Abstract
The brain is the most complex tissue in terms of cell types that it comprises, to the extent that it is still poorly understood. Single cell genome and transcriptome profiling allow to disentangle the neuronal heterogeneity, enabling the categorization of individual neurons into groups with similar molecular signatures. Herein, we unravel the current state of knowledge in single cell neurogenomics. We describe the molecular understanding of the cellular architecture of the mammalian nervous system in health and in disease; from the discovery of unrecognized cell types to the validation of known ones, applying these state-of-the-art technologies.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Single cell genomics
- single cell transcriptomics
- neurobiology
- cortex
- hippocampus
- midbrain
- somatosensory nervous system
- oligodendrocyte
- neurodegenerative diseases and neuro-oncology
1 Introduction
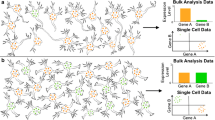
Single cells are the fundamental units of life. It has not been until recently that single-cell analysis has enabled us to interrogate the heterogeneity of complex cellular populations at ultrahigh resolution. The development of powerful single-cell genomics techniques and advances in next-generation sequencing technologies made the sequencing of thousands of cells feasible and affordable. Single-cell studies reveal information that work performed on bulk populations could not address. They provide fine-grained resolution to define cell-type heterogeneity of complex tissues and define cell states in dynamic processes with high sensitivity. The mammalian brain is the most complex organ in our body, which is thought to contain thousands of cell types [1]. However, the function of most cell types in the nervous system is still unknown. Cells have historically been classified according to location, morphology, and electrophysiological characteristics, combined with molecular markers. Cell identities and function have been assigned through the expression of marker genes [2]. Large-scale projects, such as Allen Brain Atlas (ABA) or Gene Expression Nervous System Atlas (GENSAT) provided gene expression profiles of different brain regions and cell types [3, 4]. Single-cell technologies now allow the analysis of single-cell units that form this complex tissue and to identify cell types in an unbiased manner without prior knowledge about phenotype or function. In this chapter, we review how the single-cell genomics approaches are providing novel insights into the neural cell type heterogeneity and classification. We conclude by summarizing future applications of single-cell technologies in neuroscience.
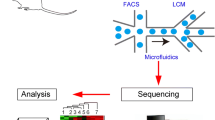
2 Single-Cell Capture and Isolation
Isolation of single cells from complex tissues is a critical step in single-cell sequencing. It is very important to preserve cellular integrity to produce a representative image of the transcriptome for phenotype inference. For solid tissues, a proteolytic enzymatic treatment, such as collagenase, papain, and trypsin, is applied to obtain single-cell solutions. Caution has to be taken as such treatments can affect cell viability, which can later impact on transcriptional profiles or causes underrepresentation of certain cell types. The main approaches in isolation of cells from tissues or cell cultures are manual or automated micropipetting, laser capture microdissection (LCM), fluorescence-activated cell sorting (FACS), microfluidics, and droplet-based separation. In the following we summarize mechanisms, applications, and their pros and cons for each technique. Manual or automated micropipette and laser capture microdissection are low-throughput methods. Isolation is performed under a microscope based on visual inspection of cellular morphology. It enables specific cell selection but requires a trained experimentalist and very time-consuming. Micromanipulation is mostly used on early embryos and LCM on tissue sections [5]. FACS, microfluidics, and droplet techniques are high-throughput methods that are based on isolating cells based on cell-specific characteristics of fluorescence markers, light scattering, and size. FACS requires a large number of cells in suspension to set up the instrument, being a downfall on samples with low quantity. FACS instrument can accurately sort cells into the center of a well of microtiter plates, ensuring cells are immersed in the lysis buffer. FACS-based systems require a minimum reaction volume increasing the costs per cell. In contrary, microfluidic systems allow the isolation of single cells in microfluidic chips with individual micro-reaction chambers and micro-mechanical valves that allow automating downstream of biochemical reactions [6]. The most popular and commercial microfluidic systems in the market is the Fluidigm C1, which can capture up to 800 cells per chip, also allowing the visualization of cells under the microscope to identify potential doublets. Afterward, the reactions in nanoliter volumes are monitored, reducing the quantity of reagents required. Microdroplet systems, an emerging new technology, such as DROP-seq [7] or commercial 10× Genomics devices, encapsulate cells in aqueous droplets in flowing oil that included cell-specific barcoding. These techniques enable the processing of thousands of cells in parallel with relative low-sequencing library preparation cost per cell.
