Abstract
This study aimed to determine the accuracy to which prone and supine MRI can be registered using biomechanical models, to help clinicians with pre-operative breast cancer treatment planning. A framework was developed for constructing a biomechanical model of the breast from diagnostic prone MRI. These images were then warped using a prone-to-supine mechanics simulation to generate an initial estimate of the tissue displacement in the supine position. This initial estimate of tissue motion was subsequently used for registering the prone MRI to pre-operative supine MRI using an image intensity-based non-rigid registration algorithm. The framework was demonstrated using MR images from two volunteers. The results showed that a two parameter breast model provided good initial estimates of tissue displacement for registering the prone and supine MR images, with registration errors less than 5 mm for mean tissue displacement magnitudes of up to 61.8 mm between the prone and supine positions.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Breast cancer is the leading cause of cancer-related death in females, affecting 1 in every 10 women worldwide. Breast conserving therapy (BCT) is the most common procedure used for treating early stage invasive breast cancers, and involves localized excision of tumorous lesions followed by radiotherapy. Clinical imaging modalities used for diagnosing the disease (e.g., MRI) are acquired with the patient positioned differently to that assumed during the actual treatment procedures. Since such procedures are not performed under image guidance, localization of tumors is challenging, especially when the stiffness of the tumor is similar to the stiffness of the surrounding tissue. This represents a significant challenge for clinicians, with the majority of studies in the literature reporting incomplete excision of tumors in 20–40% of the patients who underwent BCT [1]. This study therefore aimed to register regions of interest between diagnostic prone MRI and pre-operative supine MRI to help clinicians localize tumors during treatment procedures.
Such registration problems have typically been solved using image intensity-based non-rigid registration algorithms in a number of different organs [2]. In such approaches, a transformation is applied to one image in order to best match image intensity values in another image, using nonlinear optimization [3, 4]. However, these methods have proven to be unreliable when attempting to account for large changes in tissue motion in highly deformable organs, such as that observed in the breast between the prone and supine positions. The failure of these approaches is usually due to poor registration initialization, which can cause the locally convergent optimization algorithms used for maximizing voxel correspondence to diverge, especially if physically based constraints are not used to help constrain the problem [5].
Previous studies have therefore aimed to develop biomechanical models of the breast to provide an estimate of the tissue displacement observed between the prone and supine MRI for initializing non-rigid registration algorithms [6]. However, recent studies have observed relatively large errors in tissue displacement estimates from the biomechanical models. For example, [7] observed mean model errors between 11.5 and 39.2 mm when simulating prone to supine breast deformation, even with a model that accounted for the mechanical behavior of multiple tissues in the breast (skin, fat, fibroglandular tissues), and the pectoral muscle, on which these breast tissues sit.
These large errors may be due to the simplified frictionless boundary conditions that were applied on the posterior surface of model during the simulation. These boundary conditions were used for approximating sliding of the pectoral muscle over the ribcage that occurs due to relative changes in the position of the arm between the prone and supine positions. However, frictionless boundary conditions may not be suitable, as the extent to which the pectoral muscle slides over the breast tissues is then largely dependent on the stiffness of the breast tissues (with stiffer parameters resulting in less sliding and less stiff parameters resulting in a greater extent of sliding). In this study we aimed to determine the accuracy to which prone and supine MRI could be registered using an initial estimate of tissue displacement from a biomechanical model that directly prescribes the observed motion of the pectoral muscle between the prone and supine positions.
Section 2 describes the methodology used for solving prone-to-supine breast mechanics, and using this initial estimate for non-rigidly registering the prone and supine MRI. Section 3 describes the results of the registration procedure, and provides an analysis of the accuracy of the initial estimate of tissue displacement provided by the mechanics simulation. Potential approaches for improving the accuracy of the mechanics simulations are then discussed in Section 4.
2 Methodology
2.1 Modeling Breast Anatomy
Prone and supine MR images were acquired from two volunteers using a Siemens 1.5 T Magnetom Avanto MRI system. T2-weighted imaging sequences were acquired for each volunteer with an axial (transversal) slice orientation, and a 0.684 mm × 0.684 mm × 2.5 mm voxel size.
