Abstract
In the production of biofuels and chemicals from biomass-derived sugars, the yeast Saccharomyces cerevisiae has emerged as a key microbial host. Producing these biochemicals in yields and productivities satisfactory to be useful for establishing a cost-effective production process requires the engineering of the yeast’s metabolism. This is a challenging mission since metabolic pathways are intriguingly connected with genetic regulatory circuits, and we are just deciphering these networks. However, global technologies of systems biology in combination with the adequate design capabilities of synthetic biology, and random or rational mutagenesis through adaptive laboratory evolution have emerged to improve our understanding of basic aspects of yeast cellular processes and come up with proper metabolic engineering strategies (the systems metabolic engineering approach). In this chapter, we will review recent advances in systems metabolic engineering of S. cerevisiae for production of biofuels and commodity chemicals from lignocellulosic biomass.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Matters of pollution and the environment, as well as of petroleum economy play a key role in the increasing interest for the production of fuels and commodity chemicals from lignocellulosic biomass. Another stimulus is the fact that this type of biomass can be renewed at a rate close to 55 Petagrams (Pg) of carbon per year (Field 1998; Barber 2009), which is around twelve times higher than the ~4.5 Pg of petroleum carbon currently in use. Of the different methods for biomass conversion, the microbial fermentation of biomass-derived sugars will offer a huge diversity of molecules that can be used as biofuels and commodity chemicals (Rabinovitch-Deere et al. 2013; Straathof 2014). Nevertheless, microbes were not naturally shaped to satisfy practical requirements of commercial industrial applications. In particular, they cannot produce a single molecule with high yield (grams of product produced per gram of biomass) and productivity (grams of product produced per hour and fermentor volume). However, the scientific and technological advances in biosciences and bioengineering, as well as the progress in methods to delete and transfer genes, modulate gene expression and to study biological systems in a holistic manner, have been motivating metabolic engineers to take microbial production to its full capacity (Stephanopoulos 2007; Nielsen and Jewett 2008; Lee et al. 2011; Rabinovitch-Deere et al. 2013).
Of the different microorganisms utilized for the biosynthesis of biofuels and chemicals (Lee et al. 2011; Rabinovitch-Deere et al. 2013), this review focuses on Saccharomyces cerevisiae since this one is a repository of many scientific and technological advances allowing for systems metabolic engineering. This yeast has an unprecedented recognition in industrial fermentations, holds the generally recognized as safe (GRAS) designation, and is the repository of a large repertory of recombinant DNA technologies. Furthermore, the scientific community possesses a thorough knowledge of its biochemistry and genetics, and genomic revolution has provided with a new array of methods for studying the yeast system in a holistic manner. The combination of these factors with the advances in metabolic engineering (Bailey 1991; Stephanopoulos 1999; Nielsen et al. 2001) have been used for developing yeast strains for cost-efficient production of diverse biochemicals from biomass.
The basic operations of a biomass-to-biochemicals conversion process are shown in Fig. 1. Once harvested, lignocellulosic biomass is chopped and pretreated to disrupt the tangled structure of cellulose, hemicellulose and lignin. Pretreatment methods include diluted acid, ammonia fiber expansion, and steam explosion (Hahn-Hägerdal et al. 2006). After pretreatment, cellulose and hemicellulose are digested to release six- and five-carbon sugars, primarily glucose and xylose (Caspeta et al. 2014a). Naturally, S. cerevisiae cannot metabolize xylose which comprises up to ~30% of total lignocellulosic biomass. Byproducts such as acetic acid, furfural, hydroxyl-methyl-furfural (HMF), and phenolics can also be released during pretreatment. These chemicals impair yeast metabolism, resulting in a reduction of yields and productivities of targeted biochemicals (Palmqvist and Hahn-Hägerdal 2000; Caspeta et al. 2015). Fermentation of biomass-derived sugars to the desired product is the last step before downstream processing. High product titers (grams of product per fermentor volume) are required to reduce efforts in the last step of the process. Ethanol fermentation at high yields, titers and productivities is relatively simple since yeast naturally synthesizes this in response to the presence of glucose or low oxygen concentrations. However, restructuration of yeast metabolic and regulatory networks to overproduce other biochemicals besides ethanol is a key challenge.
Metabolic engineering is useful for overcoming yeast obstacles in biomass-to-biochemicals conversion processes. This integrates detailed biological information into graphic and mathematical representations of metabolic and regulatory networks and then used these to seek for cellular functions which constrain either, the overproduction of a biochemical, or the development of the desired performance. A standard metabolic engineering strategy is cyclic (Fig. 2), initiating with the analysis of biological information obtained from previous experiments, published data and/or in silico simulation (Park et al. 2008). Mathematical modeling of metabolic networks can be used for calculating the conversion yield of a biochemical, as well as for targeting network elements constraining its overproduction. The detected constrains are then released by modifying genetic networks via recombinant DNA technologies—including the chance to insert heterologous genes and regulatory elements (Bailey 1991; Nielsen 1998; Stephanopoulos 1999). Cycles of these activities are performed while the targeted yield or productivity is not reached. Commonly, constrains releasing tasks in cells are based on the overexpression of genes encoding the rate-limiting enzymes in the biosynthesis pathway; knockout or down-regulation of genes encoding the enzymes of competing metabolic pathways; heterologous expression of genes completing non-natural pathways in the host strain; and engineering of enzyme functions (Bailey 1991; Yang et al. 1998; Nielsen 1998; Stephanopoulos 1999).
Genome sequencing and associated technologies for functional annotation of genes certainly bring the study of S. cerevisiae to a systems level. After sequencing its genome (Goffeau et al. 1996), the first generation of DNA microarrays and associated computational algorithms for transcriptomic analysis appeared (DeRisi et al. 1997; Eisen et al. 1998). These allowed for the analysis of genes functions through global gene expression analyses of mutants and wild-type yeast strains under various internal and external changes (Gasch et al. 2000; Boer et al. 2003). Also, an extensive analysis of genes functions through gene deletions and proteins localization become available (Winzeler et al. 1999; Giaever et al. 2002; Huh et al. 2003). The abundance of basic biological information accumulated in the pre- and post-genomic era required integrative platforms, thus leading the reconstruction of the first genome-scale metabolic model (GEM) (Förster et al. 2003a). Development of GEMs was accompanied by an expansion of computational algorithms and methods which allow GEMs to perform multiple tasks (Park et al. 2009; Thiele and Palsson 2010; Osterlund et al. 2012), such as: integrating ‘omics’ data, performing metabolic flux analysis, analysis of signaling and regulatory networks, predicting cell growth and gene essentiality, comparing gene functions among different species, and seeking for target gene functions for metabolic engineering.
Frequently, the complexity of metabolic and molecular interactions in a cellular system cannot be captured in a model. When that happen, the rational application of metabolic engineering through modeling of metabolism and recombinant DNA technologies is not likely. In this case, the application of evolutionary engineering approaches, which follow nature’s engineering principles of variation and selection, is used as a complementary strategy for strain development (Sauer 2001; Dragosits and Mattanovich 2013). This method exploits in vivo recombination through evolution of populations aiming that the generated phenotype is coupled with the genotype. One can further know, through ‘omics’-based characterization, the genetic changes leading the desired phenotypic response, and transfer them to the desired microbial host (Dettman et al. 2012; Caspeta et al. 2014b). This approach is called inverse metabolic engineering.
In this chapter, we provide a short description of concepts end methods used in the systems metabolic engineering, and what this platform is useful for realizing the full potential of S. cerevisiae as a cell factory for converting biomass-derived sugars into biofuels and chemicals. We also give information about the applications of systems metabolic engineering to overproduce natural and non-natural chemicals and biofuels from biomass with this yeast.
2 Systems Metabolic Engineering Tools and Methods
Classical physiological studies and quantitative analyses of metabolism have been supporting traditional methods for targeting gene manipulations, such as: metabolic flux analysis (MFA), metabolic control analysis (MCA), thermodynamic analysis of pathways, and kinetic modeling (Nielsen 1998; Stephanopoulos 1999). The MFA approach is the simplest but very powerful method. This is based on a stoichiometric model constructed with metabolic coefficients of participating reactions. Extracellular metabolic fluxes are used for the calculation of Internal fluxes by applying mass balances in intracellular metabolites. However, the integration of measured internal fluxes with 13C-enriched carbon sources improves MFA predictions (Stephanopoulos 1999). The advent of new concepts and methods molded by the genomic age upgraded traditional MFA to a systems analysis of metabolic fluxes through GEMs.
2.1 Genome-Scale Metabolic Models (GEMs) of S. cerevisiae
As any global reconstruction of cellular metabolism, the GEMs of S. cerevisiae were reconstructed with the annotation of its genome sequence, and copious experimental evidence on metabolic reactions, pathways and associated genes (Osterlund et al. 2012). A protocol for generating high-quality GEMs has been published (Thiele and Palsson 2010). The first model draft is structured with stoichiometric reactions compiled from gene annotation data (e.g. E.C. numbers of enzyme-coding genes). Extensive information published in literature is then used to ensure the validity of the information contained in the model. The curated GEM is then examined on its ability to connect metabolic reactions through the synthesis of biomass and relevant byproducts from typical elements of the culture media. Further introduction of non-native reactions is required to represent heterologous pathways. After checking the connectivity, the GEM is converted into a computational format represented by a matrix S of stoichiometric coefficients arranged in rows and columns, representing N reactions and M metabolites. This representation enables innumerable computational biological studies, such as the valuation of network content and capabilities, testing and generation of hypotheses, phenotypes analyses, and metabolic engineering (Thiele and Palsson 2010).
Today, there are not less than ten GEMs of S. cerevisiae (Osterlund et al. 2012; Aung et al. 2013). The very first one, called iFF708 (Förster et al. 2003b), contains 1175 reactions, 584 metabolites, 3 compartments, and 708 genes (comprising ~16% of total yeast genome). Simulations with this model proved its value for predicting experimental values of the specific rates of glucose and oxygen consumption, and biomass, CO2 and ethanol production, as well as the impact of single gene deletions on cell growth and metabolic shift in anaerobic/aerobic glucose-limited continuous culture; in addition to the correlation between metabolic shift and gene expression (Famili et al. 2003). The iFF708 model was fully compartmentalized (Duarte et al. 2004) and used as a scaffold to generate the iIN800 model, which covers a larger extension of lipid metabolism, thus containing 1446 reactions, 1013 metabolites, and 800 genes (Nookaew et al. 2008). The first model containing regulatory information based on genes interactions with 55 transcription factors (TFs) was the iMH805/775 GEM. This was useful to predict growth and gene expression profiles upon deletions of TFs in different S. cerevisiae in silico and experimental strains (Herrgård et al. 2006b). The reconstruction of additional models brought the necessity to make a consensus yeast GEM called Yeast 4.0 (Dobson et al. 2010), which contains 16 compartments and 924 genes. GEM-based analyses and their utility in metabolic engineering projects are described below and in Table 1.