3 Single-Cell DNA Sequencing Methods
Only 6 pg of DNA is contained in a human diploid cell, being an insufficient material for standard DNA sequencing applications [8]. Therefore, single-cell genome sequencing requires whole-genome amplification (WGA) prior to library preparation. However, there are major challenges while generating high fidelity and unbiased WGA for adequate single-cell applications. Different approaches exist, but all have their advantages and limitations. Current WGA methodologies are based on multiple displacement amplification (MDA), polymerase chain reaction (PCR), or a combination of both methods.
MDA is based on the use of random hexamers binding to denatured DNA, where the phi29 polymerase catalyzes a strand displacement synthesis at a constant temperature [9]. The polymerase produces high DNA yield with high amplification fidelity [10]. However, compared to PCR-based techniques, it shows a significant amplification bias resulting in less evenly amplified genomes. The phi29 presents unique molecular properties, due to the ability of proofreading activity and high replication fidelity (3′ → 5′ exonuclease activity) [11]. It generates DNA amplicons of up to 10 kb in length.
PCR-based WGA methods rely on primer extension pre-amplification PCR (PEP-PCR), degenerated oligonucleotide-primed PCR (DOP-PCR), or linker-adaptor PCR (LA-PCR). During PEP-PCR, DNA is amplified with oligonucleotides of degenerate sequences using permissive thermocycling with increasing annealing temperatures [12]. In DOP-PCR, DNA is amplified with hybrid oligonucleotides containing the degenerate and unique sequences, starting with thermocycling at low annealing temperature (semi-random priming), followed by PCR at high annealing temperature (nonrandom priming) [13]. In contrary to the random priming-based methods, LA-PCR utilizes sheared or digested DNA and adaptors with universal sequences ligated to the DNA ends. The unique sequences are then used for subsequent PCR amplification [14].
A methodology that combines both MDA and PCR-based method is known as multiple annealing and looping-based amplification cycles (MALBAC). Its unique feature is the quasi-linear amplification to reduce the bias associated with nonlinear amplification methods [15]. The primers that anneal randomly to the genome contain specific sequences that allow the amplicons to form looped pre-amplification products. This looping protects previously copied segments to be further pre-amplified, therefore avoiding sequence-dependent biases by exponential amplification.
Several studies have been performed to compare WGA amplification techniques [16, 17]. They conclude that WGA from single cells presents a suitable tool for profiling copy number and structural variants or the detection of small-scale alteration, such as point mutations. Nevertheless, whole-genome DNA sequencing remains challenging due to the loss of material that causes dropout events or the introduction of sequencing errors that complicate variant calling.
4 Single-Cell RNA Sequencing Methods
The vast majority of single-cell RNA sequencing methodologies follow a similar strategy as DNA-based methods, since the typical mammalian cell contains 10 pg of RNA, but only 0.1 pg of messenger RNA. Thus profound amplification is required before sequencing libraries can be prepared. In initial steps, single cells are captured and lysed, and reverse transcription converts polyA-tailed RNA into cDNA. Then, the minute amounts of cDNA are amplified by PCR or by in vitro transcription before sequencing library preparation. The first single-cell whole-transcriptome method for mammalian cells was described by Tang [18]. Since then, many new methods have been developed that tackle different challenges and range from full-length approaches (SMART-seq [19] and SMART-seq2 [20]) to 5′-end- (STRT and STRT-C1 [21]) or 3′-end-focused (CEL-seq [22], CEL-seq2 [23], MARS-seq [24], Quartz-seq [25], and DROP-seq [7]) methods. Techniques that focus on the full-length approach rely on the Moloney murine leukemia virus (MMLV) reverse transcriptase. This enzyme presents unique properties that enable both template-switching and terminal transferase activity, resulting in the addition of a non-templated cytosine residue to the 5′ end of the cDNA. By adding a poly(G) template primer with an adapter sequence to the reaction, the enzyme can switch templates and transcribe the other strand. The resulting full-length cDNA can be amplified by PCR [19]. It is of note that all single-cell RNA sequencing methods depend on the amplification of the minute amount of starting material which can introduce technical variability and amplification bias. To correct such errors, methodologies focused on 5′- or 3′-end amplification include unique molecular indexes (UMIs) in the reverse transcription primer to label the original pool of RNA molecules [26]. Another approach to controlling this technical variability is the addition of external spike-in RNAs of known concentrations, such as the External RNA Controls Consortium (ERCC) mix [27].