Subject-specific finite element (FE) models of the left breast were created from the prone MR images of each volunteer considered in this study. In each model, separate FE meshes were created to represent the geometry of the breast tissue and pectoral muscle. The meshes were created by fitting cubic Hermite basis functions to manually segmented skin, muscle, and chest wall boundaries seen in the prone MRI (denoted as Γskin, Γmuscle, and Γchest−wall, respectively), using an iterative closest point algorithm. The skin and chest wall root-mean-squared errors (RMSE) following the fitting were 1.19 mm, 0.98 mm, and 1.20 mm, respectively, for volunteer 1 and 0.74 mm, 1.02 mm, 1.17 mm, respectively, for volunteer 2. The fitted meshes are shown in Fig. 1.
2.2 Modeling Breast Mechanics
Finite elasticity theory was used to simulate the large deformations observed during prone-to-supine breast repositioning [8]. These governing equations were solved using the finite element method in the OpenCMISS computational modeling software package [9]. The mechanical responses of the breast tissues were assumed to be isotropic and were modeled using an ideally incompressible, hyperelastic neo-Hookean constitutive relation. A linear Lagrange hydrostatic pressure field was used to enforce incompressibility of the breast tissues.
The breast and muscle tissues were each described by a different neo-Hookean stiffness parameter, namely θ breast (a homogeneous representation of the stiffness of the breast tissues, composed mainly of adipose, fibroglandular, and skin tissues) and θ muscle (representing the stiffness of the pectoral muscles). These parameters were assigned to their respective mesh elements.
Since the breast was imaged under gravity loading conditions, either the regional stress-state of the breast, or its stress-free reference configuration, is required for accurate mechanics simulations. The importance of determining this stress-free reference configuration was highlighted for the breast in [10]. In this study, a stress-free reference configuration was numerically determined, by implementing the algorithm described in [11], in the OpenCMISS numerical software package. Once the reference configuration was identified, the breast could be re-orientated to simulate the supine position.
2.2.1 Boundary and Loading Conditions
An important aspect of simulating breast tissue movement involves accounting for any relative motion between the pectoral muscles and the chest wall. This relative motion arises due to a relative change in position of the shoulder joint and the arm, to which the pectoral muscles attach, when an individual is repositioned. Previous studies have attempted to indirectly account for this sliding behavior by introducing frictionless [7] or frictional [12] contact constraints, at the muscle and chest wall interface. However, this assumes that the observed degree of sliding depends mainly on the stiffness of the breast tissues and not the actual motion of the shoulder joint and arm (which is dependent on the orientation in which the subject is positioned during imaging). Such an assumption can therefore introduce large modeling errors.
The lateral insertion points of the pectoralis major muscle or the shoulder joint were not visible within the field of view of either the prone or supine MR images of each volunteer. Therefore, the amount of sliding due to shoulder and arm motion could only be approximated in this study. This was achieved by first aligning the anterior sternocostal articulation junctions in both the prone and supine images. The axillary boundary of the pectoral muscle (defined by the dashed blue line in Fig. 1) was then identified in both the prone and supine MR images, and used to estimate the degree of sliding. Kinematic constraints were then applied on the posterior surface of the muscle to enforce the observed sliding motion along the curvature of the chest wall. A linear gradient was used to determine the amount of sliding at any point along this posterior surface, from a fixed edge at the sternal boundary of the model (representing the medial insertion point of the pectoralis major muscles) to the observed degree of sliding at the axillary boundary of the pectoral muscle.
Gravity loading was applied as a body-force acting in the direction determined from the alignment of the prone and supine MR images. The density of the breast tissue was defined as the volume weighted combination of the adipose (928 kg/m3) and fibroglandular (1035 kg/m3) tissue densities [13]. A density of 1060 kg/m3 [14] was used for the muscle tissue.
2.2.2 Mesh Convergence Analysis
An FE displacement convergence analysis was performed to ensure the simulation results were mesh resolution independent. Six geometric points were randomly embedded within the fibroglandular region of the prone breast model. The simulated positions of these material points were compared with successive refinements to the mesh. This convergence analysis was performed using the breast geometry of volunteer 2, with neo-Hookean stiffness parameters θ muscle = 106 Pa and θ breast = 190 Pa. From this analysis, a 7756 degree of freedom (DOF) mesh resolution was chosen for generating the results in the remainder of the study, as it produced a 0.14 mm maximum displacement difference of the embedded points compared with a 14,404 DOF mesh.
