Abstract
Exome sequencing has become a powerful tool in disease gene discoveries as well as diagnostic evaluations of genetic disorders. Clinical exome sequencing can identify contributing single nucleotide variants (SNVs) and some classes of copy-number variants (CNVs) in approximately 30% of unselected cases. For clinical laboratories that offer exome sequencing, it is important to establish reliable and robust NGS analysis, interpretation and reporting pipelines and procedures in order to ensure test quality, reliability and meet required turn-around-time (TAT). The clinical report for exome sequencing usually includes primary molecular findings related to the patient’s clinical phenotype, as well as opted in medically actionable secondary findings and carrier status for autosomal recessive disorders. Clinical exome sequencing, which was initially regarded as mainly the “last resort” for patients who had previously tested negative for specific genes, karyotype, and/or microarray studies, can now be utilized as an effective first tier test, usually for patients with nonspecific phenotypes or phenotypes suggesting substantial genetic heterogeneity. Additionally, the improvement of the methodologies, analysis tools, and TAT has brought new applications of exome sequencing into the genetic testing of patients, esp. prenatal patients or patients who are at high risk for potentially life-threatening conditions and are therefore considered critically ill.
Clinical NGS panel testing, exome sequencing and more rarely whole genome sequencing (WGS) currently co-exist. In the context of decreasing cost of NGS, rigorous comparisons of different clinical NGS platforms are needed in order to better understand their clinical utilities and harness the power of whole-wide NGS sequencing in the clinical arena.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction to the NGS/Exome Sequencing Process
Next-generation sequencing (NGS) allows for simultaneous interrogation of multiple genes with dramatically reduced cost of sequencing per base. As a result, NGS-based analysis has changed the landscape of research and clinical diagnostic testing. Many clinical laboratories are now able to offer a wide array of disease-targeted NGS panels and exome Sequencing. Whole genome sequencing (WGS) has also been available in a few clinical laboratories.
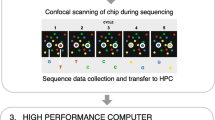
In general, the process of NGS involves breaking down patient DNA into short fragments, followed by adaptor ligation during library construction, target region enrichment (capture), and sequencing. The capture step is needed for targeted analysis of a subset of genomic regions (exome or panel sequencing, but not WGS); the process involves an additional step prior to sequencing. In this step, the targeted regions are enriched from the total genomic DNA by means of probe hybridization. Enriched libraries are then loaded on NGS sequencers to undergo sequencing in a massively parallel fashion. With NGS becoming routine practice and the availability of many open source software for the analysis of such data, certain tools are becoming standards in common bioinformatic pipelines.
2 Variant Calls and Annotations in the Bioinformatics Pipeline
Whether the sequencing occurs in the clinical or research laboratory, there are generally three broad steps to go from raw sequencing data to annotated variants (Fig. 1). The first step consists of processing and mapping reads to a reference genome. This would include trimming and removing duplicate reads, as well as local realignment and readjusting base quality scores around problematic regions. The second step would involve calling Single Nucleotide Variants (SNVs) and sort insertion-deletions (INDELs), as well as, in some laboratories, detecting copy number variations (CNVs). The third step is annotating and classifying the variation type and assigning candidate consequences to the variants. It is important to note that the choice, design and implementation of the wide assortment of available tools into a bioinformatics pipeline can significantly affect the accuracy and identification of variants. With the increasing number of possible combination of various tools, one can build many different variant-calling pipelines . Recent reports of systematic comparison of variant callers provide valuable insight and guidance to choosing variant callers when building a pipeline [1].
Although the list of tools may be long, there are invariably predominant tools that have become standards and widely used in the community. For read aligners, the major three are BWA-MEM (http://arxiv.org/abs/1303.3997), Bowtie [2] and Novoalign (http://www.novocraft.com). For SNVs and INDELs variant callers, the major ones are: GATK-HC (Genome Analysis Tool Kit HaplotypeCaller) [3], Samtools [4], Freebayes (http://arxiv.org/abs/1207.3907), Atlas [5], Platypus [6]. For classifying and annotating variants, the major three tools are: Annovar [7], SnpEff [8] and Variant Effect Predictor (VEP) [9].