5 Single-Cell Epigenomic Sequencing Methods
DNA methylome can be profiled at a single-cell level; however, technical peculiarities, such as the DNA degradation caused by the bisulfite conversion, challenge the preparation procedures. The first single-cell method to measure genome-wide 5-methylcytosine (5mC) levels utilized reduced single-cell representation bisulfite sequencing (scRBS). This technique digests the genomic DNA with restriction enzyme, prior to treatment with bisulfite. Resulting sequencing libraries are enriched in CpG methylation and, however, present limited genome coverage and a bias toward CpG-dense regions [28]. On the other hand, genome-wide DNA methylation profiling techniques provide a reasonable representation of each cell’s DNA methylome [29, 30] but increasing sequencing cost per single cell. Subsequent comparative analysis enables the modeling of epigenetic dynamics and variability in contexts, such as development or differentiation.
Histones are subjected to a wide variety of posttranslational modifications. Technically, the profiling of the histone marks at a given genomic location is performed by chromatin immunoprecipitation followed by sequencing (ChIP-seq). On single-cell level, this technique is extremely challenging due to the background noise caused by nonspecific antibody binding. To overcome this problem, immunoprecipitation was performed on a pool of single-cell chromatin that underwent prior barcoding in droplet-based systems [31]. Another layer of epigenetic regulation is chromatin structure that can also be evaluated at a single-cell level. Two independent approaches have been developed to evaluate open chromatin regions that indicate regulatory activity. Buenrostro et al. used on a microfluidic device that implements an assay for transposase-accessible chromatin sequencing (ATAC-seq) [32]. Here, hyperactive prokaryotic Tn5 transposase inserts into accessible chromatin and tags the sites with sequencing adaptors. Cusanovich et al. used a combinatorial indexing strategy, where two-level tagmentation is carried out which introduces a unique barcode to each pool [33]. This strategy allows reactions with multiple cells to increase tagmentation efficiencies, while barcode combination allows the subsequent deconvolution to single-cell level. Chromosome conformation, a higher order of epigenome regulation, can be assessed by 3C-based method that can profile genome-wide chromosome interactions (e.g., HiC methods). At a single-cell level, HiC analysis revealed cell-to-cell variability in chromosome structure and compartmentalization [34, 35].
6 Linking Different Single-Cell Genomics Strategies
The combination of different single-cell genomics methods is an actively pursued issue in the field. Certain combinations of techniques measuring two modalities from a single cell are now possible. Macaulay et al. developed “genome and transcriptome” sequencing (scG&T) enabling the assessment of gene expression level and genetic variant from the same single cell [36]. Specifically, G&T-seq allows whole-genome and whole-transcriptome amplification following the physical separation of nucleic acids (DNA and mRNA) from a single cell. G&T-seq further modified the technique to allow the simultaneous assessment of DNA methylation and gene transcription (scM&T-seq) [37]. Both methodologies will link phenotypes (defined by gene expression or epigenetic states) to their genotypes. Combined single-cell methods enable the clear assessment of cellular relationships to better understand tissue heterogeneity in normal diseased states.
6.1 Studying Neurobiology Systems Using Single-Cell Sequencing Approaches
To date single-cell studies have been used to characterize cells from complex tissues, such as lung epithelium [38], spleen [24], or the pancreas [39]. In neuronal systems, single-cell genomics identified different neural types in various regions of the mouse and human nervous system. Here sample preparation and computational analysis face specific challenges due to the complex cell morphology and strongly interconnected expression profiles. Nevertheless, single-cell analysis allowed the identification of novel cell types with unique biological properties, the inference of neural connectivity, and the association to neurological diseases.
