Abstract
Hemp (2n = 20) is an economically important crop and good model species for plant sex studying. It has the XX/XY system of sex chromosomes in which Y is longer then X. Cytogenetic studies of hemp were evidently started in the early 20th century and are continuing today. The most modern karyotype of hemp is described by formula 8 m + 1sm (SAT) + Xm/Ym for male and 8 m + 1sm (SAT) + Xm for female plants. The number of widely used cytogenetic markers (for example 5S rDNA and 45S rDNA) and species specific probes were mapped to mitotic and meiotic hemp chromosomes. The history of formation of knowledge about hemp karyotype and modern results of cytogenetic studies are discoursed in detail in this chapter.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
18.1 Introduction
Hemp (Cannabis sativa L.) is an economically important crop and one of the earliest known cultivated plants (van der Werf et al. 1996; Struik et al. 2000; Truta et al. 2007; Shahzad 2012). Hemp has dioecious nature, though monoecious cultivars have been developed. Hemp has a diploid genome (2n = 20) with a karyotype composed of nine autosomes and a pair of sex chromosomes (X and Y) (Sakamoto et al. 1998; Divashuk et al. 2014).
Although the cytogenetic studies of hemp chromosomes started relatively early, from the cytogenetic point of view Cannabis sativa is relatively poorly studied in comparison with other economically important species. This is because of the small size of the chromosomes and inability of their identification based on morphology, the lack of heterochromatin banding (C-banding) and the limitation of the methods available to cytologists early to mid last century. The downturn of research interest to Cannabis also may have contributed by the restrictions on the hemp planting set in 50–60 years of XX century in a number of countries. In the second half of the XX century cytogenetic studies of Cannabis were not almost carried out, started again only at the end of 1990s. After the introduction of Non-Psychoactive Hemp varieties the interest to its cultivation as an alternative crop increased, as well as to the molecular genetic and cytogenetic studies.
C. sativa has a relatively small genome size (0.84–0.91 pg) (Kubešová et al. 2010). The estimated haploid genome size is 818 Mb for female plants and 843 Mb for males (Sakamoto et al. 1998). Hemp have a chromosome set of 2n = 20 with XX/XY chromosome system. The chromosomes are small, their size varies from 2.6 to 3.8 µm, and they can not all be distinguished by their length and centromere position (Fig. 18.1).
The main progress was made when draft genome was published (Van Bakel et al. 2011). This promoted the hemp karyotype analyses with molecular cytogenetic approaches. By now a modern karyotype has been developed (Divashuk et al. 2014). The chromosomes X and Y and 5 of 9 autosomes of haploid chromosomes set can be clearly identified by the application of molecular cytogenetic markers by FISH. Using modern molecular cytogenetic analysis the chromosomal constitution of monoecious cultivars has been studied (Razumova et al. 2016). However, there are still no physical and genetic maps, the karyotype needs more detailed analyses including the development of modern pachytene chromosome map. In addition, the exact role of the sex chromosomes in sex determination has still not been established. Cytogenetics is becoming an important complement that has bridged the gap between genetics and genomes studies.
In this chapter we examine the achievements that have been obtained using classical and molecular cytogenetics to analyze and exploit the Cannabis sativa genome.
18.2 Classical Cytogenetic Study of Hemp Chromosomes
Cytogenetic studies of hemp were evidently started in the 1920s and it is difficult to tell who first described the hemp chromosomes. Yosito Sinotô in his review “On the Chromosome Number and the Unequal Pair of Chromosomes in Some Dioecious Plants” (1928) reported about research by Strasburger, Hirata, MacPhee and other scientists who had established that haploid set of hemp is equal to 10 (Sinotô 1928). In 1926, Breslavets (1926) described the polyploid cells (2n = 40) in hemp roots that had arisen by endomitosis. Tetraploid plants were obtained by a number of researchers (Breslavets 1932; Lindstrom 1939; Warmke and Blakeslee 1939; Nishiyama 1940), who noted fertility and vitality of these plants, the normal process of meiosis, with infrequent abnormalities in meiosis where formation of tetravalents and sexual bivalents XX, YY instead of the normal XY was found. All researchers described the hemp chromosomes as metacentric and had small size that did not allow its identification.