2.2 GEM-Based Analysis of Targets for Genetic Manipulations
Building a comprehensive GEM of a whole organism and using this in simulations has several limitations because we do not know all the biological information (Palsson 2000)—including for S. cerevisiae. However, if internal and external environmental constraints limiting particular cell behaviors are known (e.g. systems stoichiometry, maximum/minimum metabolic fluxes, enzyme kinetics, gene knockouts and knockins, regulation, and molecular diffusion), then it is possible to examine, understand and predict the genotype-phenotype relationships. In silico algorithms to evaluate cells behavior through GEM simulations were generated based on optimization techniques using constraints to improve simulations performance. Some useful methods are described, for a complete reference see Machado and Herrgård (2015).
2.2.1 GEM Analysis Metabolically Constrained
2.2.1.1 FBA
Flux balance analysis (FBA) is applied to the estimation of optimal states of metabolic fluxes attained for maximizing growth, ATP, and ethanol production using S. cerevisiae GEMs (Förster et al. 2003a; Famili et al. 2003). FBA calculations around metabolites in the S matrix is formalized as follow.
where any v that satisfies this equation is a null space of S, and is part of the flux space of solutions \((\varPhi )\). FBA requires that the objective of GEM simulations is to maximize or minimize a desired linear function \((max/min\,\,v_{j} )\), such as maximize biomass production (μ, or \(v_{biomass}\)) or minimize glucose consumption (\(v_{glucose{\text{-}}intake}\)). Hence, FBA uses linear programming (LP) to calculate internal and external metabolic fluxes to max/min the objective function, according to \(Z = c^{T} v\) (Orth et al. 2010). Where c is a vector of weights, usually with zeros and a one in the targeted reaction. Some fluxes can be constrained by limiting their upper and lower boundaries, e.g. \(\alpha_{j} \le v_{j} \le \beta_{j}\). \(\alpha_{j}\) and \(\beta_{j}\) are intake or uptake fluxes measured experimentally, or forced to zero for irreversible and disabled reactions (e.g. during catabolic repression or after gene knockout). Hence, an important application of FBA is the study of phenotypic effects of gene deletions which allow a specific \(\varPhi_{mutant}\), which can be compared with an \(\varPhi_{wild{\text{-}}type}\) calculated for a wildtype cell strain. Notice that \(S_{ij}\), \(v_{j}\), N, M, \(dx/dt = 0\) and \(v_{min} \le v_{j} \le v_{max}\) is the set of constrains required for the calculation of \(\varPhi\). FBA-based calculations are routinely performed to catch optimum cellular states. For example, these are valuable to predict gene essentially in S. cerevisiae (Famili et al. 2003; Duarte et al. 2004); also serve as the initiating point for many calculations using different algorithms.
2.2.1.2 MOMA
Maximization of growth may not apply to lab mutants, where knockouts may not impose comparable constraints. For this case, the method of minimization of metabolic adjustment (MOMA) establishes that knockout strains undergo minimal changes in metabolic fluxes \((x_{i} )\) compared to the wild type \((w_{i} )\) (Segrè et al. 2002). Hence, this method has the following minimization problem.
MOMA seeks the minimal distance between two points in \({\Upphi}\) of the wild-type and knockout strains subjected to the same set of FBA constraints, but using quadratic (QP) solver instead. Also, MOMA defines another set of constraints, \(v_{d} = 0,\forall \,d \in A\)—where d and A are the index and set of deleted reactions. The aim is to find a x in \(\Phi _{knockout}\) for which the Euclidean distance from \(\Phi _{wild{\text{-}}type}\) is minimized by \(f(x) = \frac{1}{2}x^{T} Qx + c^{T} x\). This contains linear and quadratic parts of the objective function (Q and cT).
2.2.1.3 ROOM
The regulatory on/off minimization algorithm (ROOM) is used for predicting metabolic fluxes at the steady-state after gene knockouts (Shlomi et al. 2005)—similar to FBA. ROOM establishes that cells do not evolve to cope with non-natural knockouts but regulatory mechanisms seem to minimize flux changes of knockout strain.
where, for each flux j, \(y_{j} = 1\,(1 \le j \le N)\) for a significant flux change in \(v_{j}\), and \(y_{j} = 0\) otherwise. As \(y_{j} \in \{ 0,1\}\) is an integer constrain, ROOM solves this as a mixed-integer LP (MILP) problem. Flux distributions satisfy the same set of constrains as FBA and MOMA, in addition to the following constrains.
where \(w^{u}\) and \(w^{l}\) are thresholds determining the significance of flux changes. \(\delta\) and \(\varepsilon\) are relative and absolute ranges of tolerance with values 0.03 and 0.001 for flux predictions, and 0.1 and 0.01 for lethal predictions.
FBA, MOMA and ROOM were applied to the calculation of external metabolic fluxes in two petite mutants of S. cerevisiae under respiratory deficient conditions. The iFF708 GEM was constrained with external fluxes from chemostat cultivations of these mutants (Cakir et al. 2007). Ethanol production was simulated using various objective functions, such as maximizing/minimizing oxygen consumption. GEM predictions resulted more accurate when using FBA, ROOM and MOMA, in this successive order. However, internal metabolic fluxes in central metabolism calculated over a pyruvate-carboxylase mutant were more accurately predicted with MOMA compared to FBA (Segrè et al. 2002).
2.2.1.4 OptKnock
This is the first, on purpose, framework for targeting gene knockouts which constrain the overproduction of a biochemical (Burgard et al. 2003). Using a bi-level optimization problem, OptKnock establishes one particular cellular objective (e.g. \(\hbox{max} \,v_{biomass}\)), and another consisting on maximizing the metabolic engineering objective (e.g. \(max\,\,v_{biofuel\,or\,chemical}\)). Hence, this method is based on the idea that metabolite overproduction is obligatory coupled with a cellular objective, and a combination of gene knockouts which maximize both is found to solve Sv = 0. OptKnock is similarly constrained as FBA, with the following distinctive constrains.
where \(y_{j}\) assumes 0 or 1 if a reaction j is non-active (knockout) or active, respectively. K is the number of allowable knockouts. The bi-level formulation of OptKnock is solved through MILP.
Agreement of gene knockouts predicted by OptKnock and results with mutant strains overproducing succinate, lactate, and 1,3-propanediol, confirmed the value of this algorithm for analysis of targets for gene manipulations (Burgard et al. 2003). In S. cerevisiae, this framework was used to confirm the value of targeted genes suggested by OptGene (Patil et al. 2005) and MOMA for metabolic engineering of yeast to produce vainillin (Brochado et al. 2010).
2.2.1.5 OptStrain
Through OptStrain, pathway modifications can be achieved by gene knockouts and knockins (Pharkya et al. 2004). This utilizes a comprehensive database of cellular reactions (the universal database). A combinatorial optimization is used for searching a set of non-native functions, obtained from the universal database, which is added to the GEM host to enable the synthesis of the targeted biochemical. Biochemical reactions can be also removed when they constrain the overproduction. OptStrain consists of two steps of optimization, one consisting in maximizing the biochemical yield.
where \(\Re\) is the set of substrates, \(MW_{i}\) is the molecular weight of metabolite i. In the second optimization problem, OptStrain computes the minimum number of non-native reactions required to reach the maximum yield calculated in the first step. To do that, the following objective function is stablished.
The elimination of reactions from the augmented network is performed with the OptKnock framework in the last step (Burgard et al. 2003).
This approach uses a set of non-natural reactions that, otherwise, can be added to the GEM manually. That is, whereas OptStrain can be used for targeting gene modifications in E. coli for production of vanillin (Pharkya et al. 2004), in S. cerevisiae, the heterologous reactions were incorporated in the iFF708 GEM and used OptGene, OptKnock and MOMA to seek for gene deletions (Brochado et al. 2010). The intensive computational time required by OptStrain is probably the main disadvantage of this method.
2.2.1.6 OMNI
Experimental metabolic fluxes are used in the optimal metabolic flux identification (OMNI) framework to recognize a reaction set which leads the consistency between prediction and experimental (Herrgård et al. 2006a). In order to find the optimal solution of a metabolic reaction set which match model predictions and experimental data, the problem can be formulated as a bi-level optimization problem. An outer optimization problem that searches for a set of reactions to incorporate in the model, and an inner optimization problem which computes a flux distribution to solve the FBA problem with the following objective function.
It can be seen that the bi-level optimization formulation problem (1, 0) in the OMNI is similar to that used in OptKnock. Here, E represent the exchanged, measured fluxes, F is the set of reactions that cannot be removed from the model, and D the set of reactions which can be removed from the model. The linear programming nature of the inner problem allows for the overall problem to be solved by MILP. The OMNI method can be potentially used for deciphering unnecessary reactions in a GEM which, upon deletion, increases the accuracy of model predictions and experimental fluxes. Therefore, OMNI is also reliable for analyzing evolved strains through the evolution of fluxes.
2.2.1.7 OptReg
Besides finding gene knockouts and knockins, OptReg also seeks for modulation of gene functions (Pharkya and Maranas 2006). This algorithm requires optimal metabolic fluxes of the wild-type strain calculated with FBA—it is desirable to constrain the GEM with few experimental fluxes. A min/max problem is then solved to maximize \(v_{biochemical}\), constrained as following.
\(M_{exp}\) is the set of reactions which fluxes are fixed with experimental values. Minimum and maximum values for each flux through reaction j are denoted \(v_{j,L}^{0}\) and \(v_{j,U}^{0}\). Then, modeling of genetic manipulations based on three sets of binary variables (0, 1) for each reaction j are included to all possible combinations in the model: gene downregulation \((y_{j}^{d} )\), upregulation \((y_{j}^{U} )\) and knockout \((y_{j}^{k} )\). The fluxes are calculated based on the following constrains.
where \(v_{j}^{min}\) and \(v_{j}^{max}\) are minimized and maximized fluxes according to some specific considerations. The strength parameter C contains values between 0 and 1. A unique optimum solution value to the inner primal and the dual problem can be solved by MILP—see details at Pharkya and Maranas (2006).