6.2 Transcriptome of the Mouse Cortex and Hippocampus at Single-Cell Resolution
The mammalian cerebral cortex is involved in cognitive functions such as sensorimotor integration, memory, and social behaviors. To elucidate the transcriptome of the primary somatosensory cortex (S1) and hippocampal (CA1) region, single-cell RNA-seq was performed on 3005 single cells, applying the 5′-end-focused STRT technology [40]. The resulting fine-grained characterization of cell type composition was confirmed by single-molecule RNA fluorescence in situ hybridization (RNA-FISH). The study identified nine molecularly distinct classes of cells by computational clustering, which were confirmed by the presence of specific markers, which play a functional role in the cell types. The nine clusters represented cortical and hippocampal pyramidal neurons, interneurons, oligodendrocytes, astrocytes, microglia, vascular endothelial cells, mural cells (pericytes and vascular smooth muscle cells), and ependymal cells. Exemplarily, S1 pyramidal cells were identified by the expression of Tbr1 (T-Box Brain 1), a transcription factor implicated in neuronal migration and axonal projection; and oligodendrocytes were marked by Mbp (Myelin basic protein), a major constituent of the myelin sheath. Subsequently, biclustering analysis was performed on the nine major classes, identifying a total of 47 molecularly distinct subclasses of cell types. The authors observed that the RNA content was different among cells, with neurons containing more molecules than glia and vascular cells, correlating with a higher number of detected genes. Within the cortical pyramidal neurons, the work identified seven subclasses with layer specificity. Two subtypes of hippocampal pyramidal CA1 neurons were identified, associated to high mitochondrial function, plus cells derived from the adjacent CA2 and subiculum. Sixteen subclasses of interneurons were described, with regions both in the cortex and hippocampus containing these closely related subclasses. As for non-neural diversity, two major subtypes were identified for astrocytes and immune cells. In oligodendrocytes, six populations were found, representing different stages of maturation [40]. Ependymal cells expressed a largest set of subclass-specific genes. Altogether, the single-cell transcriptome study of primary somatosensory cortex and hippocampus indicates an extensive functional specialization between cellular subclasses reflected by specific gene expression patterns.
Another study profiling the cortex by single-cell RNA-seq utilized full-length transcriptome sequencing (SMART-seq technology). The work focused on a single cortical region, the primary visual mouse cortex [41], and used a transgenic mouse line with specifically labeled cortical cells [42]. By analyzing more than 1600 single cells, the authors identified 49 transcriptomic cortical cell types assigned by cell location and marker gene expression. These included 23 GABAergic, 19 glutamatergic, and 7 nonneuronal subtypes, with the majority of marker genes also detected in the RNA-ISH data from the Allen Brain Atlas [3]. Together, both studies provide a comprehensive overview of the transcriptomic landscape in the cortex and hippocampus areas of the mouse. Importantly, the unbiased analysis of single-cell transcriptomes allowed the identification of novel cell subtypes with likely highly specialized functions in these areas.
7 Transcriptome on the Human Cortex at Single-Cell Resolution
The human brain is a highly complex tissue, and study designs are hindered by the accessibility of material mostly being postmortem. Consequently, seminal studies on neuronal tissues have been performed on nonhuman model organisms, which may not recapitulate the full molecular complexity of the human brain. To tackle this problem, the use of human cerebral organoids has proven to reflect gene expression programs of the fetal neocortex development at a single-cell level [43].
To interrogate the heterogeneity of the human cortex, Darmanis et al. evaluated single-cell transcriptomes of normal primary fetal and adult brain tissue subjected to surgery using the SMART-seq technology. Their findings demonstrate that the transcriptomic profile obtained from single cells can successfully identify all major neuronal, glial, and vascular cell types in the human cerebral cortex. Moreover, the work supplemented traditional neuronal classifications based on marker genes with the underlying transcriptome. Also, subclustering neurons revealed two distinct groups, excitatory and inhibitory neurons. These cells represent a neuronal community with unique expression signatures and a specific role in the network niche. Gene expression analysis between pre- and postnatal neurons determined considerably distinct expression patterns. For example, adult neurons displayed high expression of SNAP25 and GAD1; fetal neuronal progenitors expressed MKI67 and PAX6; and quiescent neurons revealed specific activation of DCX and TUBB3. Moreover, the work interrogated HLA expression in fetal and adult neurons, since the central nervous system (CNS) is believed to be immunologically inert. Although MCHI proteins are expressed in adult mouse brain [44], the expression of these genes in the human brain has been subject of controversial discussion, since their identification in a subpopulation of adult neurons.