2.3 Registering Prone and Supine MRI
As described in Sect. 1, image intensity-based registration techniques alone are incapable of successfully registering the large degree of breast tissue deformation observed between the prone and supine MR images, unless a suitable initial estimate of the tissue motion is provided. In this study, the nonlinear transformation, T m (θ), obtained from simulating prone-to-supine repositioning using the FE mechanics model (where θ are the constitutive parameters of the FE model) was used as this initial estimate. This was achieved by embedding prone MRI pixels into the prone model, allowing each to be assigned a unique material point. These material points were subsequently transformed using T m , and re-sampled to generate a model-simulated supine MRI. The model-simulated supine MRI and the independently acquired supine MRI were then registered using image intensity-based registration techniques. This procedure defined another nonlinear and invertible transformation, T r , which maps material points between these two images. The total tissue motion was therefore described by T = T r ·T m .
2.3.1 Implementation
In this study, the registration framework developed by [3] was used to determine the transformation T r using a NMI similarity metric. This was performed using the IRTK software package [3, 4].
The IRTK registration procedure involved both rigid and non-rigid components. The rigid component of the registration was parameterized using a 12 DOF, affine transformation (which allowed translation, rotation, shear, and anisotropic scaling). The non-rigid component of T r was represented by a multi-resolution free-form-deformation (FFD), parameterized using a lattice of control point vectors, to allow linear interpolation of the MR images. The NMI similarity metric was maximized with a conjugate gradient descent optimization algorithm to determine the optimal parameters of the rigid and non-rigid components of T r . The muscle tissue was not registered during this procedure due to the low signal quality and lack of visible features within the tissue.
2.3.2 Verifying Implementation Accuracy
The accuracy of the registration framework was verified by applying a known deformation field from a mechanics solution to the prone MR image [15]. The extent to which the applied deformation could be recovered using the registration framework was then analyzed. This approach was also used to tune the numerical parameters of the registration framework (such as the spacing between control points and number of registration iterations) to ensure the framework was suitable for registering the MR images used in this study. The results of the MRI registration verification tests indicated that the framework was capable of recovering the simulated deformation with a 3D RMSE of 0.45 mm.
2.4 Identifying Subject-Specific Mechanical Properties
The registered displacements (described by T r ) can be used to indicate the discrepancy between the model-simulated supine MRI and the independently acquired supine MRI. The constitutive parameters of the model (θ) could therefore be optimized to minimize these discrepancies. The optimal constitutive parameters (θ *) identified from this procedure therefore allow the breast model to provide the best estimate of tissue motion in the supine position, for optimally registering the model-simulated supine MRI and the independently acquired supine MRI.
A nonlinear, least squares optimization procedure was implemented to minimize the mean-squared registered displacements, Φ MSE , and thus to determine subject specific θ * as shown below:
where x i denotes the geometric location of the ith breast tissue voxel in the model-simulated supine MRI (\( \overset{\sim }{{\boldsymbol{I}}_{\boldsymbol{s}}}\left(\boldsymbol{\theta}, {\boldsymbol{x}}_{\boldsymbol{i}}\right) \)) and its corresponding registered location in the independently acquired supine MRI (\( {\boldsymbol{T}}_{\boldsymbol{r}}(\overset{\sim }{{\boldsymbol{I}}_{\boldsymbol{s}}}\left(\boldsymbol{\theta}, {\boldsymbol{x}}_{\boldsymbol{i}}\right),{\boldsymbol{I}}_{\boldsymbol{s}}) \)). While the registration was performed over the entire breast tissue region, only the pixels representing the fibroglandular tissue were used for evaluating Φ MSE. This was because many features were present in the fibroglandular tissue region, compared with the relatively featureless adipose tissue.
The optimization procedure was implemented in Matlab using the lsqnonlin optimization function. Box constraints, defining upper and lower bounds on the parameters, were manually defined during the optimization procedure to describe a feasible range of parameters.
3 Results
The optimal constitutive parameters (θ *) that minimized the registered displacements are shown in Table 1. These results indicated that the breast tissue of volunteer 2 was stiffer than volunteer 1. However, similar muscle stiffness parameters were identified for both volunteers. These parameters are similar to those obtained in a similar study that modeled only the breast and muscle tissues (θ breast * = 50 Pa, and θ muscle * = 260 Pa) [16].