Results from simulations with other algorithms have revealed the existence of synergism between reaction deletions and modulations, implying that the simultaneous application of both types of genetic manipulations produces more promising results. For example, the regulation of phosphoglucomutase activity in conjunction with the deletion of the oxygen uptake rate function and pyruvate formate lyase, yields 99.8% of maximum theoretical ethanol yield in E. coli. This yield was higher that when all the enzymes were deleted (Pharkya and Maranas 2006).
As the number of gene modifications increased due to the more objectives to cover in metabolic engineering projects, the necessity of algorithms with the ability to recognize more than three to four gene targets is clear. For example, the artemisinic acid-producing strain of S. cerevisiae required around seven gene modifications, including knockins and modulations (Ro et al. 2006).
2.2.1.8 EMILiO
The enhancing metabolism with iterative linear optimization (EMILiO) algorithm aims to meet increasing demands of the number and variety of genetic manipulations involved in metabolic engineering (Yang et al. 2011). Derived from OptKnock and OptReg, EMILiO uses a successive LP solution to individually optimize reaction fluxes, thus incrementing the scope of strain design. EMILiO identifies the optimal set of modified reactions and their optimal fluxes for overproduction of a target biochemical subjected to two objective functions, a cellular one \(max\,\,v_{j} = v_{bio} - \varepsilon \times v_{chemical/biofuel}\) and a biochemical-production one \(max\,\,v_{chemical/biofuel}\), which are constrained as following.
where the product \(\varepsilon \times v_{chemical/biofuel}\) is a small weighted minimization—\(\varepsilon = 0.001\). This algorithm also couples biochemical production with growth, where \(v_{bio}^{min}\) represents the minimum growth required for product formation and \(c_{p}\) the objective vector of the exchange fluxes of the targeted metabolites. Therefore, EMILiO is formulated as a bi-level optimization problem with additional constrains: \(w^{L} \;{\text{and}}\;w^{U}\) are slack variables for the lower and upper bounds, and \(\mu^{L} \;{\text{and}}\;\mu^{U}\) dual variables for the lower and upper flux bounds. Compared with OptReg, EMILiO is faster and obtains similar results.
2.2.1.9 GDLS
The genetic design through local search (GDLS) method also aims to meet increasing demands of the number and variety of genetic manipulations involved in metabolic engineering (Lun et al. 2009). GDLS starts with a user-defined strategy which then uses for searching better ones, limited to a maximum size (M). Best M strategies are used for another round, resulting in k additional manipulations. This approach continues until no better manipulations can be found—see Lun et al. (2009). GDLS initiates with a reduction of an FBA model. Dead-end reactions that do not carry any flux are deleted and reactions with linked metabolites are included in one reaction as following, \(S_{{ij_{1} }} v_{{j_{1} }} + S_{{ij_{2} }} v_{{j_{2} }} = 0;\;{\text{thus}}\;v_{{j_{1} }} = - S_{{ij_{2} }} /S_{{ij_{1} }} v_{{j_{2} }}\). Then, \(v_{j}\) is maximized or minimized subject to \(S_{ij} v_{j} = 0\), for \(v_{min} \le v \le v_{max}\). If \(v_{j}^{L}\) and \(v_{j}^{U}\) are the minimizing and maximizing individual fluxes, for any reaction with \(v_{j}^{U} \le v_{j}^{L}\), this is removed from model. Then GDLS looks for genetic manipulation strategies as a bi-level optimization problem and converting them to an optimization MILP problem, having \(max\,g_{j} v_{j}\) as the objective function, and the following constrains.
where G is the L × N matrix, with L genetic manipulations. g is the synthetic vector, y is the knockout vector, and C is the maximum number of knockouts. Converting the bi-level problem to a MILP problem using dual variables, results in the following constrains: \(\lambda\) is used for equality constrains, \(\upsilon \;{\text{and}}\;\mu\) for the lower and upper bounds, and \(\xi\) for \(v_{j} = 0\) if \(y_{j} = 1\), and D a scalar for ensuring that \(\xi_{j}\) is effectively unconstrained. The summaries represent interactions for each knockout strategy until the best set of manipulations is found (Lun et al. 2009). Compared with OptKnock, GDLS is ten times faster when targeting the same number of genes.
2.2.1.10 SIMUP
This is a bi-level based framework that searches for gene knockouts to allow the utilization of two carbon sources simultaneously (Gawand et al. 2013)—e.g. glucose and xylose from biomass hydrolysates. This algorithm is based on the idea that lethality of a gene knockout depend on the external nutrient conditions, without considering regulatory networks. The framework was formulated as a bi-level optimization problem to maximize the following objective functions.
where \(\mu\) is the specific growth rate of the mutant strain, and \(\mu_{WT}\) for the wild type strain. Superscripts define the growth condition—e.g. 1 for glucose and xylose, 2 for glucose and 3 for xylose. Calculations with the GEM are performed using the following constrains.
Superscripts L and U define lower and upper bounds and subscrips rev and irr are for reversible and irreversible. C is the coefficient vector of the objective function. \(K_{max}\) is the limit of reactions that can be deleted. Decision variables \(y_{j}\) have a value of 0 or 1. This is converted to a MILP problem.
2.2.2 GEM Analysis Constrained by Metabolism and Gene Expression
Besides invariant constraints that limits possible cellular behavior, such as stoichiometry, capacity, and thermodynamic limits, there are also variant constraints that limit allowable behavior, such as enzyme kinetics and regulation of gene expression which are adjustable through evolutionary processes (Palsson 2000). Both groups of constraints can be applied to narrow the possible space of solutions for the attainable distribution of metabolic fluxes.
Incorporation of gene expression data from microarrays and RNAseq technologies into GEMs can be achieved by Boolean logic equations representing the transcriptional regulatory structure—e.g. 1 for transcriptionally active gene and 0 for the opposite (Covert et al. 2001). This structure was established on the base that mRNA accumulation depends on both, a time interval and a defined environmental condition. In a metabolic reaction, X can be converted in Y, and Y may interact with the binding site of gene A which catalyzes the conversion of X. Hence, the transcription of A can be expressed as \(transformation = IF(A)\,AND\,NOT(Y)\). In reactions conditioned by the presence of both the metabolite and the enzyme, this can be represented as \(rxn = IF(X)\,AND(A)\). If the presence of all the regulated enzymes in the metabolic network is determined for a time interval, then one can stablish a set of constrains when the absence of a given enzyme transcript is found during this interval.
where \(v_{j}\) is the flux through the reaction at a given time point. After, the GEM can be converted in a problem that can be solved by FBA. This strategy is useful for calculating the effects of gene mutations and knockouts as well as for simulating gene expression profiles which can rise with new components and interactions in biological networks (Covert et al. 2001, 2004).
2.2.2.1 GIMME
The gene inactivity moderated by metabolism and expression (GIMME) algorithm uses quantitative gene expression and various metabolic objectives to calculate metabolic fluxes (Becker and Palsson 2008). This algorithm works in two steps. First, the algorithm finds maximum fluxes through a metabolic network with required metabolic functionalities (RMF), using FBA and typical constraints. The second step involves the calculation of a set of minimum available reactions that best fit the quantitative data \(\left( {min \sum {c_{i} \times \left| {v_{i} } \right|} } \right)\). Those reactions should operate above some minimum level—a percent of those found in the first step. This can be solved as a linear programing problem subjected to the following constrains.
where \(X_{i}\) is the normalized gene expression data, and \(X_{cutoff}\) is the cutoff value set by the user. Since the algorithm provides an inconsistency score indicating the consistency of gene expression data with a particular metabolic objective, this can be used to check biological experiments and as an intuitive approach in adaptive evolution and rational design of metabolic networks. Thus, this algorithm was used for targeting gene modifications to increase lactate production in E. coli strains with knockouts in the Phosphate acetyltransferase (Pta) and the aldehyde-alcohol dehydrogenase E (AdhE) and exposed to an evolution process (Becker and Palsson 2008).
2.2.2.2 E-Flux
E-Flux (flux-expression method) uses FBA to calculate maximum metabolic fluxes constrained by measured gene expression (Colijn et al. 2009). This approach modifies the typical FAB to the following optimization problem.
Maximum flux \((v_{j} )\) is constrained by gene expression according to \(maxFlux = f(G_{1} )\), \(b_{j} = f(expression\,level\,of\,genes\,associated\,to\,reaction\,j)\). Expression data is represented by \(y_{ijkg}\), which is the log transformation of the signal measured in the ith channel, jth chip, kth experimental condition, and gth gene, subjected to an error ε ijkg , thus: \(y_{ijkg} = \mu_{ij} + G_{g} + (AG)_{jg} + (DG)_{ig} + \hat{y}_{kg} + \varepsilon_{ijkg}\). Where \(\mu_{ij}\) is the average for channel j of chip i, \(G_{g}\) the effect of gene g, \((DG)_{jg}\) the effect of chip j and gene g, \((DG)_{ig}\) the effect of channel i and gene g, and \(\hat{y}_{kg}\) the effect of variety k and gene g. With E-Flux, it is generated the constraint vectors a and b from control and experimental conditions. Thereby, b is a vector weighing the magnitude of metabolic fluxes and introduce additional constrains for a given FBA-based objective function.
This method was utilized for predicting the metabolic state constrained by gene expression data, and was useful for targeting the impact of different drugs in Mycobacterium tuberculosis. This approach can be also used to calculate a consistency correlation among gene transcription and translation. In S. cerevisiae, for example, this correlation is 0.61% (Ideker et al. 2001). Therefore, additional care should be put in model predictions when using gene transcription levels.
2.2.2.3 Random sampling
Transcriptional regulation of key metabolic enzymes can be evaluated by the Random Sampling method (Bordel et al. 2010). This algorithm requires the calculation of the space of feasible solutions \((\varPhi )\) among a set of different strains or environmental conditions. Random Sampling then defines a set of possible flux distributions by randomly sampling the \(\varPhi\)—particularly in the corners. These values are used for the calculation of an average and standard deviation for every GEM reaction and then used them for the estimation of a significant change in flux.