Since there are obviously differences in the bioinformatics assumptions and rules that are codified into a given tool, it is not surprising that pipelines with different components do not always produce identical results when compared to each other. Nevertheless, since the process of aligning and calling variants are usually well integrated with variant annotation in any given complete pipeline, the real challenge is trying to understand the behavior and expected outcomes of such a pipeline in relation to the biological consequence of variants . This complexity arises, not only because of the chosen tool (static factors), but is also affected by dynamic factors, such as the transcript set used and the constantly evolving database resources in the public domain. One example is a recent report by McCarthy’s et al., which concluded that both the selection of annotation software and transcript set can affect variant classification and concordance when comparing two different tools using the same transcript set [10]. In other words, variant calling and annotation is a tight intertwinement of several major components, namely, pipeline tools, public data resources, the actual single sample of interest, and the aggregate information of all previously analyzed clinical samples.
The ability to detect copy number variants (CNVs) in NGS is becoming an important component of any analysis pipeline. Estimating copy number in from the exome data is highly dependent on both the quality and the coverage read depth of exon targets in the exome capture design. The detection is limited only to genic and exonic regions, and therefore pure intergenic and intronic CNVs may be missed. Similar to calling SNP and INDELs, it is worth noting that there is considerable variability in CNV calling algorithms, and so it’s recommended to try or combine multiple approaches . Tools like CoNVex [11], CoNIFER [12] and XHMM [13], among others, are commonly used to call CNVs from exome data. Some of these tools use normalization methods that can also take into account batch effects and other background noises that should be removed to improve signal detection of copy losses or gains. Other considerations include whether the samples of interest are part of a large (cohort case control), small (tumor/normal comparison) study, or a growing collection of samples from a clinical laboratory with varying types of phenotypes.
3 Variant Classification and Clinical Reporting for Exome Cases
Exome sequencing is a highly complex test in which a large number of rare variants are detected. Bioinformatic pipelines are built to filter variants based on frequencies and additional criteria to help reduce the number of variants to assess. As a basic criteria, a 1% minimum allele frequency (MAF) is commonly used to filter for rare variants. This wide-ranging cut off value allows for the detection of any potentially disease-causing variants. This cut off value is also relaxed for variants that have previously been reported as disease-causing in the literature. Furthermore, additional internal information can be added to reduce the number of variants to assess. This filtering process helps in reducing the number of variants to review and classify. Once filtered, the list of variants needs to be assessed based on the newly updated ACMG/AMP guidelines for variant interpretation [14]. Studies have shown that intra- and inter-laboratory discrepancies exist regarding variant classification [15], indicating the urgent need of optimizing and expanding the current framework ACMG/AMP guidelines and improving communications among laboratories.
One additional uniqueness and challenge of genome wide test such as exome sequencing as opposed to targeted testing is the unfamiliarity and ambiguity of the patient’s disorder. The phenotype of the patient is often unclear and broad. Therefore, while the variants are classified based on the ACMG guidelines, extensive genotype/phenotype correlation is also necessary in the analysis of variants from clinical exome sequencing. Thus, the challenge lies in the number of variants to assess, the assessment of the classification based on the ACMG guidelines and the uncertain phenotype/genotype correlation.
As with any other clinical genetic testing, the content of clinical exome sequencing reports follows the current CLIA regulations and requirements (42 CFR §493.1291) and CAP recommendations [16]. However, the major challenges and differences with single genes and panels reports lies in (1) the complexity of the test and its interpretation and (2) the possible discovery of secondary variants unrelated to the patient’s phenotype but that could affect the patient’s health (incidental findings) and reproductive risk (carrier status). Therefore, the development of a clear and concise report is essential to effectively communicate the clinically relevant findings back to the referring clinician and patient [17].
3.1 Key Elements of Clinical Exome Report Content and Organization
The exact content and organization of exome reports is dependent on the type of exome testing requested and the clinical laboratory issuing the report (See Table 1). Nevertheless, common key elements are essential and are as follow:
-
3.1.1 A clear and concise title indicating the most relevant molecular finding .
-
3.1.2 The clinical indication/referral for exome testing . This clinical information is essential for the clinical laboratory to provide an accurate correlation between the molecular findings and the clinical presentation of the patient during the review analysis of the exome data.
-
3.1.3 The primary molecular findings including a list of variants detected and related to the patient’s clinical phenotype, followed by an interpretation of the results. For clarity, exome reports usually contain a table with the variants’ information followed by the interpretation of the results. These variants are interpreted and categorized following the modified ACMG guidelines [14] and include pathogenic, likely pathogenic variants and VUS related to the patient’s clinical phenotype. Relevant information on the report usually include the disease associated with defects in the gene and a description of the disorder, inheritance pattern(s) of the disorder, the gene symbol, the classification, genomic coordinate, nucleotide and amino acid positions, and zygosity of the variant, if the parents are available, the inheritance from each parent is indicated and de novo and compound heterozygous variants are also indicated. If the variant has previously been reported in public databases or in the literature, references such as dbSNP, ClinVar, or PMID numbers and population frequencies are specified to help the clinician understanding the meaning of the findings. Additional information may also include (1) whether the variant has been confirmed by a second methodology such as Sanger sequencing; (2) the predicted pathogenecity of the variant based on bioinformatic algorithms such as SIFT, Polyphen2 [18, 19] and (3) the coverage depth of the gene and whether all exons of the gene were entirely covered by NGS: this information is relevant if a single heterozygous variant is detected in an autosomal recessive gene disorder , to insure that no additional variant in trans configuration may be missed by exome sequencing.