A second study analyzed the transcriptome of a postmortem brain using neuronal nuclear antigen (NeuN) to isolate single neuronal nuclei and applying the SMART-seq technology for single-cell transcriptome generation. The study focused on six classically defined Brodmann areas (BAs) with well-documented anatomical and electrophysiological properties. Single-cell transcriptome profiles identified two major classifications within the cerebral cortex, inhibitory neurons that encompass interneurons, and excitatory including pyramidal or projection neurons. Each class and their associated subtypes revealed significant cell heterogeneity among the BAs, indicating that neural composition varies profoundly among regions in the brain. Another study on the developmental cerebral cortex identified different cell types of dividing neural progenitor, radial glia, and newborn neurons to maturing neurons [45].
Taking together single-cell transcriptome studies laid the groundwork for the construction of a cellular atlas of the human cortex, being a stepping stone toward elucidating the full cellular complexity of the human brain [46].
8 Midbrain in Mouse and Human at Single-Cell Resolution
The midbrain is a portion of the central nervous system associated with vision, hearing, motor control, alertness, and temperature regulation. An initial single-cell study was performed on the midbrain dopaminergic (DA) system, to assess neuron diversity. The classical anatomical classification of the midbrain DA neurons are substantia nigra pars compacta (SNc), the ventral tegmental area (VTA), and the retrorubral area (RR). This area has its clinical importance due to its implication in Parkinson’s disease (PD), schizophrenia, attention deficit hyperactivity disorders (ADHD), obsessive-compulsive disorder, addiction, and depression. To identify molecular distinct DA neurons, early postnatal brain from mice was analyzed. Specifically, FACS separation combined with the expression analysis of 96 genes (using Fluidigm Biomark system) discovered six different types of cells (DA1A, DA1B, DA2A, DA2B, DA3A, and DA3B), which could be validated by RNA-ISH [47]. A more recent study focused on the development of the midbrain in human and mouse [48]. Although it is thought that the development of the organ in humans follows similar sequence of events as in rodent, the degree of conservation is unclear. Single-cell RNA-seq was performed on the developmental ventral midbrain and 1907 mouse cells from 271 embryos and in 1977 human cells from 10 human embryos. The analysis at different time points uncovered dopaminergic progenitor specification, neurogenesis, and differentiation. Moreover, it identified that gene expression profiles were conserved across species, which, however, showed differences in proliferation, timing, and dopaminergic neuron development.
9 Somatosensory Nervous System in Mice at Single-Cell Resolution
The somatosensory nervous system responds to mechanical, thermal, and nociceptive stimuli. The ability to perceive and discriminate these sensations is due to the existence of specialized dorsal root ganglion (DRG). The system comprises diverse neuronal subsets with distinct conduction properties and peripheral and central innervations patterns. They include small-diameter unmyelinated C-fibers, thinly myelinated Aδ-fibers, and large-diameter thickly myelinated Aα-/β-fibers [49]. DRGs were dissected from the mouse lumbar to classify the neuronal types and to reveal the complexity of this primary sensory system. Analyzing the transcriptome of 799 single cells, four clusters could be distinguished and identified as known marker genes: first, the NF cluster expressing the neurofilament heavy chain (Nefh) and parvalbumin (Pvalb); second, the PEP cluster with active substance P (Tac1), TRKA (Ntrk1), and calcitonin gene-related peptide (Calca), associated with peptidergic nociceptors; the third subtype, the NP cluster, expressing Mrgprd and P2rx3, associated with nonpeptidergic nociceptors; and, lastly, the TH clusters showing expression of tyrosine hydroxylase (Th), associated with unmyelinated neurons. Within the main populations, a total of 11 neuronal classes were identified, NF1 to NF5, NP1 to NP3, PEP1, PEP2, and TH. This dissection illustrates the diversity of sensory neuron types and their cellular complexity [50].