A significant change in gene expression between conditions can be also calculated based on \(p_{i}\) values from transcriptomics analysis \(\left( {Z_{j}^{ex} = \pm inver\,f\left( {1 - p_{i} /2} \right)} \right)\). Comparison between both values allows the identification of enzymes showing a significant correlation between expression and metabolic flux \(\left( {P = \phi \left( {Z_{j}^{flux} } \right)\phi \left( {Z_{j}^{exp} } \right)} \right)\). The classification of enzymes according to their regulation can be as follow: enzymes showing transcriptional regulation, posttranscriptional regulation and metabolic regulation.
Transcriptomic data from different experiments consisting in gene knockouts and diauxic growth in S. cerevisiae were used along with the iFF708 GEM to analyze gene expression programs under these conditions. This kind of analysis allowed for the identification of a group of genes regulated by certain transcription factors (Bordel et al. 2010; Österlund et al. 2013; Caspeta et al. 2014b).
2.2.2.4 MADE
The metabolic adjustment by differential expression (MADE) is a method to map expression data onto a metabolic network, using non-arbitrary expression thresholds (Jensen and Papin 2011). Base on the statistical significance of an increase (I; 1), decrease (D; −1) or constancy (C; 0) of gene expression, MADE calculates a vector of binary expression states \((x_{1} \ldots x_{n} )\), \(x_{i} \in \{ 0,1\}\), for n conditions. This vector is partitioned into three sets I, D and C, and the optimization objective is the weighted sum:
MADE then maximize the sum of the objective function: \(max\sum\nolimits_{i = 1}^{n - 1} {f_{i \to i + 1} (x)}\)
This results in a mixed inter problem solved by MILP. MADE was used to construct a set of models that better reflect adjustments in the metabolism when the S. cerevisiae transits from fermentative to glycerol-respiration, attaining 98.7% of possible changes in expression (Jensen and Papin 2011).
2.2.2.5 PROM
The probabilistic regulation of metabolism (PROM) requires a GEM, a regulatory network structure based on gene-TFs interactions, abundant gene expression data, and information about enzymatic regulation by metabolites (Chandrasekaran and Price 2010). This establishes the possibility to represent gene states and gene-TF interactions—e.g. gene A is active when the regulating TF B is off or \(P(A = 1|B = 0)\). This is evaluated with abundant microarray data—e.g. the P of gene A to be ON is 0.8, if this appears 80% of the time in microarray experiments when TF B is knocked out. These gene state values are used to constrain metabolic fluxes in a GEM using FBA method—e.g. \(v_{max}\) trough gene A is \(0.8xv_{max} ;{\text{uperbound}} = Pxv_{max}\). When the constrains have been set in the GEM, the optimal solution space for the desired objective function is solved as a LP problem using FBA. Predictions of E. coli growth rate upon different knockout strains were accurately predicted with PROM—correlation coefficient of 0.96.
2.2.2.6 TIGER
The toolbox for integrating genome-scale metabolism, expression, and regulation (TIGER) uses Boolean or multilevel rules to stated arrangements of the relation between gene-TF-metabolite in the form: TF B and (not metabolite M) then gene D, then the constrain set \(A = (1, 0, 1)\) is stablished for this state (Jensen et al. 2011). A is converted to an inequality \(( - 1 \le 2x + 2y - 4I_{1} \le 3)\). The structure of simulations then gets stablished as following.
The inequality constrain A is converted to an indicator of reaction participation \((R_{i} )\). Thus, \(v_{j}^{min} R_{i} \le v_{j} \le v_{j}^{max} R_{j}\). Boolean rules are converted to a MILP problem.
Variations on GIMME and MADE algorithms can be made for contextualizing specific models with global expression data using TIGER. The flexibility of TIGER for using Boolean or multilevel rules formats allows more accurate descriptions of cellular functions, such as reactions with isozymes and protein quaternary structures, and hence flux control and transcriptional regulation. The authors used these features to identify and solve inconsistencies within existing transcriptional regulatory networks in the GEM iND750 of S. cerevisiae.
2.3 Inverse Metabolic Engineering Through Adaptive Evolution
Adaptive laboratory evolution (ALE) combined with whole-genome sequencing and global analysis has become a compelling strategy to study the biological basis of evolution (Dettman et al. 2012). Combined with ‘Omics’ technologies, the evolutionary engineering can lead to a comprehensive understanding of the basis of microbial evolution (Dragosits and Mattanovich 2013). These can serve for a rational application of recombinant DNA to generate the desired phenotype in an anticipated cell host. This approach is called inverse metabolic engineering.
Compared with procedures to generate temporal tolerance, ALE experiments generate heritable tolerance phenotypes. Spontaneous mutations in microbial populations occur at a rate close to 0.0033 per genome (Drake 1991). Thus, microbial evolution can be applied to populations exceeding 1011 cells per liter and continuous evolution can be more effective than step-wise approaches (Sauer 2001). Mutations can be appearing because of single-nucleotide polymorphisms, DNA rearrangements and horizontal DNA transfer (Arber 2000). The number of mutations can change with cell type and hardness of environmental condition, for instance, metabolic stress, stationary phase or high temperature (Sauer 2001; Caspeta et al. 2014b). During adaptive laboratory evolution, the fitness can be measured by competition (Sauer 2001).
where \(x_{i}\) and \(x_{j}\) are cell densities of two populations, competitive fitness of one strain can be quantified by the selection coefficient \(S_{ij}\).
3 Systens Metabolic Engineering for Biomass-to-Biochemicals Conversion with S. cerevisiae
The application of methods described above for metabolic engineering of S. cerevisiae for the production of biofuels and chemicals from biomass-based sugars is discussed below.
3.1 Ethanol Overproduction
Thus far, one of the most important applications of the systems metabolic engineering is to generate gene modifications to reduce glycerol formation during ethanol production. Glycerol works as the yeast’s predominant sink of the NADH accumulated during aerobic growth, and its formation restores the redox balance in the cytosol under anaerobic conditions, and when the electron transport chain (ETC) is damaged (Van Hoek et al. 1998). Since NADH is used in ETC as a proton donor to move electrons and produces ATP, an important factor in the GEM is to accurately reproduce the amount of ATP produced from the movement of two electrons in the ETC—the P/O ratio. Using the iFF708 GEM, the formation of ATP in the glycolysis pathway and ETC can be distinguished for calculating the amount of ATP produced per NADH during glucose consumption. Using the exometabolome of cultivations in chemostats, it is possible to fix the NADH oxidized during glycerol synthesis and ATP production through ETC. The P/O value was calculated in 1.04 (Famili et al. 2003), which is similar to 0.95 previously reported (Verduyn et al. 1991) ~12.5 mol of ATP per mole of glucose. Ethanol and glycerol productivities in chemostats were accurately calculated with the constrained iFF708 (Famili et al. 2003). The same model was only able to reproduce ethanol and glycerol yields in batch cultivations when gene expression data constrained GEM calculations (Akesson et al. 2004). Incorporation of transcriptomic data also improved the prediction of internal fluxes and the metabolic adjustments during the transition from glucose to glycerol (Akesson et al. 2004; Jensen and Papin 2011)—e.g. from fermentation to respiration. In the latter case, MADE based analysis of metabolic fluxes matched 83.5% of gene expression transitions during the diauxic growth.
Since the accumulation of NADH stimulates glycerol synthesis, one criterion used for reducing its formation was to use this cofactor in the synthesis of ethanol or biomass (Bro et al. 2006). To evaluate strategies, the iFF708 GEM was constrained with gene knockouts/knockins for activating or inactivating metabolic fluxes of glycerol synthesis or the accumulation of NADH. In silico evaluation of strategies resulted in the elimination of glycerol formation and an increase of 10% in ethanol yield. The best strategy consisted in the insertion of the non-phosphorylating, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase gene (GapN). Implementation of this strategy resulted in a decrease of 40% in glycerol accumulation, but ethanol yields just increased by 3%, whereas growth remained unaffected. Reducing the expression of the NAD-dependent glycerol-3-phosphate dehydrogenase (Gpd1), a key enzyme of glycerol synthesis, showed a 44–61% reduction of glycerol yield, 2–7% increase of ethanol yield, and 20% reduction of biomass during anaerobic fermentation (Pagliardini et al. 2013). However, Gpd1 mutants were most susceptible to stress probably because of a reduction in the ATP yield or an inefficient response to cell-wall damage stress.
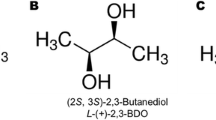
Experimental data for oxygen and glucose uptake kinetics from fed-batch cultivations, as well as frontiers parameters (e.g. initial and final volume, biomass concentration) were used to constrain the iND750 GEM (Hjersted et al. 2007). The limited model was used for calculating the dynamic of ethanol productivity under various cultivation strategies: switching dissolved oxygen concentrations from 50-to-0% DO at air saturation, cultivation media with glucose, xylose and 50%/50% glucose/xylose, and changing the ethanol inhibition constant. These simulations served for a dynamical screening of gene modifications to increase ethanol productivity. Interestingly, this strategy predicts the 4% increment in ethanol production with gene modifications used by Bro et al. (2006). Furthermore, new gene insertions were proposed to increase ethanol yield by 8%. In both studies, authors concluded that conversion of NADH to NADPH and its further utilization for ethanol and biomass synthesis can lead to decrease glycerol formation. For instance, cytosolic NADPH consumption for biomass synthesis is ~4.8 mmol/gDW. In another study, the overexpression of the NADPH-dependent, modified Bdh1 which enzyme product catalyzes the oxidation of (R,R)-2,3-butanediol to (3R)-acetoin decreased glycerol production (Celton et al. 2012). The insertion of an alternative oxidase (AOX) or the overexpression of a NADH oxidase also reduces the accumulation of glycerol, but also the production of ethanol (Vemuri et al. 2007). Higher negative effects were observed with AOX which insertion affected mitochondrial functions including downregulation of the mitochondrial inner membrane ADP/ATP translocator (AAC1).
In silico knockout of Gdh1 (NADP-dependent glutamate dehydrogenase) and overexpression of Gdh2 (NAD-dependent glutamate dehydrogenases) in the iMM904 GEM was used to calculate external metabolic fluxes. This in silico strain reduced NADPH consumption during ammonium assimilation for xylose fermentation in recombinant yeast strains carrying Xyl1 and Xyl2 (Mo et al. 2009). These results showed that ethanol production from glucose and xylose need opposite metabolic engineering strategies when xylose is metabolized via the two-step reduction-oxidation pathway.