-
3.1.4 Reports of medically actionable secondary findings and carrier status . Exome sequencing may detect secondary findings such as pathogenic variants known to affect the health of an individual and potentially medically actionable and pathogenic variants in autosomal recessive disorder predicting the carrier status of the individual. Among clinical exome cohorts, medically actionable variants are detected in about 3–4% of patients referred for exome [20, 21]. ACMG recommends the report of known pathogenic variants (and expected pathogenic for some genes) from 56 (version 1) and 59 (version 2) genes mostly related to cardiovascular disorders and cancer for which treatment or medical recommendations are available [22, 23]. The guidelines are of tremendous help for clinical laboratories. In the meantime, several differences in reporting variants in genes outside the “ACMG59” and approaching of returning results among clinical laboratories still exist. First, regarding the definition of medically actionable variants, while pathogenic variants in the 59 ACMG recommended genes are analyzed and returned, additional pathogenic variants in specific genes considered medically actionable by the clinical laboratory may be returned when encountered during data analysis [20, 21]. For instance, Tarailo-Graovac et al. detected an individual who was compound heterozygosity for two pathogenic variants in CFTR but had no reported clinical phenotype of the disorder [21]. While this gene may be on the return carrier list by some clinical laboratories, this type of findings was considered as medically actionable. Second, regarding the reporting, while the ACMG guidelines originally recommended the mandatory return of the actionable variants, the guideline has been modified and laboratories have chosen over time to give an option to opt in or out to fulfill the patient’s request. Overall, it was estimated that over 90% of the patients choose to receive medically actionable variants [24]. Finally, regarding the mechanism of return, some laboratories report medically actionable variants for the proband only and request additional consent and counseling for the parents to obtain knowledge of their status. An alternative approach is to report these variants for proband and parents at the same time, for which opt-in authorization and counselling is necessary prior to testing to address the concerns of the family. Other laboratories report these findings on a separate report, available to the patient and the parents. In all cases, these medically actionable variants warrantee genetics counseling and recommendations in addition to the primary findings related to the clinical phenotype in patients.
-
Carrier status was not included in the ACMG recommendations for reporting of incidental findings. Currently there is no official guideline for carrier status reporting specifically for exome sequencing. Different clinical laboratories choose to report a different set of gene for carrier status, based on disease severity, frequency and recommendations from professional societies such as ACMG and ACOG. For instance, pathogenic variants in CFTR may be reported because of the high prevalence of the disorder in population of European descent. Similarly, hemoglobin S may also be considered for return due to the high frequency in populations at risk for sickle cell disease . Other laboratories choose not to report any carrier findings and recommend a separate universal carrier screening if needed by the patient. Thus the report of carrier status is not consistent among clinical laboratories in the exome sequencing context.
-
3.1.5 Methodologies and recommendations based on the molecular findings . Because of the constant improvement of technologies and variations in variant annotations and variant calling algorithms between clinical laboratories, methodologies are necessary to fully comprehend the test. Limitations of the exome test should be also mentioned such as triplet nucleotide disorders, large deletion and duplications and further testing should be recommended if indicated.
3.2 Additional Optional Report Categories
In addition to the primary and secondary findings, some clinical laboratories have chosen to report on pharmacogenomic variants, including known pathogenic variants VKORC1/CYP2C9 for instance that can alter warfarin metabolism and known pathogenic variants in CYP2C19, that can alter Plavix metabolism . Moreover, mitochondrial sequencing may be available as part of exome testing. Thus, clinicians need to be aware of the variations between the type of exome test and variation between laboratories, when ordering a whole exome sequencing test.
In the case of trio exome analysis, de novo variants and compound heterozygous variants in genes unknown to cause a disorder or in genes unrelated to the patient’s clinical presentation are reported in an additional table. If the exome does not provide a molecular diagnosis, this additional information may become relevant in the future as more genes causing disease are discovered.
3.3 Special Cases
The organization and content of exome reports will vary based on the type of exomes ordered. Examples of specific exome test reports are mentioned below and compared in Table 1.