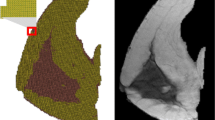
The supine shape determined using the biomechanical model with the optimal set of model constitutive parameters (θ *) is shown in Fig. 2a. Figure 3a shows an example of axial slice of the model-simulated supine MRI obtained from transforming the prone MRI using T m (θ *). Figure 3b–f shows the results of the procedure used to register the model-simulated supine MRI and the independently acquired supine MRI. The differences in image intensities before and after registration are shown in Fig. 3e, f, respectively. These results showed good agreement between the registration-simulated supine MRI and the independently acquired supine MRI. A quantitative comparison between these images was also performed by comparing distances between the centroid of eight manually segmented landmarks identified in these images that were distributed throughout the breast tissue. The results of this analysis are shown in Table 2, and also indicated good agreement between the images.
The simulated supine shape of the breast from the mechanics solution solved using the optimal constitutive parameters (θ *). Each red arrow indicates the error between the model-simulated nipple position (indicated by the green sphere) and its location identified from the independently acquired supine MRI
MRI registration results for a transverse section of the FE model, simulated with the optimal tissue parameters (θ *). The FE model-simulated supine MRI and independently acquired supine MRI are shown in (a) and (b), respectively. The registered displacements described by T r are indicated by the yellow arrows in (c), and the resulting registration-simulated MRI is shown in (d). The differences in image intensities before registration and after registration are shown in (e) and (f), respectively
The displacements described by T r were then used to provide an indication of the accuracy of the models. These displacements are described in Table 3. The magnitudes of these displacements are also shown on the simulated supine geometry in Fig. 4 to indicate the distribution of the errors within the breast tissue.
The results in Fig. 4 indicate large 3D registered displacement magnitudes were observed near the nipple position of the model. This was identified to be due to the model over-estimating the tissue motion near the nipple in both volunteers (as seen in Figs. 2 and 3c) in the axial (x–y) plane. The results in Table 3 also indicate that the registered displacements were the largest along the caudal-cranial direction (z-axis). Registered displacements of up to 12 mm were observed towards the caudal boundary (−z) in volunteer 1, and up to 23 mm towards the cranial boundary (+z) in volunteer 2.
4 Discussion
The results described in this study indicated that the two parameter models with the improved boundary conditions provided a good initial estimate of the tissue motion for registering the prone and supine MRI. These models showed mean modeling errors (identified from MRI registration) of 9.1 and 19.4 mm for volunteer 1 and 2, respectively, compared with mean modeling errors between 11.5 and 39.2 mm for the six parameter model considered in [7] (identified by tracking the displacement of nine fiducial markers placed on the breast surface). However, the results in this study highlighted that relatively large errors were present in certain regions of the model.
The largest registered displacements between the model-simulated supine MRI and the independently acquired supine MRI were along the cranial-caudal axis (z-axis). The RMS alignment errors between the sternum landmarks in the prone and supine MRI (2.29 mm and 2.40 mm for volunteers 1 and 2, respectively) were an order of magnitude smaller than these registered displacements. It was therefore unlikely that these discrepancies were due to errors in aligning the prone and supine MR images. Furthermore, the direction of the registered displacements along the z-axis for volunteer 1 was different to that observed in volunteer 2. These additional displacements may be due to different motion of the shoulders in the z-axis (cranial-caudal direction), during repositioning from the prone-to-supine positions, which could not be accounted for in the model. However, further investigation is required to confirm this hypothesis.
Large errors were also observed near the nipple position of the model. This observation maybe due to the lack of a stiffer skin layer in the model which, if present, may resist motion of the nipple towards the axilla, and help improve the accuracy of the models.
The tissue landmarks used for assessing the accuracy of the registration were only labelled by one human rater. Obtaining labels from multiple raters would allow the accuracy of the segmentation to be estimated using probabilistic methods such as Simultaneous Truth And Performance Level Estimation (STAPLE) [17]. The use of Hausdorff distance-based automatic segmentation approaches may also help assess the alignment of structures within the breast, particularly in the regions where fibroglandular tissue is present [18].
The breast is supported by Cooper’s ligaments that extend from the skin into the pectoral muscle fascia. It is unclear if these structures contribute significantly when simulating the supine position as the ligaments would likely be in compression and therefore unable to carry load. However, their influence is likely more significant when determining the unloaded state of the breast from the prone position (where the ligaments are in tension). Further investigations are required to determine the influence of these ligaments during prone to supine repositioning.