Exometabolome of the wild-type and Gdh1/Gdh2 mutant strains was incorporated as constraints in this model and calculations of internal fluxes were performed. Results were consistent with intracellular metabolite levels and fluxes previously reported. In this study, predictions with FBA, MOMA and linear MOMA perform similarly (Mo et al. 2009). However, under similar simulations using exometabolome of nuclear petit yeast mutants, the iFF708 GEM predicted better results with FBA and ROOM than with MOMA (Cakir et al. 2007). The compartmentalization of both GEMs is the only difference in calculations.
3.2 Xylose Utilization
Xylose comprises ~30% of total biomass-based fermentable sugars but is not naturally metabolized by S. cerevisiae. There are two distinct approaches to introduce this metabolic function in the yeast. One consisting in the heterologous expression of Xyl1 and Xyl2 coding for the NADH-preferring xylose reductase and the NADPH-preferred xylose dehydrogenase from Pichia stipitis; and another consisting in the expression of the xylose isomerase (XylA) from bacteria (Jeffries 1985). The XYL1/XYL2 strategy is disadvantageous since the yeast cannot easily deal with the redox balance. To find the constraints that limit xylose utilization, FBA was used to calculate metabolic fluxes with a model of xylose metabolism including central metabolism and a P/O ratio of one (Jin and Jeffries 2004). This study ended with the incorporation of the xylulokinase activity (Xyl3) in the Xyl1/Xyl2 background. FBA based calculations predicted that maximum ethanol production in this strain could be reached under oxygen-limited conditions, a fact that was proved experimentally.
An inverse metabolic engineering strategy was also used to seek for constraints limiting xylose utilization in a XYL1/XYL2 mutant strain. DNA fragments of a genomic library of P. stipitis were used to complement this strain (Jin et al. 2005). Serial dilutions of strain populations carrying the complementary gene were used to enrich the population with individuals having the useful gene function. 16 colonies were selected, and their plasmids sequenced. 10 out of the 16 strains harbored plasmids with the Xyl3 gene, and one with high homology to S. cerevisiae Tal1 encoding the transaldolase, an enzyme of the non-oxidative pentose phosphate pathway. Yeast strains with Xyl1, Xyl2, Xyl3 and PsTal1 insertions increase xylose consumption and ethanol production by 100% and 70% compared with the parental strain. Similarly, the overexpression of a native xylulokinase (Xks1) in a recombinant yeast carrying Xyl1/Xyl2 increased xylose consumption which was then improved by chemical mutagenesis and adaptive evolution over 60 days (Liu and Hu 2010).
Anaerobic fermentation of xylose to ethanol via the two steps strategy was possible with the insertion of the Xks1 gene (Eliasson et al. 2000). However, the resulting strain was unable to grow in the absence of oxygen. Adaptive laboratory evolution of this strain over 460 generations on, consecutively, aerobic, microaerobic and anaerobic serial cultivations allowed for the generation of strains able to utilize xylose under anaerobic conditions (Sonderegger and Sauer 2003). Transcriptomics and metabolic flux analyses of these strains cultivated in chemostats with xylose and xylose-glucose under aerobic conditions and xylose-glucose under anaerobic conditions, suggested that cytosolic NADPH formation and NADH consumption enabled a high flux through the two-step oxidoreductase reactions (Sonderegger et al. 2004). Anaerobic fermentations were not improved probably because the absence of a NADH sink or by an increased production of ATP—similar results were found in (Wasylenko and Stephanopoulos 2015). Complementation of the pathway can be achieved by reducing acetate to ethanol through the activity of the acetylating acetaldehyde dehydrogenase (AadH) into S. cerevisiae, which served as a sink for NADH excess. This strategy also increased ethanol production (Wei et al. 2013).
Adaptive laboratory evolution of a respiratory deficient, Xyl1/Xyl2 yeast strain, lacking the cytochrome C oxidase subunit IV was used as a strategy to increase growth rate and ethanol production under anaerobic conditions. The specific growth rate, ethanol yield, and xylitol yield of the evolved strain on xylose were 0.06 1/h, 0.39 g/g, and 0.13 g/g, respectively (Peng et al. 2012). An S. cerevisiae strain over-expressing genes of the non-reductive pathway of xylose utilization and the non-oxidative PPP, was evolved through a three steps evolution strategy (aerobic, anaerobic and xylose-limited chemostat). This approach allowed for the generation of a strain with a high xylose consumption (1.86 g/gDCW/h) and ethanol conversion yield (0.41 g/g) (Zhou et al. 2012). Combining different media compositions with mixtures of glucose, xylose and arabinose, the use of adaptive evolution can be also used to generate yeast strains capable of producing ethanol at a yield of 0.43 g/g of total sugars under anaerobic fermentation (Wisselink et al. 2009). The parental yeast strains overexpressed XylA from Piromyces sp., endogenous genes of the PPP and Xks1, and Lactobacillus plantarum AraA, AraB, and AraD genes.
Insertion of XylA in S. cerevisiae avoids the utilization of pyridine nucleotide cofactors during xylose consumption. According to a 13C-FBA performed in a central metabolism model, yeast strain carrying XylA had a low distribution of metabolic fluxes in the non-oxidative PPP and did not show a full carbon catabolic repression typical of glucose fermentation (Wasylenko and Stephanopoulos 2015)—mutation of Hxt7 and Gal2 in a XylA mutant strains also generated glucose-insensitive phenotypes (Farwick et al. 2014). Higher concentrations of NADH were observed in xylose consumer strains growing under anaerobic conditions, whereas energy charge remained similar (Wasylenko and Stephanopoulos 2015). Lower metabolic fluxes in the last tree reactions of glycolysis seemed to limit the production of ethanol with xylose under anaerobic conditions. Therefore, the productivity and yield of anaerobic conversion of xylose to ethanol were 6% lower compared with 12% increase observed with glucose under anaerobic and aerobic conditions. Compared with aerobic conditions, glycerol accumulation increased in cultivations with glucose and xylose under anaerobic conditions.
3.3 Tolerance to Toxic Byproducts and Temperature
Adaptive laboratory evolution has been successfully used for generating yeast strains tolerant to furfural, HMF and acetate (Liu et al. 2005; Heer and Sauer 2008). Increased tolerance to 30 and 60 mM of furfural and HMF were observed in yeast strains isolated from ALE experiments (Liu et al. 2005). These strains also increased glucose consumption. Two isolated strains efficiently transform HMF to 2,5-bis-hydroxymethylfuran and one transformed furfural into furfuryl alcohol (Liu et al. 2005). Evolved strains reduced the lag phase of growth suggesting that furfural conversion into its alcohol is the mechanism for yeast adaptation to this byproduct (Liu et al. 2005; Heer and Sauer 2008). Evolution of the industrial strain ethanol-red of S. cerevisiae in spruce hydrolysate and high temperature resulted in the selection of strains capable of converting spruce hydrolysates into ethanol with high efficiency (Wallace-Salinas and Gorwa-Grauslund 2013). Compared with evolved strains selected with furfural and HMF alone, which increased the conversion of these into their alcohols, these strains tolerance did not rely on higher reductase activities, but rather on a higher thermotolerance.
Microarray analysis of strains with different abilities to tolerate HMF permitted the identification of 15 reductase/dehydrogenase genes, whose overexpression in poor resistance yeast strains generated tolerance to HMF (Petersson et al. 2006). Among them, the overexpression of NADH/NADPH dependent Adh6, which converts HMF into 5-hydroxymethylfurfuryl alcohol, resulted in the highest increment in HMF transformation, and tolerance. Proteomic and transcriptomic analyses of S. cerevisiae cultivated with furfural showed the downregulation of genes involved in glycerol synthesis, changed the expression of alcohol dehydrogenases, and reduced the levels of cytosolic NADH (Lin et al. 2009), suggesting an increased demand in redox potential for transformation of furfural into its alcohol. This demand seems to cause the long lag phase of ethanol production in cultivations with furfural (Liu et al. 2005). Besides the changes in redox metabolism, yeast subjected to furfural also change expression levels of genes involved in oxidative and salt stress as well as the TFs, Msn2/Msn4, Yap1, and Hsf1 which regulate different stress responses (Lin et al. 2009). Overexpression of Yap1 and Msn2 highly correlated with the increase in yeast tolerance to furfural and HMF (Lin et al. 2009; Sasano et al. 2012). The former results were generated in cultivations with glucose as a carbon source but were also reproduced in cultivations with xylose as a carbon source (Ask et al. 2013).
The iFF708 GEM was complemented with metabolic equations for oxidative and reductive conversion of furfural into furfuryl alcohol, and constrained with experimental data from fed-batch fermentations in glucose-xylose media containing furfural. Dynamic FBA was carried out with the model and the results from simulations showed increasing fluxes though PPP, TCA cycle and serine-proline synthesis to replenish the extra consumption of NADPH (Pornkamol and Franzen 2015).
Many of the thermo-tolerant phenotypes produced trough metabolic engineering methods, have been generated by inverse metabolic engineering through adaptive evolution and multi ‘omics’ analyses. Yeast strains with improved high-temperature tolerance can be isolated after several hundreds of generations in serial cultivations at high temperatures (Cakar et al. 2012; Yona et al. 2012; Caspeta et al. 2014b). Genome sequencing and multi-‘omics’ analyses drove the identification of gene rearrangements responsible for the improved performance (Yona et al. 2012; Caspeta et al. 2014b). Remarkably, deleterious mutations in just one gene (Erg3) allowed for the parental strain to obtain 85% of the tolerant phenotype observed in seven evolved strains (Caspeta et al. 2014b). Complete and segmental duplications of chromosome III were also detected in the genome sequence and transcriptomics analyses of evolved strains (Yona et al. 2012; Caspeta et al. 2014b). From genes comprising the chromosome III, Hcm1 and Rrt12 encoding a TF involved in chromosome segregation and a probable subtilisin-family protease, partially recovered thermo-tolerance in the parental strain (Yona et al. 2012).