3.3.1 Prenatal Trio Exome
This report includes all variants related to the prenatal indications as well as variants in disease genes unrelated to the prenatal indications but likely to cause significant disorders during childhood. Because of the nature of this test, the incidental findings are reported after birth as requested. Although this test is a trio test, de novo and compound heterozygous variants in non-disease causing genes are not reported as these will not facilitate a clinical diagnosis.
3.3.2 Adult Screening Exome
This test is offered by several laboratories to individuals, usually in good health, with no significant abnormal clinical presentation. Reports include the IF, carrier status and pathogenic findings in adult conditions.
3.4 Variants Usually Not Included in Exome Reporting
Variants not reported in exome reports consist of clinically irrelevant variants including: (1) Variants in disease genes not related to the patient’s clinical phenotype; (2) Benign and likely benign variants; and (3) Variants in genes unknown to cause Mendelian disorders including susceptibility genes. These variants may be available in secondary reports available from the clinical laboratories and may help in future diagnosis. For instance, if an exome does not provide a molecular diagnosis, variants located in genes unknown to cause disease at the time of the report may become clinically relevant later as new genes causing disorders are discovered.
Additionally, pathological findings in adult neurological degenerative disorders including Huntington, Alzheimer and Parkinson’s diseases, are usually not reported unless the patient is an adult referred for testing with one of these specific clinical phenotype, although specific gene testing is recommended in this particular case.
3.5 Additional Considerations and Challenges
Regarding the delivery and communication of clinical exome reports, clinical laboratories have primarily issued PDF reports, easily printable and delivered to the referring clinic. Due to the complex nature of the exome sequencing, the return of results needs more dynamic interactions between the referring physician and the clinical laboratory. An interactive web-based reporting portal with hyperlinks to the relevant web-based clinical and genomic information will help the physicians better understand the information provided in the exome reports. In addition, the interactive web-based reporting system should also enable better communication and interaction between the physician and clinical laboratory.
4 Clinical Utility of Whole Exome Sequencing
In an analysis of 500 patients evaluated in a medical genetics clinic setting, Shashi et al. (2014) reported that conventional diagnostic evaluation (i.e. clinical exam, biochemical testing, CGH array, and phenotype-directed sequencing) failed to establish a specific etiology in approximately 50% of patients with suspected genetic disorders [25]. This statistic is a testament to the great challenge of genetic diagnosis which is complicated by the rarity of many genetic syndromes and by the potential for phenotype and locus heterogeneity to obscure the causative gene. Without a diagnosis, patients are left with uncertainties about disease progression and long-term prognosis , may be ineligible for medically-indicated social services, and are often subject to additional and potentially invasive diagnostic testing (e.g. muscle biopsy).
Recent studies have demonstrated an inarguable role for exome sequencing in the diagnostic assessment of such unsolved cases. Using a proband-only approach , Yang et al. (2014) reported a diagnosis rate of 25% among 2000 consecutively tested patients for whom traditional approaches failed to elucidate a genetic etiology [20]. A combination of trio-based and proband-only testing yielded a diagnosis in 26% of 814 patients evaluated by exome sequencing and reported by Lee et al. (2014) [26]. In a smaller cohort of Canadian patients, Sawyer et al. (2016) identified a pathogenic variant in a known disease gene in 29% of cases, and this specifically in patients who were previously extensively evaluated and nearing the end of a protracted diagnostic odyssey [27]. In an unselected cohort of 500 patients, exome sequencing detected a positive or likely positive result in a recognized disease gene in 30% of patients (Farwell et al. 2015) [28]. Thus, although the process of exome sequencing data acquisition and variant analysis may differ between clinical laboratories, the general approach of sequencing the exome consistently yields a diagnosis in at least one out of every four patients tested.
Several groups have examined the effect of genetic diagnosis by exome sequencing on subsequent patient management. Valencia et al. (2015) reviewed in detail the first 40 pediatric exome cases performed at a single institution [29]. Consistent with other reports, the overall diagnostic rate was 30%. All patients who received a molecular diagnosis were considered meaningfully impacted by the result in that exome sequencing brought an end to the diagnostic odyssey and enabled disorder-specific genetic counseling. In addition, variants detected by exome sequencing resulted in a targeted treatment plan in three patients, an altered approach to clinical management in one patient, and disorder-specific surveillance in four patients [29]. Thevenon et al. (2016) similarly studied 43 patients with intellectual disability or epileptic encephalopathy at a single institution who underwent exome sequencing analysis [30]. Fourteen patients received a molecular diagnosis; in two cases this enabled prenatal testing and in two cases disease management was altered by the exome findings. In a third study, six of 105 patients diagnosed by exome sequencing were reported to have had a dramatic change in management as a consequence of the exome result (Sawyer et al. 2016) including, for example, a patient whose diagnosis was modified from infantile myofibromatosis to fibrodysplasia ossificans progressiva by exome sequencing resulting in discontinuation of chemotherapy [27].