About the same time, with the establishment of the haploid number of chromosomes, studies on the mechanism of sex determination in hemp were started. McPhee (1924) gave a detailed description of the hemp meiosis stages, but morphologically could not identify the sex chromosomes, as well as Strasburger twenty years earlier (from McPhee 1924, Strasburger 1900).
At metaphase I of meiosis a pair of heteromorphic sex chromosomes were described by Sinotô (1928). Schaffner in the studies on sex determination in Cannabis on various environmental conditions (Schaffner 1919, 1921, 1923) insisted on epigamic mechanism of sex determination and directly denies the role of sex chromosomes: «The mere fact that sex determination and segregation usually do not at all coincide with fertilization of reduction in the higher plants and also not in most lower forms, and that such coincidence is confined to a comparatively few out of many types of sexual cycles, made it plain that those botanists who were seeking an explanation of sex determination and sex segregation in a Mendelian formula of homozygous and heterozygous chromosome or factor constitutions were not only following a delusion, but attempting to establish an hypothesis of sexuality that would result in nothing except a contradiction of the most evident phenomena» (Schaffner 1923, p. 225). However, Hirata (1927) did not agree with him and reported about the XY-mechanism of determination of sex in hemp. In his study, he found a pair of unequal chromosomes in the meiotic preparations of one of the two studied cultivars. Breslavets (1932) identified heteromorphic sex chromosomes: large chromosome was referred to as X and small chromosome as Y. Referring to the discrepancy between the results obtained in different studies, some authors suggested that Hemp varieties can differ in the presence or absence of the sex chromosomes in the karyotype (Hoffmann 1938). Elizabeth L. Mackay (1939) refuted these data and reported that XY chromosomes appeared to be present in all male plants, and the Y-chromosome is still regarded as the smallest chromosome in the karyotype. Yamada in (1943) also reported about the unequal pair of chromosomes (Yamada 1943). Lindsay, in his review of the sex chromosomes in plants, discussed sex determination mechanism in Cannabis. He believed that, despite the appropriateness of using the term “sex chromosomes”, they are not the chromosomes that directly affect the formation of sex, but only chromosomes that carry some of the factors that can contribute to the formation of male and female flowers under the influence of certain environmental conditions (Lindsay 1930).
The role of Y chromosome is not clear, considering the experiments on polyploid plants (Warmke and Davidson 1944), in which phenotypically female plants were shown to have XXY and XXXY genotypes. In addition, in hemp spontaneous monoecious forms of plants occur. Although monoecious forms are rare and are more likely to be an exception, it was shown that the transformation of one form to another is achieved by the influence of various kinds of biologically active substances, as well as some chemical compounds (e.g., carbon monoxide II – CO). By now, breeders have developed the monoecious hemp varieties, but monoecity is not stable due to contamination by cross hybridization with monoecious plants, and all such varieties have a tendency to return to dioecy.
Hoffman (1952) suggested that plants of either sex may have karyotype XX, XY, and even YY. Therefore, a plant with a male habit and female flowers may have the XX sex chromosomes. Like the Westergaard (1958), Hoffman thought that the Y chromosome is less active than X. However, while Westergaard supported the balance theory of sex determination in hemp, Hoffman suggested multifactor hereditary mechanisms. Based on a series of crosses between monoecious and dioecious hemp plants Dierks and von Sengbusch assumed the mortality of the YY genotype (Dierks and von Sengbusch 1967).
First karyotype and pachytene map was developed only in 1964 by Menzel (1964) This study was done on the monoecious hemp varietie ‘Kentucky’ and several dioecious plants of unknown origin. The author failed to identify the sex bivalent at the pachytene stage, although it was well visualized at the diakinesis stage. At the same time, all the dioecious male habit plants with the male flowers had the XY genotype, while female and monoecious—XX, regardless of what kind of flowers they carried.
