic detection of seed points and/or centerlines. The enhancement step is often applied to enhance vessel structures to obtain better segmentation results. Enhanced images and provided seed points/centerlines are then used in the segmentation process. Sometimes there is a fourth step in which segmented coronary tree is used to extract centerlines or detect and quantify stenoses.
This paper shows the effect of vessel enhancement on the coronary artery segmentation results. Firstly, a Hessian-based vessel filter is applied to input CTA images. Secondly, Otsu thresholding is applied to get the thresholds that discriminate between vessel and non-vessel regions. Finally, these thresholds are then used in a region growing process for segmenting the vessel region.
The rest of the paper consists of four sections. Section 2 reviews some related work. Section 3 explains the vessel enhancement approach for coronary artery. Section 4 shows the results and evaluation of applying vessel enhancement on coronary arteries. Section 5 concludes the proposed method and provides possible improvements for achieving better results.
2 Related Work
Hennemuth et al. [6] proposed a three-step coronary segmentation approach. First, the algorithm was initialized by a single seed point placed in ascending aorta. This point was used for coronary ostium detection. Second, the detection of ostium was done by segmenting aorta and analyzing the geometry of its cross-sections. Finally, coronary arteries were segmented using 3D region growing based on Boskamp et al. [7]. Starting from detected coronary origins, neighboring voxels were checked for satisfying some merging criteria. These criteria were a lower threshold, an adaptive upper grey threshold and also a gradient threshold to check the grey level difference in neighboring voxels.
Metz et al. [8] proposed a four-step approach for coronary segmentation and centerline extraction. First, the algorithm was initialized by one seed point at the beginning of the vessel to be segmented. Second, an enhancement step was performed using vessel enhancing diffusion (VED) filter developed by Manniesing et al. [9], which was a smoothed version of Frangi vessel filter [10]. Third, the three main coronaries were segmented using greyscale-based region growing. Starting from vessel beginning, voxels were merged based on predefined greyscale thresholds. Finally, six sub-iteration thinning algorithm was applied to extract centerlines of the coronary tree.
Bock et al. [11] proposed a three-step coronary segmentation approach. First, the algorithm was initialized by a seed point in ascending aorta. Second, this seed point was used to track aorta and detect coronaries. Finally, coronaries were segmented by applying a progressive region growing algorithm. The growing process depended on two thresholds (a lower and an upper threshold) that were initially defined based on the gray value of coronary seeds’ neighboring voxels. These thresholds were continuously updated due to intensity changes along the vessel which allowed for detecting the small distal parts of vessel.
Öksüz et al. [12] proposed a five-step approach for coronary artery analysis. First, the algorithm was initialized by two seed points at the origins of left and right coronaries. Second, a thresholding at \(-400\) HU and 500 HU was performed to remove pulmonary vessels and calcifications, respectively. Third, Hessian-based Frangi Vesselness filter was applied to enhance vessel (tubular) structures. Fourth, coronary arteries were segmented according to the intensity of voxels. Voxels were added to vessel region based on a similarity merging criterion which is the absolute difference between the gray value of the current voxel and the average gray value of the current segmented region. This approach required extra seed points in both left and right coronaries during segmentation for completing non-segmented parts. Finally, obtained coronary tree was used for detection and quantification of stenoses.
Chen et al. [13] proposed a three-step coronary segmentation approach. First, the algorithm was initialized with a seed point placed at the beginning of the vessel to be segmented. Second, input CTA images were enhanced by removing pulmonary vessels and high-intensity regions like calcifications, stents or pacemakers using morphological closing and thresholding at 676 HU, respectively. Finally, coronaries were segmented using a minimum path-based region growing (MP-RG) that used the minimum path searching process as a merging criterion.