The diagnostic yield of exome sequencing may be further increased if the testing context permits careful patient selection, rigorous phenotyping, and functional analyses. In a recent study of 41 deeply-phenotyped patients with intellectual disability and suspected metabolic disease evaluated with proband or trio-based exome , Tarailo-Graovac et al. (2016) found a molecular diagnosis in a remarkable 68% of patients [21]. However, this high diagnostic rate was contingent upon the establishment of two novel disease genes and the recognition of phenotypic expansion associated with 22 known disease genes; functional studies were performed to provide evidence of pathogenicity for variants in a subset of these genes. These exome sequencing results were reported to have altered or influenced subsequent clinical management in 44% of patients. While not feasible in a high throughput clinical setting, this work demonstrates that comprehensive phenotypic assessment together with the time allowance to pursue new gene discovery and the availability of resources to functionally address questions of phenotypic expansion may greatly augment the solve rate achievable by exome sequencing.
Although in most cases a clinical approach based on syndrome recognition abets the diagnostic process, it can also be a source of bias as the true phenotypic spectrum of many genetic disorders is not known. In addition, clinicians often operate under the assumption of Occam’s razor – that the simplest explanation is the most likely. Studies of clinical exome cases have demonstrated the power of comprehensive sequencing to address diagnostic holes that may result from unavoidable clinician bias. For example, Farwell et al. (2015) specifically describe several cases in which autosomal recessive inheritance was suspected on the basis of a family history of consanguinity however de novo dominant events were ultimately detected by exome sequencing [28]. In 362 families tested by exome sequencing, Sawyer et al. (2016) found causative variants in established disease genes in 26 patients who escaped diagnosis because of atypical disease presentation [27]. Yang et al. (2014) found bona fide pathogenic variants in two disease-associated genes in 23 patients in their cohort resulting in blended and likely convoluted phenotypes, and also reported somewhat counterintuitively that X-linked disorders were found in equal numbers of male and female patients [20]. These scenarios underscore the value of unbiased genetic analysis in the diagnostic evaluation of unsolved cases.
The benefit of exome sequencing in augmenting diagnostic yield and providing medically-actionable information should be weighed against the potential cost to the individual and effect on societal healthcare expenditures . Individual costs may be financial or may come in the form of increased anxiety and/or additional medical surveillance following detection of a variant of uncertain clinical significance. To minimize personal cost and justify exome sequencing in a resource-limited context, judicious application of the test to those patients most likely to benefit is essential. The American College of Medical Genetics and Genomics policy statement on the Clinical Application of Genomic Sequencing (2012) suggests consideration of exome sequencing in affected patients with (1) non-diagnostic clinical features for whom a genetic etiology is likely (i.e. positive family history), (2) a disorder characterized by substantial locus heterogeneity, (3) a defined genetic disorder for which a molecular diagnosis has not been established by existing assays; and (4) for prenatal evaluation in cases where a clear diagnosis remains elusive after conventional genetic testing [https://www.acmg.net/staticcontent/ppg/clinical_application_of_genomic_sequencing.pdf]. The application of exome sequencing may also be appropriate for patients suspected of having a genetic condition for which no clinically-validated assay exists. The appropriate use of exome sequencing also requires recognition that exome sequencing as a methodology does not detect all forms of genetic variation. For example, single nucleotide variants and small insertion/deletion events (<10 bp) are reliably identified on exome sequencing whereas trinucleotide repeats, copy number variants, large insertion/deletion events, structural variants, aneuploidy, and epigenetic changes are not (Biesecker et al. 2014) [31]. In addition, technical limitations hinder complete coverage of the exome and in most cases, a small subset of genes lack the depth of coverage required for rigorous diagnostic assessment of that region. As such, ensuring coverage of key genes through online tools and acquiring a basic knowledge of the mechanism of gene disruption for disorders high on the differential diagnosis are important considerations prior to exome testing.