At the end of 20-th century attempts to develop the hemp karyotype were undertaken by several research groups and the most significant results were obtained by Sakamoto et al. (1998) and Srivastava et al. (1999). Sakamoto et al. (1998) developed a karyotype where the Y chromosome is described as the longest chromosome with heterochromatic arm that is intensively stained by Giemsa and shows bright fluorescence when stained with DAPI. The authors also suggested that the Y chromosome carries satellite that was not confirmed by later research (Srivastava et al. 1999; Divashuk et al. 2014). Srivastava et al. (1999) analyzed metaphase chromosomes of Cannabis sativa L. var. indica (Lam.) and suggested the presence of satellites on one pair of autosomes only (chromosome 3). In this paper, it was also noted that the sex chromosomes are submetacentric, with Y longer than X, and autosomes (with exception for submetacentric chromosome 1) are metacentric and difficult to distinguish from each other. It seems that all the contradictions relating to the sex chromosomes of Cannabis sativa can be attributed to a small and very similar size of the chromosomes in its karyotype, and the lack of classical cytogenetic markers for its identification.
18.3 Molecular Cytogenetics in Hemp
The development of fluorescent in situ hybridization (FISH) techniques has made a huge contribution to karyotyping and analyzing genome organization in many plant and animal species (De Jong et al. 1999; Figueroa and Bass 2010; Iovene et al. 2011). Molecular cytogenetic maps of cultivated plants have great practical and research value. Fluorescent in situ hybridization techniques have been used mainly for mapping repetitive DNA sequences, multicopy gene families and, recently, for mapping of low or single-copy sequences (Heslop-Harrison and Schwarzacher 2011; Kirov et al. 2014). FISH offer new opportunity not only for reliable chromosome identification, structural and functional chromosome analyses, but also for evaluation physical genome distances and the integration of genetic and physical maps (Kirov et al. 2015).
The application of FISH analyses to the study of the hemp chromosomes yielded new data on the features of its karyotype. Firstly, FISH on hemp was used by Sakamoto et al. (2000) and Sakamoto et al. (2005). The male associated DNA sequences (MADC1, MADC3, MADC4) were used as probes in FISH experiments. It was demonstrated that MADC3 and MADC4 probes show more intense fluorescence signal on chromosome Y. Furthermore, a signal of the MADC4 probe with similar intensity was detected on one pair of autosomes, so that, according to the authors, it can be used as a cytogenetic marker for this pair. On the remaining chromosomes the MADC3 and MADC4 probes showed equally dispersed signals typical for retroelements. In contrast to the previous two probes, the MADC1 probe showed signal in the terminal part of the long arm of chromosome Y only. Because the MADC1 sequence was classified as the LINE-like retroelements, the data on its FISH mapping give reason to assume that the formation of the Y chromosome was accompanied by the accumulation of this sub-type of retroelements. However, it should be noted that the low frequency of MADC1 signals (probably due to the small size of the locus) does not make it possible to use it as a reliable cytogenetic marker. Several male associated DNA sequences (SCARopa08, C11Komp, C11Seq, AAT_330Komp, 330_GW) were used in other FISH experiments (Riedel 2005). In these experiments, the C11Komp and 330_GW probes showed uniform distribution of the signals on all chromosomes, while SCARopa08 and AAT_330Komp probes showed more intense signals on one chromosome (probably, on Y). The S11Seq probe was localized on one chromosome pair. Furthermore, Riedel in this study localized 5S rDNA and 45S rDNA to different chromosomes pairs. Unfortunately, in this study neither karyotyping nor chromosome identification was carried out.