A possible disadvantage of thermotolerance based on Erg3 mutations is that the related yeast strains are deficient in the electron transport chain and ATP synthesis. Therefore, they displayed their thermal niche to higher temperatures and are inefficient to synthesize biomass under aerobic conditions. In fact, thermotolerant yeast strains (TTs) showed similar behavior to the parental yeast growing under anaerobic conditions (Caspeta and Nielsen 2015). Molecular responses in evolved strains cultivated under optimal (30 °C) and high temperatures (40–50 °C) were analyzed using the iIN800 GEM constrained by exofluxome and with a thermodynamic model coupling protein folding-unfolding thermodynamics and growth kinetics (Caspeta and Nielsen 2015). TTs displaced their thermal niche while keeping high tolerance to higher temperatures than the wild type strain. Also, TTs showed a preemptive response to high temperature when cultivated at 30 °C (Caspeta et al. 2016). These responses limited the growth of evolved yeasts in cultivations at optimal temperature.
3.4 Production of Biochemicals
Genome-scale metabolic modeling with the iFF708 GEM and FBA method were used to set up metabolic engineering strategies to increase the production of succinic acid (Agren et al. 2013). Objective functions included: growth maximization with limiting glucose uptake, oxygen uptake was unconstrained for aerobic and microaerobic processes, and under anaerobic conditions, this was constrained to the minimum (0.016 mmol/gDCW/h). Glucose uptake rate was constrained with experimental data, and maintenance ATP was set to 1 mmol/gDCW/h. According with model predictions, the top three single gene deletions include Mdh1, Oac1 and Dic1 coding the mitochondrial malate dehydrogenase, a mitochondrial inner membrane transporter, and a mitochondrial dicarboxylate carrier, respectively. Model simulations also detected that succinate production is sensitive to the oxygen uptake rate, and it is more sensitive for Mdh1 mutant than the Mdh1/Rip1 (Rip1 coding the ubiquinol-cytochrome-c reductase) double mutant. Transcriptional analysis of Dic1 mutant suggested that electron transport chain, ATP synthesis, sterol transport and metabolic processes for energy formation are coupled with succinate formation. Targeting gene modifications for succinate overproduction was also carried out with the OptGene algorithm and iFF708 GEM (Patil et al. 2005). The results guided the modification of a yeast strain with gene knockouts including Sdh3 (cytochrome b subunit of succinate dehydrogenase complex), Ser3p/Ser33 (3-phosphoglycerate dehydrogenase isoenzymes) (Otero et al. 2013). A maximum yield of 0.14 g/g biomass was obtained. Further evolution in media containing glycine leads the generation a strain which accumulates 0.69 g/g biomass. Transcriptomics analysis of the evolved strain lead the identification of Icl1 (isocitrate lyase) as a target to increase succinate production in the evolved strain. Incorporation of this mutations increased succinate production to 0.9 g/g biomass (Otero et al. 2013).
The production of the non-natural compound vanillin was evaluated in S. cerevisiae. Reactions for vanillin production from protocatechuate and formaldehyde were introduced in the iFF708 GEM. The set of gene knockouts for overproduction of vanillin were predicted with OptGene (Patil et al. 2005)—an algorithm that derives from OptKnock. GEM simulations predicted that deletion of pyruvate decarboxylase and glutamate dehydrogenase activities could improve vanillin production to 90% of maximum theoretical value. In a complementary study, the iFF708 GEM combined with OptGene, FAB and MOMA served to find five reactions to convert 3-dehydroshikimate, a natural intermediate in aromatic amino acids biosynthesis, into vanillin β-D-glucoside (VG) (Brochado et al. 2010). OptGene was used for predicting metabolic engineering targets. MOMA was used as the biological objective function with wild type flux distributions spanning three modes of yeast physiological responses. The optimality of the targeted genes was verified with OptKnock. Based on this and their previous analysis, two gene candidates, Pdc1 (Pyruvate decarboxylase) and Gdh1 (Glutamate dehydrogenase), were selected for strain construction. Compared with the reference strain, Pdc1/Gdh1 mutant strains produced 40% more vanillin, whereas ethanol yield was similar and protocatechuic acid yield increased.
The production of terpenoids with recombinant S. cerevisiae has been also targeted. The iMM904 GEM was used with FBA/MOMA analyses to find metabolic engineering targets for increasing terpenoids production (Sun et al. 2014). A set of single mutations predicted in simulations were tested, showing that mutation of Alt2, Hxk2 and Sor1 resulted in the highest titer of amorphadiene (55 mg/L)—the artemisinic acid precursor. Amorphadiene production at concentrations >40 g/L was reached with recombinant yeast strains overexpressing every enzyme of mevalonate pathway, using fed-batch cultivations (Westfall et al. 2012). Incorporation of additional three heterologous enzymes was needed to convert amorphadiene into the antimalarial drug artemisinin. Since terpenoids derivatives have applications as biofuels, its overproduction in yeast also became strategic. Terpenoids can serve as drop-in fuels and can substitute gasoline, diesel, and kerosene (Rabinovitch-Deere et al. 2013). Deletion of Hfd1 together with the expression of an alkane biosynthesis pathway resulted in the production of the alkanes tridecane, pentadecane, and heptadecane (Buijs et al. 2015). Metabolic flux analysis of a reduced model with 69 reactions of central metabolism was used to calculate yields for terpenoids production using the pyruvate-glyceraldehyde-3-phosphate (DXP) pathway and mevalonate pathway (MVA). Although carbon balances favor terpenoids production via DXP, further reduction occurs when redox and energy is considered (Gruchattka and Kayser 2015).
GEM simulations combined with industrial process analysis can be used for the selection of biosynthetic routes which allow the economical synthesis of the desired target molecule. For instance, this approach was useful to identify advanced biofuels as more efficient fuels in terms positive energy balances and production costs (Caspeta and Nielsen 2013). With this approach, it was also identified that the most promising route for 3-Hydropropionic acid (3HP) synthesis is the β-alanine biosynthetic route (Borodina et al. 2015). With this in mind, a yeast strain expressing the heterologous pathway for β-alanine synthesis from Bacillus cereus and its subsequent conversion into 3HP was engineered. This strain produced 3HP at a titer of 13.7 ± 0.3 g/L, and 0.14 C-mol/C-mol yield. Adaptive laboratory evolution and genome sequence of 3HP production strains at pH 3.5 revealed that mutations in Sfa1 gene encoding S-(hydroxymethyl)-glutathione dehydrogenase increased tolerance to 50 g/L of 3HP, suggesting that detoxification of 3-hydroxypropionic aldehyde via glutathione is the main factor (Kildegaard et al. 2014).
Esterification of fatty acids (FA) is another source of biofuels with properties similar to diesel. A strategy to increase the accumulation of FA involves the deactivation of beta-oxidation pathway and the increase of steryl-esters degradation (Valle-Rodríguez et al. 2014). A broader approach also involved the overexpression of TCA enzymes to replenish acetyl-CoA pull and deletion of Pox1 (encoding the fatty-acyl coenzyme A oxidase). This strategy led the accumulation of FA at concentrations up to 10.4 g/L (Zhou et al. 2016). Lipid metabolism in S. cerevisiae plays a key role in many cellular functions. To get insight on lipid metabolism, the measurement of 5636 mRNAs, 50 metabolites, 97 lipids, and 57 13C-reaction fluxes were performed (Jewett et al. 2013). The results were mapped into the iIN800 GEM, which was used to map network topologies of lipid metabolism and regulation. Results suggested that sterols are mainly regulated at the transcriptional level, whereas FA synthesis at the metabolic level. Using a GEM of Yarrowia lipolytica and multi-‘omics’ analysis, including RNAseq, metabolic profiling and lipidomics, it was found that lipid accumulation does not involve transcriptional regulation, and is associated with regulation of amino acids synthesis (Kerkhoven et al. 2016). Finally, the manipulation of structural and regulatory genes of lipid metabolism, including the overexpression of Acc1, the deletion of Ino1, and the overexpression of Rpd3 induced the production of 1-hexadecanol from xylose in a yeast strain carrying Xyl1, Xyl2 and Xyl3 (Feng et al. 2015). Adaptive evolution of these strains on xylose as a sole carbon source improved 1-hexadecanol production to a final concentration of 1.2 g/L (Guo et al. 2016).
4 Concluding Remarks
The yeast S. cerevisiae is a very tractable microorganism with a long record of useful applications in classical and modern industrial fermentations. This is hitherto the workhorse in winemaking, brewing and baking, as well as in the production of different pharmaceuticals and fuel ethanol from sugarcane and starch. The capabilities that highly positioned this yeast in these applications were however not fully appropriate under practical situations of the cost-efficient, biomass-based processes. Most importantly, the yeast did not metabolize pentoses and had an undesired limited tolerance. Also, a key challenge was to find metabolic and genetic regulatory conditions leading to the synthesis of other molecules besides ethanol in the presence of glucose, including non-natural chemicals.
Through metabolic engineering, yeast capabilities have been improved to fit practical applications of the biomass-to-biochemicals conversion processes. The current progress in the procedures for deciphering genomes, transcriptomes, proteomes, fluxomes and metabolomes, along with mathematical and computational tools, synthetic biology and evolutionary engineering, have led to a new set of technology platforms for metabolic engineering of S. cerevisiae in a holistic manner. These platforms have been providing concepts and methods to partially resolve the obstacles for cost-efficient production of chemicals and biofuels from biomass.
Many more examples of how the holistic understanding of S. cerevisiae’s biology has impacted metabolic engineering have been seen up today compared with a few years ago (Nielsen and Jewett 2008). During this period, there has been an explosion of new in silico systems biology methods for mapping detailed phenotypes and for targeting gene modifications for metabolic engineering purposes (Machado and Herrgård 2015). Combined with the advances in sequencing and synthesis of whole genomes and high-throughput technologies, these in silico methods have provided a valuable platform for increasing the production of desired chemicals and improving yeast behavior under commercial-process environments. A key challenge is to find or generate ideal metabolic and regulatory networks for supporting both, cell growth and product formation. As systems metabolic engineering becomes more robust regarding better predictions with genome-scale simulations, we will see that this challenge will be overcome in the near future.