The sensitivity of exome sequencing may be further enhanced when a parent-child trio-based approach is used. Trio exome readily identifies de novo events and provides upfront phase data for variants found in autosomal recessive genes . Lee et al. (2014) compared 410 trio exome cases with 338 proband-only cases. The diagnostic rate was significantly higher in the trio exome cohort (31% vs. 22%, p = .003) although this was not a randomized comparison [26]. In addition to improved diagnostic yield, a second benefit of the trio-based approach is reduced turnaround time, which permits exome reporting in a time frame suitable for prenatal diagnosis and testing of critically-ill patients. Carss et al. (2014) performed a proof of principle study in which trio exome sequencing was performed on 30 fetuses and neonates with structural anomalies detected on prenatal ultrasound [32]. Three de novo likely causative variants were detected yielding a diagnostic rate of 10%. Drury et al. (2015) examined the utility of proband-only and trio-based exome sequencing for diagnosis of fetuses with abnormal ultrasound findings [33]. Return of results did not occur during pregnancy but pertinent findings were shared with families afterward. A definitive diagnosis was established by proband exome sequencing in 2 of 14 cases (14%) and by trio exome in 3 of 10 cases (30%). In the largest study to date, Normand et al. (unpublished data) reviewed 92 cases of prenatal WES performed on fetal samples obtained by amniocentesis/chorionic villus sampling or on products of conception. In 15 of 42 probandonly cases and 21 of 50 trio cases (~39% overall) a molecular diagnosis was ascertained by WES, suggesting a promising role for WES in improving prenatal diagnosis.
For critically-ill patients, studies have also evaluated the utility of whole genome sequencing (WGS) as WGS does not require a capture step and can be performed expeditiously. Fifty-seven percent of 35 acutely ill patients with heterogeneous clinical phenotypes reported by Willig et al. (2015) were found to have a causative variant on rapid trio-based WGS [34]. Soden et al. (2014) employed a rapid WGS protocol in 15 patients with primarily neurological phenotypes from neonatal or pediatric intensive care units ; a molecular diagnostic rate of 73% was achieved in this cohort [35]. Notably, the fastest time to final report for rapid WGS in this study was 6–10 days, suggesting that trio-based exome sequencing (which can be performed clinically with a turnaround time of 2–3 weeks) is a reasonable alternative to WGS in critically-ill patients. Meng et al. (unpublished data) reviewed 40 patients tested clinically with critical or time-sensitive exome sequencing. The median turnaround time was 12.8 days and a potential or partial diagnosis was established in 52.5% of patients. In at least 14 cases, the results of exome sequencing influenced subsequent patient care decisions, demonstrating the utility and feasibility of exome sequencing in the critical care setting.
5 Exome Sequencing Versus NGS Panel Versus WGS
In order to select the most appropriate test for each patient, from single-gene to whole genome sequencing, it is essential for clinicians to understand the strengths, limitations, and diagnostic indications for each test. It is important to note that, even in the era of NGS technology, the traditional approach of single-gene testing still holds great utility for many disorders. Single-gene testing is preferred for patients who present with distinctive clinical findings that point to a particular Mendelian genetic disorder, for which the causative gene has been established. On the other hand, for disorders associated with wide clinical variability and genetic locus heterogeneity, a multigene panel approach or the whole exome or genome sequencing approach may provide greater benefit over single-gene tests.
Gene panel testing is preferred for patients who present with disorders associated with multiple causative genes, and/or present with a phenotype that cannot clearly point to one disorder. An advantage of targeted gene panels over whole exome and whole genome sequencing is comprehensive sequence coverage because these panels are often combined with complementary technologies such as Sanger sequencing or long range PCR to fill gaps that NGS fails to cover (due to high GC content, sequence homology, repetitive sequences, etc.). Some panels are also complemented with aCGH to simultaneously detect exon-level copy-number changes in targeted genes . Another advantage of targeted gene panels is better depth of coverage that provides greater confidence in variants detected, and shorter turnaround time.
Selecting the most appropriate gene panel can be a challenge for ordering physicians. Because clinical laboratories may use different stringencies for gene inclusion, the number of genes incorporated into a panel may vary significantly among laboratories even for the same clinical indications (Xue Y et al. Genet Med. 2015) [36]. Therefore, it is essential to know which genes show strong disease association and are therefore more relevant to the patient phenotype versus those that were linked with the disease based only on association studies or single studies. It is also important to note that addition of newly-identified disease genes may take time before they are added to existing panels. Many laboratories may also decide against adding new genes if not cost effective. Therefore, some laboratories have shifted to performing whole exome sequencing and limiting the analysis to genes associated with a particular phenotype and filling up the gaps with Sanger sequencing.
Clinical whole exome sequencing is currently indicated for patients who have either remained undiagnosed after single- or multi-gene panel testing, or for disorders with extreme heterogeneity and clinical variability that multigene testing is deemed less cost effective. Although exomes are intended to cover all protein coding regions of the genome, certain genomic regions (e.g. repetitive regions, high GC regions) decrease the performance of assay. Clinical exome sequencing usually has slightly lower coverage (usually up to 95–98%) than clinical NGS panels.