The first modern hemp karyotype was developed by Divashuk et al. (2014) using FISH with a number of DNA probes as cytogenetic markers. The karyotype formula is 2n = 20 with 8 m + 1sm (SAT) + Xm/Ym for male and 8 m + 1sm (SAT) + Xm for female plants. The 5S rDNA signal was localized to a single chromosome pair (the middle part of the short arm of chromosome 8). The 45S rDNA signal was detected on the other chromosome pair (the terminal part of the short arm of submetacentric chromosome 9). Besides, the subtelomeric repeat CS-1 has also been localized. This repeat we isolated using genome sequence data (Van Bakel et al. 2011). The CS-1 signals were localized to both arms of chromosomes 1, 2, 3, 5, 6, 7, 8, and X. In chromosomes 4 and chromosome Y the CS-1 signal was observed only on the short arm, and in chromosome 9 on long arm. As in the earlier studies (Sakamoto et al. 2005; Sakamoto et al. 1998; Sakamoto et al. 2000), it was detected that the Y chromosome is highly heterochromatic and intensely stained by DAPI. In contrast to Sakamoto et al. 2000 opinion, it was proved that the Y chromosome does not carry satellite. Furthermore, Divashuk et al. (2014) statistically confirmed the statement of Srivastava et al. (1999) regarding that the Y chromosome is longer than X and at the same time disproved that metacentric chromosome 3 carries satellite, claiming that it is submetacentric chromosome 9, instead. The application of the CS-1 probe in FISH experiments with meiotic chromosomes at metaphase I stage enabled Divashuk et al. (2014) to show the orientation of the X and Y chromosomes in the sex bivalent and location of pseudoautosomic region (PAR) (Fig. 18.2). It was found that the PAR is located on non heterochromatic part of short arm of chromosome Y and colocalized with CS-1. Recently the disposition of PAR was confirmed using the self-GISH method (Razumova et al. in preparation).
The CS-1 probe was used in FISH study of the sex chromosome status of monoecious and dioecious hemp cultivars (Razumova et al. 2016). It was conclusively proved the absence of the Y chromosome in the studied karyotypes of monoecious cultivars ‘Gentus’, ‘Diana’, ‘Ingreda’, ‘Margo’, ‘Tzivilsky Skorospeliy’ and ‘Rigs’ (Chuvashian Research Institute of Agriculture, Tsivilsk, Russia) and ‘Maria’, ‘Kubanka’ (P.P. Lukyanenko Krasnodar Research Institute of Agriculture, Krasnodar, Russia). The high level of inter- and intracultivar karyotype variations was shown. In dioecious and monoecious cultivars, 10 cytotypes were identified differing by the presence of the Y chromosome and the distribution of CS-1 signals on chromosomes 2 and 9.
The data of hemp genome sequencing project (Van Bakel et al. 2011) can be used to develop more chromosome specific molecular cytogenetic markers. We isolated several sequences that are suitable for cytogenetic analyses. For example, the CS-154 tandem repeat shows chromosome specific location (Fig. 18.3a). The single copy DNA sequences such as genes can be also physically mapped on hemp chromosomes. We used single copy fraction of scaffold 20878_8 to map it on hemp chromosomes. This sequence showed clear signal on a single pair of homologous chromosomes (Fig. 18.3b). The resolution of physical mapping of single copy sequences on mitotic chromosomes is limited due to their high compactization. It is difficult to establish the order of sequences arrangement on chromosome which are located adjacent to each other.
At pachytene stage in prophase I the meiotic chromosomes as much as 15 times less condensed than mitotic metaphase chromosomes (De Jong et al. 1999; Zhong et al. 1996). Often in species with small chromosomes only the use of pachytene chromosomes make it possible to physically associate genetic linkage groups with particular chromosomes (Zhang et al. 2010). This make pachytene chromosomes attractive for molecular cytogenetic analysis of plant genomes organization. In many plants as well as in hemp, the pachytene chromosomes are characterized by well-defined structure and can be accurately identified based on their morphology. For Cannabis sativa pachytene chromosome map was created by Menzel (1964). In our studies, we use hemp pachytene chromosomes for FISH mapping of different DNA sequences. The DAPI (4’,6-diamidino-2-phenylindole) stained chromosomes can be identified (Fig. 18.4a). The satellite chromosome 9 can be identified not only by the fluorescent signal of 45S rDNA, but also by a pronounced DAPI positive area near satellite (Fig. 18.4a). The FISH results of 5S rDNA hybridization to pachytene chromosomes is shown in Fig. 18.4b. The signal is localized to chromosome 8.