References
Agren R, Otero JM, Nielsen J (2013) Genome-scale modeling enables metabolic engineering of Saccharomyces cerevisiae for succinic acid production. J Ind Microbiol Biotechnol 40:735–747. doi:10.1007/s10295-013-1269-3
Akesson M, Förster J, Nielsen J (2004) Integration of gene expression data into genome-scale metabolic models. Metab Eng 6:285–293. doi:10.1016/j.ymben.2003.12.002
Arber W (2000) Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev 24:1–7. doi:10.1111/j.1574-6976.2000.tb00529.x
Asadollahi MA, Maury J, Patil KR et al (2009) Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng 11:328–334. doi:10.1016/j.ymben.2009.07.001
Ask M, Bettiga M, Mapelli V, Olsson L (2013) The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnol Biofuels 6:22. doi:10.1186/1754-6834-6-22
Aung HW, Henry SA, Walker LP (2013) Revising the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism in the consensus model of yeast metabolism. Ind Biotechnol (New Rochelle NY) 9:215–228. doi:10.1089/ind.2013.0013
Bailey J (1991) Toward a science of metabolic engineering. Science (80-) 252:1668–1675. doi:10.1126/science.2047876
Barber J (2009) Photosynthetic energy conversion: natural and artificial. Chem Soc Rev 38:185–196. doi:10.1039/b802262n
Becker SA, Palsson BO (2008) Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol 4:e1000082. doi:10.1371/journal.pcbi.1000082
Boer VM, de Winde JH, Pronk JT, Piper MDW (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278:3265–3274. doi:10.1074/jbc.M209759200
Bordel S, Agren R, Nielsen J (2010) Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput Biol 6:e1000859. doi:10.1371/journal.pcbi.1000859
Borodina I, Kildegaard KR, Jensen NB et al (2015) Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine. Metab Eng 27:57–64. doi:10.1016/j.ymben.2014.10.003
Bro C, Knudsen S, Regenberg B et al (2005) Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol 71:6465–6472. doi:10.1128/AEM.71.11.6465-6472.2005
Bro C, Regenberg B, Förster J, Nielsen J (2006) In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng 8:102–111. doi:10.1016/j.ymben.2005.09.007
Brochado AR, Matos C, Møller BL et al (2010) Improved vanillin production in baker’s yeast through in silico design. Microb Cell Fact 9:84. doi:10.1186/1475-2859-9-84
Buijs NA, Zhou YJ, Siewers V, Nielsen J (2015) Long-chain alkane production by the yeast Saccharomyces cerevisiae. Biotechnol Bioeng 112:1275–1279. doi:10.1002/bit.25522
Burgard AP, Pharkya P, Maranas CD (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84:647–657. doi:10.1002/bit.10803
Cakar ZP, Turanli-Yildiz B, Alkim C, Yilmaz U (2012) Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res 12:171–182. doi:10.1111/j.1567-1364.2011.00775.x
Cakir T, Efe C, Dikicioglu D et al (2007) Flux balance analysis of a genome-scale yeast model constrained by exometabolomic data allows metabolic system identification of genetically different strains. Biotechnol Prog 23:320–326. doi:10.1021/bp060272r
Caspeta L, Nielsen J (2013) Economic and environmental impacts of microbial biodiesel. Nat Biotechnol 31:789–793. doi:10.1038/nbt.2683
Caspeta L, Nielsen J (2015) Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. MBio. doi:10.1128/mBio.00431-15
Caspeta L, Caro-Bermúdez MA, Ponce-Noyola T, Martinez A (2014a) Enzymatic hydrolysis at high-solids loadings for the conversion of agave bagasse to fuel ethanol. Appl Energy 113:277–286. doi:10.1016/j.apenergy.2013.07.036
Caspeta L, Chen Y, Ghiaci P et al (2014b) Biofuels. Altered sterol composition renders yeast thermotolerant. Science 346:75–78. doi:10.1126/science.1258137
Caspeta L, Castillo T, Nielsen J (2015) Modifying yeast tolerance to inhibitory conditions of ethanol production processes. Front Bioeng Biotechnol 3:184. doi:10.3389/fbioe.2015.00184
Caspeta L, Chen Y, Nielsen J (2016) Thermotolerant yeasts selected by adaptive evolution express heat stress response at 30 °C. Sci Rep 6:27003. doi: 10.1038/srep27003
Cautha SC, Gowen CM, Lussier FX, et al (2013) Model-driven design of a Saccharomyces cerevisiae platform strain with improved tyrosine production capabilities. 12th IFAC symposium on computer applications in biotechnology. December, Mumbai, India
Celton M, Goelzer A, Camarasa C et al (2012) A constraint-based model analysis of the metabolic consequences of increased NADPH oxidation in Saccharomyces cerevisiae. Metab Eng 14:366–379. doi:10.1016/j.ymben.2012.03.008
Chandrasekaran S, Price ND (2010) Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci USA 107:17845–17850. doi:10.1073/pnas.1005139107
Colijn C, Brandes A, Zucker J et al (2009) Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput Biol 5:e1000489. doi:10.1371/journal.pcbi.1000489
Covert MW, Knight EM, Reed JL et al (2004) Integrating high-throughput and computational data elucidates bacterial networks. Nature 429:92–96. doi:10.1038/nature02456
Covert MW, Schilling CH, Palsoon B (2001) Regulation of gene expression in flux balance models of metabolism. J Theor Biol 213:73–88. doi:10.1006/jtbi.2001.2405
DeRisi JL, Iyer VW, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. doi:10.1126/science.278.5338.680 (80)
Dettman JR, Rodrigue N, Melnyk AH et al (2012) Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol Ecol 21:2058–2077. doi:10.1111/j.1365-294X.2012.05484.x
Dobson PD, Smallbone K, Jameson D et al (2010) Further developments towards a genome-scale metabolic model of yeast. BMC Syst Biol 4:145. doi:10.1186/1752-0509-4-145
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Fact 12:64. doi:10.1186/1475-2859-12-64
Drake JW (1991) A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA 88:7160–7164
Duarte NC, Herrgård MJ, Palsson BØ (2004) Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 14:1298–1309. doi:10.1101/gr.2250904
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95:14863–14868. doi:10.1073/pnas.95.25.14863
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B (2000) Anaerobic xylose fermentation by gecombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386. doi:10.1128/AEM.66.8.3381-3386.2000
Famili I, Forster J, Nielsen J, Palsson BO (2003) Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc Natl Acad Sci USA 100:13134–13139. doi:10.1073/pnas.2235812100
Farwick A, Bruder S, Schadeweg V et al (2014) Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc Natl Acad Sci USA 111:5159–5164. doi:10.1073/pnas.1323464111
Feng X, Lian J, Zhao H (2015) Metabolic engineering of Saccharomyces cerevisiae to improve 1-hexadecanol production. Metab Eng 27:10–19. doi:10.1016/j.ymben.2014.10.001
Field CB (1998) Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240. doi:10.1126/science.281.5374.237 (80-)
Förster J, Famili I, Fu P (2003a) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13:244–253
Förster J, Famili I, Palsson BO, Nielsen J (2003b) Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae. OMICS 7:193–202. doi:10.1089/153623103322246584
Gasch AP, Spellman PT, Kao CM et al (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257. doi:10.1091/mbc.11.12.4241
Gawand P, Hyland P, Ekins A et al (2013) Novel approach to engineer strains for simultaneous sugar utilization. Metab Eng 20:63–72. doi:10.1016/j.ymben.2013.08.003
Ghosh A, Zhao H, Price ND (2011) Genome-scale consequences of cofactor balancing in engineered pentose utilization pathways in Saccharomyces cerevisiae. PLoS ONE 6:e27316. doi:10.1371/journal.pone.0027316
Giaever G, Chu AM, Ni L et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi:10.1038/nature00935
Goffeau A, Barrell BG, Bussey H et al (1996) Life with 6000 Genes. Science 274:546–567. doi:10.1126/science.274.5287.546 (80)
Gruchattka E, Kayser O (2015) In vivo validation of in silico predicted metabolic egineering strategies in yeast: disruption of α-ketoglutarate dehydrogenase and expression of ATP-citrate lyase for terpenoid production. PLoS ONE 10:e0144981. doi:10.1371/journal.pone.0144981
Guo W, Feng X (2016) OM-FBA: integrate transcriptomics data with flux balance analysis to decipher the cell metabolism. PLoS ONE 11:e0154188. doi:10.1371/journal.pone.0154188
Guo W, Sheng J, Zhao H, Feng X (2016) Metabolic engineering of Saccharomyces cerevisiae to produce 1-hexadecanol from xylose. Microb Cell Fact 15:24. doi:10.1186/s12934-016-0423-9
Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund MF et al (2006) Bio-ethanol–the fuel of tomorrow from the residues of today. Trends Biotechnol 24:549–556. doi:10.1016/j.tibtech.2006.10.004
Heer D, Sauer U (2008) Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol 1:497–506. doi:10.1111/j.1751-7915.2008.00050.x
Herrgård MJ, Fong SS, Palsson BØ (2006a) Identification of genome-scale metabolic network models using experimentally measured flux profiles. PLoS Comput Biol 2:e72. doi:10.1371/journal.pcbi.0020072
Herrgård MJ, Lee B-S, Portnoy V, Palsson BØ (2006b) Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res 16:627–635. doi:10.1101/gr.4083206
Hjersted JL, Henson MA, Mahadevan R (2007) Genome-scale analysis of Saccharomyces cerevisiae metabolism and ethanol production in fed-batch culture. Biotechnol Bioeng 97:1190–1204. doi:10.1002/bit.21332
Huh W-K, Falvo JV, Gerke LC et al (2003) Global analysis of protein localization in budding yeast. Nature 425:686–691. doi:10.1038/nature02026
Ideker T, Thorsson V, Ranish JA et al (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929–934. doi:10.1126/science.292.5518.929
Jeffries T (1985) Emerging technology for fermenting xylose. Trends Biotechnol 3:208–212. doi:10.1016/0167-7799(85)90048-4
Jensen PA, Papin JA (2011) Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics 27:541–547. doi:10.1093/bioinformatics/btq702
Jensen PA, Lutz KA, Papin JA (2011) TIGER: toolbox for integrating genome-scale metabolic models, expression data, and transcriptional regulatory networks. BMC Syst Biol 5:147. doi:10.1186/1752-0509-5-147
Jewett MC, Workman CT, Nookaew I et al (2013) Mapping condition-dependent regulation of lipid metabolism in Saccharomyces cerevisiae. G3 (Bethesda) 3:1979–1995. doi:10.1534/g3.113.006601