Whole exome sequencing typically uncovers approximately 20,000–50,000 variants per exome (Gilissen C et al. Eur J Hum Genet 2012) [37], and identifying the causal variant(s) thus can be a challenge. Computational tools have been developed that aid in the automation of variant prioritization, however, up to hundreds of variants still require careful manual inspection and curation. In addition, there is a growing concern about the potential of this test to identify incidental findings and the how to appropriately communicate them to patients (Kiltzman R, et al. JAMA 2013) [38]. As an important side note, when ordering exome sequencing, it is very useful to provide all clinical findings to the clinical molecular geneticist to help with variant interpretation.
Despite these limitations , exome sequencing has demonstrated great success as both a gene discovery and diagnostic tool. Large studies on the clinical utility of whole exome sequencing on a range of disorders have reported an overall molecular diagnostic rate of approximately 25–28% [20, 26, 39], with the yield higher for trio exomes than proband exomes (Lee H et al. JAMA 2014) [26]. Patients who have had whole exome sequencing are commonly children, since many genetic conditions present during childhood. In a report by Yang et al., the highest rate of a positive diagnosis was in a group of patients with a nonspecific neurological disorder (Yang Y, et al. JAMA 2014) [20].
Whole genome sequencing is considered to be the most comprehensive genetic test to date, covering approximately 98% of the genome [40, 41]. Because whole genome sequencing does not require an enrichment step, it generates a more uniform coverage of the genome over exome sequencing. Also, longer reads available for whole genome sequencing allows for better calling of copy number variations, rearrangements and other structural variations. In a report by Gilissen et al., whole genome sequencing was applied to patients with severe intellectual disability and their unaffected parents and reached a diagnostic yield of 42% (Gilissen C et al. Nature 2014) [41].
Despite the rapidly falling costs of sequencing, widespread application of whole genome sequencing to clinical diagnostics has been hampered by challenges in data analysis and relatively high costs of infrastructure needed to store, manage and analyze whole genome data. With the majority of causative variants identified so far in Mendelian disease occurring in coding regions, whole exome sequencing currently appears to be a more cost-effective approach and more practical alternative to whole genome sequencing (Teer JK, Mullikin JC. Hum Mol Genet.2010) [42]. Additionally, because variation in noncoding regions is less well understood than variation in the coding region, it is more difficult to predict which variants might be relevant to a trait of interest in whole genome datasets.
References
Hwang, S., Kim, E., Lee, I., Marcotte, E.M.: Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci. Rep. 5, 17875 (2015)
Langmead, B., Salzberg, S.L.: Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9(4), 357–359 (2012)
Van der Auwera, G.A., Carneiro, M.O., Hartl, C., et al.: From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 43, 11.10.11–33 (2013)
Li, H.: A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27(21), 2987–2993 (2011)
Challis, D., Yu, J., Evani, U.S., et al.: An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinf. 13, 8 (2012)
Rimmer, A., Phan, H., Mathieson, I., et al.: Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 46(8), 912–918 (2014)
Yang, H., Wang, K.: Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 10(10), 1556–1566 (2015)
Cingolani, P., Platts, A., Wang le, L., et al.: A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6(2), 80–92 (2012)
McLaren, W., Pritchard, B., Rios, D., Chen, Y., Flicek, P., Cunningham, F.: Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 26(16), 2069–2070 (2010)
McCarthy, D.J., Humburg, P., Kanapin, A., et al.: Choice of transcripts and software has a large effect on variant annotation. Genome Med. 6(3), 26 (2014)
Amarasinghe, K.C., Li, J., Halgamuge, S.K.: CoNVEX: copy number variation estimation in exome sequencing data using HMM. BMC Bioinf. 14(Suppl 2), S2 (2013)
Krumm, N., Sudmant, P.H., Ko, A., et al.: Copy number variation detection and genotyping from exome sequence data. Genome Res. 22(8), 1525–1532 (2012)
Fromer, M., Moran, J.L., Chambert, K., et al.: Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 91(4), 597–607 (2012)
Richards, S., Aziz, N., Bale, S., et al.: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17(5), 405–424 (2015)