9.1 Olfactory and Retina Neurogenesis in Mice at Single-Cell Resolution
Odor perception is the detection of odorants by olfactory receptors (ORs), located on olfactory sensory neurons (OSNs) in the epithelium of the nose. These receptors are seven-transmembrane domain G protein-coupled receptor, encoded by a large multi-gene family. In mice, odor detection is mediated by 1000 odorant receptors genes (Olfrs) and 350 pseudogenes [51]. Hanchate et al. evaluated how developing OSNs select Olfr for expression using single cell RNA-seq. Eighty-five single-cell transcriptomes were analyzed using an unsupervised algorithm that determines cellular state of differentiation in “pseudotime” which models the dynamics in gene expression during development [52]. The predicted trajectory reflected developmental progression from progenitors, precursors, and immature OSNs to mature OSNs. The results were confirmed using stage-specific markers (progenitor, Ascl; precursor, Neurog1; immature OSN, Gap43; mature OSN, Omp). Subsequently, four olfactory sensory transduction molecules downstream of odorant receptors, Gna1, Adcy3, Cnga2, and Cnga4, supported the conclusions. Expression of Olfr appeared at a late precursor to early immature OSN stage, presenting low levels of multiple Olfrs. During subsequent development, the expression of single highly expressed Olfr overtakes the expression of the other family members. Further, coexpressed Olfrs overlap in zones of the nasal epithelium, suggesting regional biases. Hence, mature neurons express single Olfrs at elevated levels leading to two hypotheses to explain the phenomenon: First is the “winner-takes-all,” when an Olfr becomes dominant and overtakes the other expressed Olfrs. A second model based on the selection of one single Olfr independently of Olfrs initially expressed [53].
The retina represents another excellent system to study neuronal diversity. The retina contains five neuronal classes defined by morphological, physiological, and molecular features that include retinal ganglion, bipolar, horizontal, photoreceptor, and amacrine. Droplet-based single-cell transcriptome sequencing was applied on 44,808 cells from the retina of a 14-day-old mouse to create a molecular atlas of retinal cells. Thirty-nine transcriptional distinct retinal cell populations were identified by unsupervised computational analysis. These matched known types and identified additional subpopulations corresponding to astrocytes (associated with the retinal ganglion cell axons exiting the retina), resident microglia, endothelial cells (intra-retinal vasculature), pericytes, and fibroblast. A further focus lied on the 21-amacrine subtypes as it represents the most morphologically diverse neuronal class since most lack a clear molecular marker profile. Single-cell analysis classified subpopulations in inhibitory (using GABA or glycine as neurotransmitter), excitatory (release of glutamate), and undefined cell types that do not express GABAergic, glycinergic, nor glutamatergic markers [7].
9.2 Oligodendrocyte Heterogeneity in the Mouse Central Nervous System at Single-Cell Resolution
Oligodendrocytes have been considered as a functionally homogenous population in the central nervous system (CNS). Using single-cell RNA sequencing on ten regions of the anterior-posterior and dorsal-ventral axis of the mouse juvenile and adult CNS, 13 distinct cell populations were identified. Here, clustering, differential expression, and pseudotime analysis led to the identification of the transcriptional continuum between oligodendrocyte populations. These were oligodendrocyte precursor cells (OPCs), differentiation-committed oligodendrocytes (COPs), newly formed oligodendrocytes (NFOL1 and NFOL2), myelin-forming oligodendrocytes (MFOL1 and MFOL2), mature oligodendrocytes (MOL1 to MOL6), and vascular and leptomeningeal cells (VLMCs). OPCs coexpressed Pdgfra and Cspg4, while COPs lacked the expression of the genes, but expressed Neu4, Sox6, Bmp4, and Gpr17. NFOL1 and NFOL2 expressed genes involved in early stages of differentiation. MFOL1 and MFOL2 showed activity of genes involved in the myelin formation. MOL1 to MOL6 expressed late oligodendrocytes differentiation genes (Klk6 and Apod) and genes present in myelinating cells (Trf and Pmp22). VLMCs were identified as a second of Pdgfra population with low levels of Cspg4 and high levels of laminins and collagens, concluding a transcriptional continuum between different oligodendrocyte populations across multiple regions of the CNS. From a translational perspective, the identification of these cell types could provide a new vision into the etiology of myelin disorders [54].