Zhou et al. [14] proposed a four-step coronary segmentation approach. First, the algorithm was initialized with two pints at the origins of left and right coronaries. Second, heart region was extracted using thresholding at \(-970\) HU followed by adaptive expectation maximization (EM) and morphological opening operation with a spherical structuring element. Third, Coronary vessels were enhanced using a multi-scale coronary artery response (MS CAR) method by Zhou et al. [15]. Finally, coronary artery tree was segmented using a dynamic 3D rolling balloon region growing (RBG). In this method, a sphere with center at the pre-defined seed point and a 15 mm diameter was moved from vessel start to its end. The voxels inside that sphere were checked to be vessel or non-vessel voxels using an adaptive mean-value-based merging criterion.
3 Methods
In this work, coronary artery tree is segmented in two steps given the CTA datasets and their centerlines. The first step main purpose is to extract the blood vessels which is done by applying Hessian-based vesselness filter. The second step is to segment the coronary artery from enhanced CTA using Otsu thresholding.
3.1 Vessel Enhancement
Input CTA images are enhanced by applying a multi-scale Hessian-based vesselness filter proposed by Sato et al. [16]. Many previous methods have integrated multi-scales and multi-orientations to refine line structures with different widths and orientations [17,18,19], but they was insufficient to be applied to 3D medical images. Sato et al. extended these previous methods to suit 3D images. The filter uses a line filter where the line structure represents the blood vessel to be extracted and enhanced. It aims to refine tubular structures (blood vessels) by calculating a vesselness measure for each voxel in the image. The ideal 3D line is defined as
where I(x, y, z) is the calculated line value at position xyz and \(\sigma \) is the standard deviation that represents the width of the line. The maximum second derivative of that line has a value equals to zero and its direction is identical to z-axis. The second derivative value at any point in the center region of the line cross-section has a negative value.
The analysis of vessel structures is based on the eigenvectors and eigenvalues of the Hessian matrix which consists of the second order gradients of the image,
where the second order gradients of an image \(I_{(x,y,z)}\) are \(I_{xx}, I_{xy}\) and so on. The eigenvectors of H are \(\lambda _{1}\), \(\lambda _{2}\) and \(\lambda _{3}\) while \(e_{1}\), \(e_{2}\) and \(e_{3}\) are their corresponding eigenvalues. A voxel is said to be vessel (gives highest vesselness measure) if the maximum eigenvalue \(\lambda _{1}\) is zero and the direction of its eigenvector \(e_{1}\) is identical to the z-axis as the conditions of the ideal line are satisfied (\(\lambda _{1}\cong 0\) and \(\lambda _{2}\cong \lambda _{3}\ll 0\)).
Figure 2 shows the effect of applying the Hessian-based vessel filter on two CTA slices.
3.2 Coronary Segmentation
Coronary lumen is segmented using Otsu-based region growing. Otsu’s method constructs a histogram for the input image and iteratively tests each gray value for being an optimal threshold. The optimal threshold minimizes the within-class (intra-class) variance which results in maximizing the between-class (inter-class) variance. The between-class (inter-class) variance is:
where \(q_{1}(t)\) and \( q_{2}(t)\) are class probabilities given by:
and
t is the selected threshold and L is the number of bins in the used histogram. The individual class variances are:
and
where \(\mu _{1}(t)\) and \(\mu _{2}(t)\) are class means and given by
and
Otsu method can be applied to get one threshold (single-thresholding) or extended to obtain more than one thresholds (multi-thresholding). In the application of coronary artery, segmentation methods are often interested in high-intensity regions, as coronary vessel regions are brighter than surrounding tissues due to the contrast material (radiographic dye) injection. So, coronary regions are defined with the high-valued thresholds obtained from Otsu.
Figure 3 shows the output of applying the proposed method on three different datasets (dataset00, dataset02 and dataset15).
4 Experimental Results
The proposed method is applied to the 18 training CTA datasets that are publicly available in the Coronary Artery Stenoses Detection and Quantification Evaluation Framework [20, 21]. The method also uses the provided centrelines that were automatically extracted by Yang et al. [22].