5 Conclusions
This chapter describes a biomechanical modeling framework for simulating breast tissue motion from the prone-to-supine orientation to help clinicians register tissue motion between diagnostic prone MRI and pre-operative supine MRI, for example, for treatment of planning procedures. The framework was demonstrated using MR images from two volunteers. A relatively large displacement of the pectoral muscle was observed between the prone and the supine positions in both volunteers. This motion was accounted for in the models by directly prescribing the observed motion of the muscle during the prone-to-supine simulations. The results showed that a two parameter breast model provided good initial estimates of tissue displacement for registering the prone and supine MRI, with registration errors less than 5 mm for mean tissue displacement magnitudes of up to 61.8 mm.
References
Pleijhuis RG et al (2009) Obtaining adequate surgical margins in breast-conserving therapy for patients with early-stage breast cancer: current modalities and future directions. Ann Surg Oncol 16(10):2717–2730
Sotiras A, Davatzikos C, Paragios N (2013) Deformable medical image registration: a survey. IEEE Trans Med Imaging 32(7):1153–1190
Rueckert D et al (1999) Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging 18(8):712–721
Schnabel J et al (2001) A generic framework for non-rigid registration based on non-uniform multi-level free-form deformations. In: Niessen W, Viergever M (eds) Medical image computing and computer-assisted intervention 2001. Springer, Berlin Heidelberg, pp 573–581
Lee AWC et al (2010) Breast image registration by combining finite elements and free-form deformations. In: Martí J et al (eds) Digital mammography. Springer, Berlin/Heidelberg, pp 736–743
Hipwell JH et al (2016) A review of biomechanically informed breast image registration. Phys Med Biol 61(2):R1
Han L et al (2014) A nonlinear biomechanical model based registration method for aligning prone and supine MR breast images. IEEE Trans Med Imaging 33(3):682–694
Babarenda Gamage TP et al (2012) Patient-specific modeling of breast biomechanics with applications to breast cancer detection and treatment. In: Gefen A (ed) Patient-specific modeling in tomorrow’s medicine. Springer, Berlin Heidelberg, pp 379–412
Bradley C et al (2011) OpenCMISS: a multi-physics & multi-scale computational infrastructure for the VPH/Physiome project. Prog Biophys Mol Biol 107(1):32–47
Eiben B, Vavourakis V, Hipwell JH, Kabus S, Lorenz C, Buelow T, Hawkes DJ (2014) Breast deformation modelling: comparison of methods to obtain a patient specific unloaded configuration. In: Proceedings of SPIE 9036, Medical Imaging 2014: image-guided procedures, robotic interventions, and modeling, pp 903615-1–903615-8. doi:10.1117/12.2043607
Rajagopal V et al (2007) Determining the finite elasticity reference state from a loaded configuration. Int J Numer Methods Eng 72(12):1434–1451
Lee AWC (2011) Breast image fusion using biomechanics. PhD thesis, University of Auckland
Johns PC, Yaffe MJ (1987) X-ray characterisation of normal and neoplastic breast tissues. Phys Med Biol 32(6):675–695
Urbanchek MG et al (2001) Specific force deficit in skeletal muscles of old rats is partially explained by the existence of denervated muscle fibers. J Gerontol A Biol Sci Med Sci 56(5):B191–B197
Schnabel JA et al (2003) Validation of nonrigid image registration using finite-element methods: application to breast MR images. IEEE Trans Med Imaging 22(2):238–247
Babarenda Gamage TP et al (2012) Modeling prone to supine breast deformation under gravity loading using heterogeneous finite element models. In: Nielsen PMF, Wittek A, Miller K (eds) Computational biomechanics for medicine. Springer, New York, pp 29–38
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23(7):903–921
Garlapati RR et al (2015) Towards measuring neuroimage misalignment. Comput Biol Med 64:12–23
Acknowledgements
The authors are grateful for financial support from the New Zealand Government Ministry for Business, Innovation and Employment (MBIE) and the University of Auckland Foundation. The authors thank Duane Malcolm for his contributions to this study.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Gamage, T.P.B., Baluwala, H.Y., Nash, M.P., Nielsen, P.M.F. (2017). Registration of Prone and Supine Breast MRI for Breast Cancer Treatment Planning. In: Wittek, A., Joldes, G., Nielsen, P., Doyle, B., Miller, K. (eds) Computational Biomechanics for Medicine. Springer, Cham. https://doi.org/10.1007/978-3-319-54481-6_11
Download citation
DOI: https://doi.org/10.1007/978-3-319-54481-6_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-54480-9
Online ISBN: 978-3-319-54481-6
eBook Packages: EngineeringEngineering (R0)