10 Neurodegenerative Diseases at Single-Cell Resolution
Neurodegenerative disease is the progressive loss of function and structure of neurons, including their cell death. Diseases such as Parkinson’s, Alzheimer’s, Huntington’s, and amyotrophic lateral sclerosis result from neurodegenerative processes. At a single-cell level, Poulin et al. determined subclasses of DA, focusing on abnormalities in Parkinson’s disease. Specifically, DA1A neurons, located in the ventral tier of the substantia nigra, are most vulnerable to this disease. To determine the molecular peculiarities, the study evaluated the susceptibility of DA1A neurons to the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), commonly used to mimic pathological features of parkinsonism in mice [47]. The analysis of single cells revealed a decrease in Aldh1a1+ expression, previously reported to be downregulated in the disease [55], providing insights into disease-related cellular degeneration [48]. Cell replacement therapy is a promising avenue toward treatment of Parkinson’s disease [56], and the use of transplantation of human fetal midbrain tissue, containing dopaminergic neurons, could be used as a therapeutic approach. In this regard, human pluripotent stem cell (hPSC)-derived dopaminergic neurons have been proven to recover behavior in animal models with Parkinson’s disease [57]. La Manno et al. performed single-cell RNA-seq on human embryonic stem cell (hESCs) and human-induced pluripotent stem cells (hiPSCs) to evaluate their molecular composition. This approach could be used to assess the quality of stem cells for cell replacement therapy [48]. In Alzheimer’s disease (AD), one of the most common forms of dementia, aneuploidy (aberrant chromosome copy numbers) has thought to be involved in the pathogenesis of this disease. To address this, single-cell whole-genome sequencing was performed on frontal cortex neurons from healthy individuals and from AD patients, concluding that aneuploidy does not play a role in the pathogenesis of this disease [58].
11 Neuro-oncology at Single-Cell Resolution
Cancer is a heterogeneous disease with molecular characteristics that depend on the tissue of origin. Tumors evolve from a single cell, due to the accumulation of genetic and epigenetic alterations. During the progression of a tumor, additional variations appear giving rise to different cell subpopulations and the related tumor subclonal structure. Tumor heterogeneity has many implications for clinical management since different tumor clones play different roles in disease initiation, progression, metastasis, and drug resistance [59]. Current strategies analyzing bulk tumor samples to determine tumor composition lack resolution and are insufficient to recapitulate the clonal structures of the tumors. Single-cell genomics strategies present a suitable solution to define subclonal tumor structures to unprecedented resolution. Single-cell DNA-seq was performed on glioblastoma, a common primary brain tumor with a high degree of cellular heterogeneity [60], to depict clonal diversity. The study revealed convergent evolution of EGFR mutations in different subclones from the same primary tumors [61], further supporting the complex structure tumors and related difficulties in therapeutic intervention. Single-cell RNA-seq on glioblastoma showed that cancer cells display a large range of intermediate phenotypes that do not fall into distinct classes of epithelial and/or mesenchymal cell types [62]. To characterize the cellular diversity within IDH-mutant tumors, single-cell RNA-seq was performed on oligodendroglioma and astrocytoma [63]. Both entities shared a developmental hierarchy, with most cells differentiating along two glia lineages. Tumor cells derived from a set of proliferating cancer stem cells, supporting the cancer stem model, an important finding for the management of the disease.
12 Conclusions and Future Directions
The cell type complexity of the brain is widely unknown and intensively debated [64]. To understand the cellular heterogeneity, cell types have to be profoundly characterized and phenotyped for functional interpretation. Single-cell techniques present a key technology to illuminate the biological complexity in a normal context and during diseases, and fundamental questions about cell identities are now being answered for the first time. The international community has gathered together to create the Human Cell Atlas project, creating a comprehensive reference map of human cells as a basis for understanding human biology and its perturbation leading to diseases.
References
Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9.
Holmberg J, Perlmann T. Maintaining differentiated cellular identity. Nat Rev Genet. 2012;13:429–39.
Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76.
Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–25.
Frumkin D, Wasserstrom A, Itzkovitz S, Harmelin A, Rechavi G, Shapiro E. Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues. BMC Biotechnol. 2008;8:17.
Reece A, Xia B, Jiang Z, Noren B, McBride R, Oakey J. Microfluidic techniques for high throughput single cell analysis. Curr Opin Biotechnol. 2016;40:90–6.
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–14.
Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–60.
Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–9.
Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, et al. Whole-genome multiple displacement amplification from single cells. Nat Protoc. 2006;1:1965–70.