Segmentation results are evaluated with three different metrics: Dice Coefficient, Mean Surface Distance (MSD) and Max Surface Distance (MAXSD). Values of the three metrics are reported separately for healthy (H) and diseased (D) vessel segments.
Table 1 shows the quantitative evaluation of the segmentation results compared with three human expert observers. Applying Otsu thresholding segmentation after enhancement gives better results than segmenting without enhancement.
5 Conclusion and Future Work
In this paper, a coronary artery segmentation method is proposed to segment coronaries in CTA datasets. The method is applied in two steps: enhancement and segmentation. Hessian-based analysis filtering has a good effect on segmentation results by enhancing vessel regions and making them more obvious which give the next steps like segmentation a chance to achieve better results. Otsu thresholding combined with region growing to segment vessel regions in enhanced images. They are applied at each centerline point with different thresholds to cope with intensity variations along the vessel.
The segmentation approach can be redeveloped to obtain higher accuracy segmentation results. Two ways of modifications may be applied at different stages: Enhancement and Segmentation. The first suggested modification is a better enhancement technique as enhancement has a direct effect on resulted segmentation. The second suggested modification is to replace Otsu segmentation method with another method, that can achieve better results on defining vessel regions, like K-Means and Fuzzy C-Means clustering approaches. For each modification, segmentation results should be obtained and quantitatively evaluated to be compared with previous results.
References
World Health Organization. The top 10 causes of death (2014)
Tavakol, M., Ashraf, S., Brener, S.J.: Risks and complications of coronary angiography: a comprehensive review. Global J. Health Sci. 4(1), 65 (2012)
Meinel, F.G., Bayer II, R.R., Zwerner, P.L., De Cecco, C.N., Schoepf, U.J., Bamberg, F.: Coronary computed tomographic angiography in clinical practice: state of the art. Radiol. Clin. North Am. 53(2), 287–296 (2015)
Pugliese, F., Hunink, M.M., Gruszczynska, K., Alberghina, F., Malag, R., van Pelt, N., Mollet, N.R., Cademartiri, F., Weustink, A.C., Meijboom, W.B., Witteman, C.L.: Learning curve for coronary CT angiography: what constitutes sufficient training? Radiology 251(2), 359–368 (2009)
Lesage, D., Angelini, E.D., Bloch, I., Funka-Lea, G.: A review of 3D vessel lumen segmentation techniques: models, features and extraction schemes. Med. Image Anal. 13(6), 819–845 (2009)
Hennemuth, A., Boskamp, T., Fritz, D., Khnel, C., Bock, S., Rinck, D., Scheuering, M., Peitgen, H.O.: One-click coronary tree segmentation in CT angiographic images. In: International Congress Series, vol. 1281, pp. 317–321. Elsevier (2005)
Boskamp, T., Rinck, D., Link, F., Kummerlen, B., Stamm, G., Mildenberger, P.: New vessel analysis tool for morphometric quantification and visualization of vessels in CT and MR imaging data sets. Radiographics 24(1), 287–297 (2004)
Metz, C., Schaap, M., Van Der Giessen, A., Van Walsum, T., Niessen, W.: Semi-automatic coronary artery centerline extraction in computed tomography angiography data. In: IEEE 4th International Symposium on Biomedical Imaging: From Nano to Macro, pp. 856–859. IEEE (2007)
Manniesing, R., Viergever, M.A., Niessen, W.J.: Vessel enhancing diffusion: a scale space representation of vessel structures. Med. Image Anal. 10(6), 815–825 (2006)