In general, it can be noted that pachytene chromosomes of hemp differ morphologically by well distinguishable heterochromatin and using DAPI-staining in conjunction with cytogenetic markers make it possible to develop a modern pachytene map, which, in its turn, opens up opportunities for the physical mapping of genes.
18.4 Conclusion
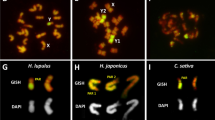
In this chapter, an attempt has been made to provide an overview of the role of classical and molecular cytogenetics in genome characterization of Cannabis sativa L. By the FISH technology we demonstrated that different types of DNA such as repetitive sequences and low- and single copy genes can be mapped on hemp chromosomes. Also, hemp is the promising object to study sex chromosome organization and evolution. It is therefore of interest to find out whether the C. sativa X and Y are homologous to X and/or Y of the related species (Humulus lupulus and Humulus japonicus) and it is a task for the future research. Our results of FISH experiments on hemp open new window to assist its full genome assembly.
References
Breslavets L (1926) Polyploide mitosen bei Cannabis sativa L. Ber Deutsch Bot Ges 44:498–502
Breslavets L (1932) Polyploide Mitosen bei Cannabis sativa L. II. Planta 17(3):644–649
De Jong JH, Fransz P, Zabel P (1999) High resolution FISH in plants–techniques and applications. Trends Plant Sci 4(7):258–263
Dierks W, von Sengbusch R (1967) Studium der Vererbung des Geschlechts und des Wuchstyps beim Hanf. Der Züchter 37(1):12–15
Divashuk MG, Alexandrov OS, Razumova OV, Kirov IV, Karlov GI (2014) Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS ONE 9(1):e85118
Figueroa DM, Bass HW (2010) A historical and modern perspective on plant cytogenetics. Briefings in Functional Genomics, elp058
Heslop-Harrison JS, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Plant J 66(1):18–33
Hirata K (1927) Sex determination in hemp (Cannabis sativa L.). J Genet 19(1):65–79
Hoffmann W (1938) Das Geschlechtsproblem des Hanfes in der Ziichtung. Zeitschr. Zticht. Reihe A. Pflanzenzticht. 22:453–468
Hoffmann W (1952) Die vererbung der geschlechtsformen des hanfes (Canabis sativa L.) Der Züchter 22(4−5):147–158
Iovene M, Cavagnaro PF, Senalik D, Buell CR, Jiang J, Simon PW (2011) Comparative FISH mapping of Daucus species (Apiaceae family). Chromosome Res 19(4):493–506
Kirov I, Van Laere K, De Riek J, De Keyser E, Van Roy N, Khrustaleva L (2014) Anchoring linkage groups of the Rosa genetic map to physical chromosomes with Tyramide-FISH and EST-SNP markers. PLoS ONE 9(4):e95793
Kirov IV, Van Laere K, Khrustaleva LI (2015) High resolution physical mapping of single gene fragments on pachytene chromosome 4 and 7 of Rosa. BMC Genet 16(1):1
Kubešová M, Moravcová L, Suda J, Jarošík V, Pyšek P (2010) Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia 82:81–96
Lindsay RH (1930) The chromosomes of some dioecious angiosperms. Am J Bot: 152–174
Lindstrom EW (1939) Abstracts of papers presented at the 1938 meetings of the Genetics Society of America. Genetics, 24(1):65–111 (p 88)
Mackay EL (1939) Sex chromosomes of Cannabis sativa. Am J Bot: 707–708
McPhee HC (1924) Meiotic cytokinesis of Cannabis. Bot Gaz: 335–341
Menzel MY (1964) Meiotic chromosomes of monoecious Kentucky hemp (Cannabis sativa). Bull Torrey Bot Club: 193–205
Nishiyama I (1940) Studies on artificially produced polyploid plants. III. Meiosis in tetraploid hemp. Bot Zool 8:47–52
Razumova OV, Alexandrov OS, Divashuk MG, Sukhorada TI, Karlov GI (2016) Molecular cytogenetic analysis of monoecious hemp (Cannabis sativa L.) cultivars reveals its karyotype variations and sex chromosomes constitution. Protoplasma 253(3):895–901