Jin Y-S, Jeffries TW (2004) Stoichiometric network constraints on xylose metabolism by recombinant Saccharomyces cerevisiae. Metab Eng 6:229–238. doi:10.1016/j.ymben.2003.11.006
Jin Y-S, Alper H, Yang Y-T, Stephanopoulos G (2005) Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl Environ Microbiol 71:8249–8256. doi:10.1128/AEM.71.12.8249-8256.2005
Kerkhoven EJ, Pomraning KR, Baker SE, Nielsen J (2016) Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. npj Syst Biol Appl 2:16005. doi:10.1038/npjsba.2016.5
Kildegaard KR, Hallström BM, Blicher TH et al (2014) Evolution reveals a glutathione-dependent mechanism of 3-hydroxypropionic acid tolerance. Metab Eng 26:57–66. doi:10.1016/j.ymben.2014.09.004
Lee JW, Kim HU, Choi S et al (2011) Microbial production of building block chemicals and polymers. Curr Opin Biotechnol 22:758–767. doi:10.1016/j.copbio.2011.02.011
Lin F-M, Qiao B, Yuan Y-J (2009) Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl Environ Microbiol 75:3765–3776. doi:10.1128/AEM.02594-08
Liu E, Hu Y (2010) Construction of a xylose-fermenting Saccharomyces cerevisiae strain by combined approaches of genetic engineering, chemical mutagenesis and evolutionary adaptation. Biochem Eng J 48:204–210. doi:10.1016/j.bej.2009.10.011
Liu ZL, Slininger PJ, Gorsich SW (2005) Enhanced biotransformation of furfural and hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol 121–124:451–460
Lun DS, Rockwell G, Guido NJ et al (2009) Large-scale identification of genetic design strategies using local search. Mol Syst Biol 5:296. doi:10.1038/msb.2009.57
Machado D, Herrgård M (2015) Co-evolution of strain design methods based on flux balance and elementary mode analysis. Metab Eng Commun 2:85–92. doi:10.1016/j.meteno.2015.04.001
Mo ML, Palsson BO, Herrgård MJ (2009) Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol 3:37. doi:10.1186/1752-0509-3-37
Nielsen J (1998) Metabolic engineering: techniques for analysis of targets for genetic manipulations. Biotechnol Bioeng 58:125–132. doi:10.1002/(SICI)1097-0290(19980420)58:2/3<125:AID-BIT3>3.0.CO;2-N
Nielsen (2001) Metabolic engineering. App Microbiol Biotechnol 55:263–283. doi:10.1007/s002530000511
Nielsen J, Jewett M (2008) Impact of systems biology on metabolic engineering of Saccharomyces cerevisiae. FEMS Yeast Res 64:34–50
Nookaew I, Jewett MC, Meechai A et al (2008) The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst Biol 2:71. doi:10.1186/1752-0509-2-71
Orth JD, Thiele I, Palsson BØ (2010) What is flux balance analysis? Nature Biotechnol 28:245–248. doi:10.1038/nbt.1614
Osterlund T, Nookaew I, Nielsen J (2012) Fifteen years of large scale metabolic modeling of yeast: developments and impacts. Biotechnol Adv 30:979–988. doi:10.1016/j.biotechadv.2011.07.021
Österlund T, Nookaew I, Bordel S, Nielsen J (2013) Mapping condition-dependent regulation of metabolism in yeast through genome-scale modeling. BMC Syst Biol 7:36. doi:10.1186/1752-0509-7-36
Otero JM, Cimini D, Patil KR et al (2013) Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS ONE 8:e54144. doi:10.1371/journal.pone.0054144
Pagliardini J, Hubmann G, Alfenore S et al (2013) The metabolic costs of improving ethanol yield by reducing glycerol formation capacity under anaerobic conditions in Saccharomyces cerevisiae. Microb Cell Fact 12:29. doi:10.1186/1475-2859-12-29
Palmqvist E, Hahn-Hägerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: Inhibitors and mechanisms of inhibition. Bioresour Technol 74:25–33. doi:10.1016/S0960-8524(99)00161-3
Palsson B (2000) The challenges of in silico biology. Nat Biotechnol 18:1147–1150. doi:10.1038/81125
Park JH, Lee SY, Kim TY, Kim HU (2008) Application of systems biology for bioprocess development. Trends Biotechnol 26:404–412. doi:10.1016/j.tibtech.2008.05.001
Park JM, Kim TY, Lee SY (2009) Constraints-based genome-scale metabolic simulation for systems metabolic engineering. Biotechnol Adv 27:979–988. doi:10.1016/j.biotechadv.2009.05.019
Patil KR, Rocha I, Förster J, Nielsen J (2005) Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinform 6:308. doi:10.1186/1471-2105-6-308
Peng B, Shen Y, Li X et al (2012) Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab Eng 14:9–18. doi:10.1016/j.ymben.2011.12.001
Petersson A, Almeida JRM, Modig T et al (2006) A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455–464. doi:10.1002/yea.1370
Pharkya P, Maranas CD (2006) An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Eng 8:1–13. doi:10.1016/j.ymben.2005.08.003
Pharkya P, Burgard AP, Maranas CD (2004) OptStrain: a computational framework for redesign of microbial production systems. Genome Res 14:2367–2376. doi:10.1101/gr.2872004
Pornkamol U, Franzen CJ (2015) Dynamic flux balancing elucidates NAD(P)H production as limiting response to furfural inhibition in Saccharomyces cerevisiae. Biotechnol J 10:1248–1258. doi:10.1002/biot.201400833
Rabinovitch-Deere CA, Oliver JWK, Rodriguez GM, Atsumi S (2013) Synthetic biology and metabolic engineering approaches to produce biofuels. Chem Rev 113:4611–4632. doi:10.1021/cr300361t
Ro DK, Paradise EM, Ouellet M, et al (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943. doi:10.1038/nature04640
Sasano Y, Watanabe D, Ukibe K, Inai T, Ohtsu I, Shimoi H et al (2012) Overexpression of the yeast transcription activator Msn2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J Biosci Bioeng 113:451–455. doi:10.1016/j.jbiosc.2011.11.017
Sauer U (2001) Evolutionary engineering of industrially important microbial phenotypes. In: Nielsen J, Eggeling L, Dynesen J et al (eds) Metabolic engineering. Springer, Berlin, pp 129–169. doi:10.1007/3-540-45300-8_7
Segrè D, Vitkup D, Church GM (2002) Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci USA 99:15112–15117. doi:10.1073/pnas.232349399
Shlomi T, Berkman O, Ruppin E (2005) Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci USA 102:7695–7700. doi:10.1073/pnas.0406346102
Sonderegger M, Sauer U (2003) Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol 69:1990–1998. doi:10.1128/AEM.69.4.1990-1998.2003
Sonderegger M, Jeppsson M, Hahn-Hagerdal B, Sauer U (2004) Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Appl Environ Microbiol 70:2307–2317. doi:10.1128/AEM.70.4.2307-2317.2004
Stephanopoulos G (1999) Metabolic fluxes and metabolic engineering. Metab Eng 1:1–11. doi:10.1006/mben.1998.0101
Stephanopoulos G (2007) Challenges in engineering microbes for biofuels production. Science 315:801–804. doi:10.1126/science.1139612
Straathof AJJ (2014) Transformation of biomass into commodity chemicals using enzymes or cells. Chem Rev 114:1871–1908. doi:10.1021/cr400309c
Sun Z, Meng H, Li J et al (2014) Identification of novel knockout targets for improving terpenoids biosynthesis in Saccharomyces cerevisiae. PLoS ONE 9:e112615. doi:10.1371/journal.pone.0112615
Thiele I, Palsson BØ (2010) A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5:93–121. doi:10.1038/nprot.2009.203
Valle-Rodríguez JO, Shi S, Siewers V, Nielsen J (2014) Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid ethyl esters, an advanced biofuel, by eliminating non-essential fatty acid utilization pathways. Appl Energy 115:226–232. doi:10.1016/j.apenergy.2013.10.003
Van Hoek P, Van Dijken JP, Pronk JT (1998) Effect of specific growth rate on fermentative capacity of baker’s yeast. Appl Environ Microbiol 64:4226–4233
Vemuri GN, Eiteman MA, McEwen JE et al (2007) Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104:2402–2407. doi:10.1073/pnas.0607469104
Verduyn C, Stouthamer AH, Scheffers WA, van Dijken JP (1991) A theoretical evaluation of growth yields of yeasts. A Van Leeuwenhoek 59:49–63. doi:10.1007/BF00582119
Wallace-Salinas V, Gorwa-Grauslund M (2013) Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol Biofuels 6:151. doi:10.1186/1754-6834-6-151
Wasylenko TM, Stephanopoulos G (2015) Metabolomic and (13)C-metabolic flux analysis of a xylose-consuming Saccharomyces cerevisiae strain expressing xylose isomerase. Biotechnol Bioeng 112:470–483. doi:10.1002/bit.25447
Wei N, Quarterman J, Kim SR et al (2013) Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat Commun 4:2580. doi:10.1038/ncomms3580
Westfall PJ, Pitera DJ, Lenihan JR et al (2012) Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA 109:E111–E118. doi:10.1073/pnas.1110740109
Winzeler EA, Shoemaker DD, Astromoff A et al (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906
Wisselink HW, Toirkens MJ, Wu Q et al (2009) Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol 75:907–914. doi:10.1128/AEM.02268-08
Yang L, Cluett WR, Mahadevan R (2011) EMILiO: a fast algorithm for genome-scale strain design. Metab Eng 13:272–281. doi:10.1016/j.ymben.2011.03.002
Yang Y-T, Bennett GN, San K-Y (1998) Genetic and metabolic engineering. Electron J Biotechnol 1:20–21. doi:10.4067/S0717-34581998000300003
Yona AH, Manor YS, Herbst RH et al (2012) Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci USA 109:21010–21015. doi:10.1073/pnas.1211150109
Zhou H, Cheng J-S, Wang BL et al (2012) Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 14:611–622. doi:10.1016/j.ymben.2012.07.011
Zhou YJ, Buijs NA, Zhu Z et al (2016) Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun 7:11709. doi:10.1038/ncomms11709
Acknowledgements
LC thanks the Secretaría de Educación Pública (SEP) which financed his integration into the Autonomous University of Morelos State through the Programa para el Desarrollo Profesional Docente (PRODEP) (grant no. DSA/103.5/14/10703). TC thanks the National Council of Science (CONACyT) for the postdoctoral scholarship.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Caspeta, L., Castillo, T. (2017). Systems Metabolic Engineering of Saccharomyces cerevisiae for Production of Biochemicals from Biomass. In: Gosset, G. (eds) Engineering of Microorganisms for the Production of Chemicals and Biofuels from Renewable Resources. Springer, Cham. https://doi.org/10.1007/978-3-319-51729-2_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-51729-2_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-51728-5
Online ISBN: 978-3-319-51729-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)