Frangi, A.F., Niessen, W.J., Vincken, K.L., Viergever, M.A.: Multiscale vessel enhancement filtering. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 130-137. Springer, Heidelberg (1998)
Bock, S., Khnel, C., Boskamp, T., Peitgen, H.O.: Robust vessel segmentation. In: Medical Imaging, pp. 691539–691539. International Society for Optics and Photonics (2008)
Öksüz, İ., Ünay, D., Kadıpaşaoğlu, K.: A hybrid method for coronary artery stenoses detection and quantification in CTA images. In: MICCAI Workshop 3D Cardiovascular Imaging: A MICCAI Segmentation (2012)
Chen, Y., Cao, Q., Zhuang, Z., Yang, Z., Luo, L., Toumoulin, C.: 3-D coronary vessel extraction using a novel minimum path based region growing. In: International Conference Image Analysis and Recognition, pp. 502–509. Springer, Heidelberg (2013)
Zhou, C., Chan, H.P., Chughtai, A., Kuriakose, J., Agarwal, P., Kazerooni, E.A., Wei, J.: Computerized analysis of coronary artery disease: performance evaluation of segmentation and tracking of coronary arteries in CT angiograms. Med. Phys. 41(8), 081912 (2014)
Zhou, C., Chan, H.P., Chughtai, A., Patel, S., Hadjiiski, L.M., Wei, J., Kazerooni, E.A.: Automated coronary artery tree extraction in coronary CT angiography using a multiscale enhancement and dynamic balloon tracking (MSCAR-DBT) method. Comput. Med. Imaging Graph. 36(1), 110 (2012)
Sato, Y., Nakajima, S., Atsumi, H., Koller, T., Gerig, G., Yoshida, S., Kikinis, R.: 3D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. In: CVRMed-MRCAS 1997, pp. 213–222. Springer, Heidelberg (1997)
Koller, T.M., Gerig, G., Szekely, G., Dettwiler, D.: Multiscale detection of curvilinear structures in 2-D and 3-D image data. In: Proceedings of IEEE Fifth International Conference on Computer Vision, pp. 864–869. IEEE (1995)
Sato, Y., Chen, J., Yamamoto, S., Tamura, S., Harada, N., Shiga, T., Harino, S., Oshima, Y.: Measuring microcirculation using spatiotemporal image analysis. In: Computer Vision. Virtual Reality and Robotics in Medicine, pp. 302–308. Springer, Heidelberg (1995)
Sato, Y., Chen, J., Zoroofi, R.A., Harada, N., Tamura, S., Shiga, T.: Automatic extraction and measurement of leukocyte motion in microvessels using spatiotemporal image analysis. IEEE Trans. Biomed. Eng. 44(4), 225–236 (1997)
Coronary Artery Stenoses Detection and Quantification Evaluation Framework. http://coronary.bigr.nl/stenoses/about.php
Kirişli, H.A., Schaap, M., Metz, C.T., Dharampal, A.S., Meijboom, W.B., Papadopoulou, S.L., Cramer, M.J.: Standardized evaluation framework for evaluating coronary artery stenosis detection, stenosis quantification and lumen segmentation algorithms in computed tomography angiography. Med. Image Anal. 17(8), 859876 (2013)
Yang, G., Kitslaar, P., Frenay, M., Broersen, A., Boogers, M.J., Bax, J.J., Reiber, J.H., Dijkstra, J.: Automatic centerline extraction of coronary arteries in coronary computed tomographic angiography. Int. J. Cardiovasc. Imaging 28(4), 921–933 (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this paper
Cite this paper
Shams, M., Hamad, S., Salem, M.A.M., Shedeed, H.A. (2018). Coronary Artery Vessel Tree Enhancement in Three-Dimensional Computed Tomography Angiography. In: Hassanien, A., Shaalan, K., Gaber, T., Tolba, M. (eds) Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2017. AISI 2017. Advances in Intelligent Systems and Computing, vol 639. Springer, Cham. https://doi.org/10.1007/978-3-319-64861-3_25
Download citation
DOI: https://doi.org/10.1007/978-3-319-64861-3_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-64860-6
Online ISBN: 978-3-319-64861-3
eBook Packages: EngineeringEngineering (R0)