Amendola, L.M., Jarvik, G.P., Leo, M.C., et al.: Performance of ACMG-AMP variant-interpretation guidelines among nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet. 98(6), 1067–1076 (2016)
Gulley, M.L., Braziel, R.M., Halling, K.C., et al.: Clinical laboratory reports in molecular pathology. Arch. Pathol. Lab. Med. 131(6), 852–863 (2007)
Dorschner, M.O., Amendola, L.M., Shirts, B.H., et al.: Refining the structure and content of clinical genomic reports. Am. J. Med. Genet. C Semin. Med. Genet. 166C(1), 85–92 (2014)
Kumar, P., Henikoff, S., Ng, P.C.: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4(7), 1073–1081 (2009)
Adzhubei, I.A., Schmidt, S., Peshkin, L., et al.: A method and server for predicting damaging missense mutations. Nat. Methods. 7(4), 248–249 (2010)
Yang, Y., Muzny, D.M., Xia, F., et al.: Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 312(18), 1870–1879 (2014)
Tarailo-Graovac, M., Shyr, C., Ross, C.J., et al.: Exome sequencing and the Management of Neurometabolic Disorders. N. Engl. J. Med. 374(23), 2246–2255 (2016)
Green, R.C., Berg, J.S., Grody, W.W., et al.: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 15(7), 565–574 (2013)
Kalia, S.S., Adelman, K., Bale, S.J., et al.: Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 19(2), 249–255 (2016)
Smith, L.A., Douglas, J., Braxton, A.A., Kramer, K.: Reporting incidental findings in clinical whole exome sequencing: incorporation of the 2013 ACMG recommendations into current practices of genetic counseling. J. Genet. Couns. 24(4), 654–662 (2015)
Shashi, V., McConkie-Rosell, A., Rosell, B., et al.: The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet. Med. 16(2), 176–182 (2014)
Lee, H., Deignan, J.L., Dorrani, N., et al.: Clinical exome sequencing for genetic identification of rare mendelian disorders. JAMA. 312(18), 1880–1887 (2014)
Sawyer, S.L., Hartley, T., Dyment, D.A., et al.: Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin. Genet. 89(3), 275–284 (2016)
Farwell, K.D., Shahmirzadi, L., El-Khechen, D., et al.: Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 17(7), 578–586 (2015)
Valencia, C.A., Husami, A., Holle, J., et al.: Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: a pediatric center’s experience. Front Pediatr. 3, 67 (2015)
Thevenon, J., Duffourd, Y., Masurel-Paulet, A., et al.: Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin. Genet. 89(6), 700–707 (2016)
Biesecker, L.G., Green, R.C.: Diagnostic clinical genome and exome sequencing. N. Engl. J. Med. 370(25), 2418–2425 (2014)
Carss, K.J., Hillman, S.C., Parthiban, V., et al.: Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum. Mol. Genet. 23(12), 3269–3277 (2014)
Drury, S., Williams, H., Trump, N., et al.: Exome sequencing for prenatal diagnosis of fetuses with sonographic abnormalities. Prenat. Diagn. 35(10), 1010–1017 (2015)
Willig, L.K., Petrikin, J.E., Smith, L.D., et al.: Whole-genome sequencing for identification of mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir. Med. 3(5), 377–387 (2015)
Soden, S.E., Saunders, C.J., Willig, L.K., et al.: Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 6(265), 265ra168 (2014)
Xue, Y., Ankala, A., Wilcox, W.R., Hegde, M.R.: Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet. Med. 17(6), 444–451 (2015)
Gilissen, C., Hoischen, A., Brunner, H.G., Veltman, J.A.: Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 20(5), 490–497 (2012)
Klitzman, R., Appelbaum, P.S., Chung, W.: Return of secondary genomic findings vs patient autonomy: implications for medical care. JAMA. 310(4), 369–370 (2013)
Yang, Y., Muzny, D.M., Reid, J.G., et al.: Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 369(16), 1502–1511 (2013)
Lupski, J.R., Reid, J.G., Gonzaga-Jauregui, C., et al.: Whole-genome sequencing in a patient with Charcot-Marie-tooth neuropathy. N. Engl. J. Med. 362(13), 1181–1191 (2010)
Gilissen, C., Hehir-Kwa, J.Y., Thung, D.T., et al.: Genome sequencing identifies major causes of severe intellectual disability. Nature. 511(7509), 344–347 (2014)
Teer, J.K., Mullikin, J.C.: Exome sequencing: the sweet spot before whole genomes. Hum. Mol. Genet. 19(R2), R145–R151 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Chiang, T., Leduc, M., Tokita, M., Santiago-Sim, T., Yang, Y. (2017). Exome Sequencing in the Clinical Setting. In: Wong, LJ. (eds) Next Generation Sequencing Based Clinical Molecular Diagnosis of Human Genetic Disorders. Springer, Cham. https://doi.org/10.1007/978-3-319-56418-0_14
Download citation
DOI: https://doi.org/10.1007/978-3-319-56418-0_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-56416-6
Online ISBN: 978-3-319-56418-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)