Riedel M (2005) In situ Hybridisierung an Hanf (Cannabis sativa L.) Chromosomen (Doctoral dissertation)
Sakamoto K, Akiyama Y, Fukui K, Kamada H, Satoh S (1998) Characterization; genome sizes and morphology of sex chromosomes in hemp (Cannabis sativa L.). Cytologia 63:459–464. doi:10.1508/cytologia.63.459
Sakamoto K, Ohmido N, Fukui K, Kamada H, Satoh S (2000) Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol Biol 44(6):723–732
Sakamoto K, Abe T, Matsuyama T, Yoshida S, Ohmido N, Fukui K, Satoh S (2005) RAPD markers encoding retrotransposable elements are linked to the male sex in Cannabis sativa L. Genome 48(5):931–936
Schaffner JH (1919) Complete reversal of sex in hemp. Science: 311–312
Schaffner JH (1921) Influence of environment on sexual expression in hemp. Bot Gaz: 197–219
Schaffner JH (1923) The time of sex determination in plants. Ohio J Sci v23 n5 (September–October 1923), 225–240
Shahzad A (2012) Hemp fiber and its composites—a review. J Compos Mater 46:973–986
Sinotô Y (1928) On the chromosome number and the unequal pair of chromosomes in some dioecious plants. Proc Imperial Acad 4(4):175–177
Srivastava P, Srivastava S, Verma MK, Mishra SK (1999) Karyological Studies in Root-Tip Cells of Cannabis sativa var. indica. Cytologia 64(4):435–440
Struik PC, Amaducci S, Bullard MJ, Stutterheim NC, Venturi G, Cromack HTH (2000) Agronomy of fibre hemp (Cannabis sativa L.) in Europe. Ind Crop Prod 11:107–118
Truta E, Olteanu N, Surdu S, Zamfirache MM, Oprica L (2007) Some aspects of sex determinism in hemp. Analele Stiintificeale Universitatii ‘‘Alexandru Ioan Cuza’’, Sectiunea Genetica si Biologie Moleculara VIII:31–39
Van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE (2011) The draft genome and transcriptome of Cannabis sativa. Genome Biol 12:R102. doi:10.1186/gb-2011-12-10-r102
van der Werf HMG, Mathijssen EWJM, Haverkort AJ (1996) The potential of hemp (Cannabis sativa L.) for sustainable fibre production: a crop physiological appraisal. Ann Appl Biol 129:109–123
Warmke HE, Blakeslee AF (1939) Carnegie Institution of Washington, Department of Genetics, Cold Spring Harbor, N.Y. Effect of polyploidy upon the sex mechanism in dioecious plants
Warmke HE, Davidson H (1944) Polyploidy investigations. Polyploid investigation. Yearb. Carnegie Institution of Washington, 43:135–139
Westergaard M (1958) The mechanism of sex determination in dioecious flowering plants. Advances in Genetics 9:217–281
Yamada I (1943) The sex chromosome of Cannabis sativa L. Seiken Ziho. 2:64–68
Zhang W, Wai CM, Ming R, Yu Q, Jiang J (2010) Integration of genetic and cytological maps and development of a pachytene chromosome-based karyotype in papaya. Trop Plant Biol 3(3):166–170
Zhong XB, de Jong JH, Zabel P (1996) Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res 4(1):24–28
Acknowledgements
The reported study was funded by RFBR according to the research project # 15-04-06244a.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Karlov, G.I., Razumova, O.V., Alexandrov, O.S., Divashuk, M.G., Kroupin, P.Y. (2017). Classical and Molecular Cytogenetics of Cannabis Sativa L.. In: Chandra, S., Lata, H., ElSohly, M. (eds) Cannabis sativa L. - Botany and Biotechnology. Springer, Cham. https://doi.org/10.1007/978-3-319-54564-6_18
Download citation
DOI: https://doi.org/10.1007/978-3-319-54564-6_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-54563-9
Online ISBN: 978-3-319-54564-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)