Garmendia C, Bernad A, Esteban JA, Blanco L, Salas M. The bacteriophage phi 29 DNA polymerase, a proofreading enzyme. J Biol Chem. 1992;267:2594–9.
Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci U S A. 1992;89:5847–51.
Cheung VG, Nelson SF. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc Natl Acad Sci U S A. 1996;93:14676–9.
Troutt AB, McHeyzer-Williams MG, Pulendran B, Nossal GJ. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc Natl Acad Sci U S A. 1992;89:9823–5.
Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6.
de Bourcy CFA, De Vlaminck I, Kanbar JN, Wang J, Gawad C, Quake SR. A quantitative comparison of single-cell whole genome amplification methods. PLoS One. Public Library of Science; 2014;9:e105585.
Huang L, Ma F, Chapman A, Lu S. Xie XS. Methodology and Applications: Single-Cell Whole-Genome Amplification and Sequencing; 2015.
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–82.
Ramsköld D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–82.
Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013 Nov;10:1096–8.
Islam S, Kjällquist U, Moliner A, Zajac P, Fan J-B, Lönnerberg P, et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat Protoc. 2012;7:813–28.
Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Rep. 2012;2:666–73.
Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. [Internet]. 2016 [cited 2016 Nov 14];17:77. Available from: http://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-0938-8
Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science. 2014;343:776–9.
Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31.
Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Meth.; 2014;11:163–6.
Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, et al. The External RNA Controls Consortium: a progress report. Nat Methods. 2005;2:731–4.
Guo H, Zhu P, Wu X, Li X, Wen L, Tang F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–35.
Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–20.
Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schnegger A, Klughammer J, et al. Single-Cell DNA Methylome Sequencing and Bioinformatic Inference of Epigenomic Cell-State Dynamics. Cell Rep. 2015;10:1386–97.
Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–72.
Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–90.
Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–4.
Nagano T, Lubling Y, Yaffe E, Wingett SW, Dean W, Tanay A, et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015;10:1986–2003.
Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64.
Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–22.
Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13:229–32.
Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–5.
Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394.e3.
Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–42.
Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–46.
Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8
Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci. 2015;201520760
Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci. 2007;104:6828–33.
Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–8.
Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112:7285–90.
Poulin J-F, Zou J, Drouin-Ouellet J, Kim K-YA, Cicchetti F, Awatramani RB. Defining Midbrain Dopaminergic Neuron Diversity by Single-Cell Gene Expression Profiling. Cell Rep. 2014;9:930–43.
La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 2016;167:566–580.e19.
Abraira VE, Ginty DD. The Sensory Neurons of Touch. Neuron. 2013;79:618–39.
Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Publ Gr. 2014;18
Niimura Y, Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005;346:13–21.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6.
Hanchate NK, Kondoh K, Lu Z, Kuang D, Ye X, Qiu X, et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350:1251–5.
Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–9.
Grünblatt E, Riederer P. Aldehyde dehydrogenase (ALDH) in Alzheimer’s and Parkinson’s disease. J Neural Transm. 2016;123:83–90.
Barrow TR. Cell replacement therapy in Parkinson’s disease. Biosci. Horizons. Oxford University Press. 2015;8:hzv002-hzv002.
Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, et al. Generation of Regionally Specified Neural Progenitors and Functional Neurons from Human Embryonic Stem Cells under Defined Conditions. Cell Rep. 2012;1:703–14.
van den Bos H, Spierings DCJ, Taudt A, Bakker B, Porubský D, Falconer E, et al. Single-cell whole genome sequencing reveals no evidence for common aneuploidy in normal and Alzheimer’s disease neurons. Genome Biol. 2016;17:116.
Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–53.
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710.
Francis JM, Zhang C-Z, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, et al. EGFR Variant Heterogeneity in Glioblastoma Resolved through Single-Nucleus Sequencing. Cancer Discov. 2014;4:956–71.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401.
Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–13.
Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–68.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Guillaumet-Adkins, A., Heyn, H. (2017). Single-Cell Genomics Unravels Brain Cell-Type Complexity. In: Delgado-Morales, R. (eds) Neuroepigenomics in Aging and Disease. Advances in Experimental Medicine and Biology(), vol 978. Springer, Cham. https://doi.org/10.1007/978-3-319-53889-1_20
Download citation
DOI: https://doi.org/10.1007/978-3-319-53889-1_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-53888-4
Online ISBN: 978-3-319-53889-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)