Abstract
To mitigate global climate change caused partly by the use of fossil fuels, the production of fuels and chemicals from renewable biomass has been attempted. The conversion of various sugars from renewable biomass into biofuels by engineered baker’s yeast (Saccharomyces cerevisiae) is one major direction which has grown dramatically in recent years. As well as shifting away from fossil fuels, the production of commodity chemicals by engineered S. cerevisiae has also increased significantly. The traditional approaches of biochemical and metabolic engineering to develop economic bioconversion processes in laboratory and industrial settings have been accelerated by rapid advancements in the areas of yeast genomics, synthetic biology, and systems biology. Together, these innovations have resulted in rapid and efficient manipulation of S. cerevisiae to expand fermentable substrates and diversify value-added products. Here, we discuss recent and major advances in rational (relying on prior experimentally-derived knowledge) and combinatorial (relying on high-throughput screening and genomics) approaches to engineer S. cerevisiae for producing ethanol, butanol, 2,3-butanediol, fatty acid ethyl esters, isoprenoids, organic acids, rare sugars, antioxidants, and sugar alcohols from glucose, xylose, cellobiose, galactose, acetate, alginate, mannitol, arabinose, and lactose.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
As human society has grown and developed, our demand for fuels and commodity chemicals has accelerated. This demand has manifested as many different outputs for both fuels and chemicals. For fuels, we have two major categories: transportation fuels and non-transportation fuels. Here we mainly discuss transportation fuels, which are currently primarily derived from non-renewable fossil fuels. These hydrocarbons, such as coal, petroleum, or natural gas, are processed into gasoline, ethanol, jet fuel, or other specialized products [1]. Approximately 80% of energy use by humans is derived from fossil fuels, with up to 58% consumed for transportation [2, 3]. Because the rate of natural production of fossil fuels has for decades been increasingly outpaced by humanity’s usage, renewable alternatives for transportation fuels are considered a societal necessity [1].
As with fuels, many non-fuel chemicals are produced using non-renewable fossil fuel feedstocks. This petrochemical-based system is non-renewable and, as with fuels, an alternative method of production is needed to allow for continued advancement of human society. In particular, the petrochemical industry produces chemicals used in nearly every industry on Earth. Many bulk chemicals, such as ethylene and propylene, are produced in the 1–100 million annual tons range [4]. The specific uses of these chemicals can vary greatly: in some cases, such as artemisinic acid, only one major use is currently considered (as a precursor to an antimalarial drug) [5], whereas other chemicals, such as lactic acid, have numerous uses, including as a plastic precursor or as a food preservative [6]. Collectively, reliable and sustainable industrial production of chemicals is a necessity for ongoing human progress.
The finite supply of fossil fuels [7, 8], the risks associated with harvesting hard-to-obtain fossil fuels [9–11], and the concerns about manmade climate change related to fossil fuel use [12–15] have collectively pushed researchers and governments toward producing fuels and chemicals from renewable biomass by engineered microbes [16, 17]. Although many microbes have been studied for the production of renewable fuels and chemicals, yeasts, Saccharomyces cerevisiae in particular, have served as major platform microbes for many of these studies.
S. cerevisiae, also known as brewer’s yeast, is a well-studied microorganism, even beyond its traditional use for the production of beer and other fermented foods and beverages [18]. Extensive tools exist for the manipulation and engineering of yeasts [19–22]. These tools have allowed for harnessing the native ability of S. cerevisiae to grow in minimal medium, their generally recognized as safe (GRAS) designation, and their tolerance to low pH and acidic conditions [23, 24]. With these tools and inherent physiological advantages, scientific advances for the production of fuels and chemicals from biomass by S. cerevisiae have improved dramatically in recent years. In this review we discuss these recent developments as they relate to feedstock utilization as well as production of fuels and chemicals with additional insight on the future economic outlook of these processes.
2 Yeast Fermentation Technologies
With modern metabolic engineering techniques improvements following their advent in the 1970s and the more recent development of synthetic biology procedures, yeast engineering technologies have grown dramatically [16]. Many yeast engineering approaches follow a scheme known as the “Design, Build, Test, and Learn” cycle [25, 26]. This scheme first requires a target outcome or goal. For example, a target goal could be to produce ethanol from the pentose sugar xylose by engineered S. cerevisiae, which natively are unable to ferment xylose.
Once the desired outcome is determined, a parental yeast strain, often a wild-type strain, is selected as the target organism to be engineered. The steps for engineering the parental strain are as follow: (1) Designing the specific yeast engineering steps, including plasmids and transformation protocols, (2) Building the engineered strain by introduction of target genetic perturbations, (3) Testing the newly-developed strain, often involving fermentation and sampling, and (4) Learning from the new strain (Fig. 1). The new knowledge obtained from this process can then be factored into the design of the next strain and the cycle can repeat until the target outcome is reached. This systematic approach has led to significant advances in the development of engineered S. cerevisiae capable of fermenting novel substrates for the production of fuels and chemicals. Although not all studies explicitly state this four-step process, the general concept is applicable in many cases.
2.1 Major Objectives and Feedstocks for Yeast Fermentations
The first-generation biofuels from cornstarch or sugarcane juice have been industrialized for decades; however the food-vs-fuel conflict has limited its further expansion [27]. Ethanol is considered the major and most highly produced of the first-generation biofuels. Despite the food-vs-fuel concerns, 23.8 billion gallons of ethanol are produced annually, primarily from cornstarch or sugarcane juice [28]. Transitioning from first-generation biofuel feedstocks (cornstarch and sugarcane juice) to second-generation feedstocks (lignocellulosic biomass) is a key objective of modern yeast fermentation research.
The second-generation biofuels from non-food lignocellulosic biomass, which is a renewable carbon source, has offered an excellent opportunity to address the food-vs-fuel issue [29, 30]. Lignocellulosic hydrolysates obtained from corn stover [31], bagasse [32, 33], sorghum biomass [34], and marine plants [35, 36] after pretreatment and hydrolysis contain substantial amounts of hexoses (six-carbon, C6 sugar) and pentoses (five-carbon, C5 sugar) which can be used as renewable carbon sources for the production of bioethanol and other value-added products (Fig. 2). Lignocellulosic hydrolysates are commonly composed of ~70% cellodextrins and glucose and 30% xylose [37], although this can vary by biomass source and processing protocol. Marine hydrolysate sugar compositions can vary wildly: as a percent of total solids, red algae hydrolysates can be composed of ~18% glucose, ~30% total of galactose/xylose/arabinose, and ~8% mannose; green algae hydrolysates can consist of ~8% glucose, 6% total of galactose/xylose/arabinose, and 5% mannose; finally, brown algae can be composed of 6–7% glucose and between 2% (Sargassum fulvellum) and 30% (Laminaria japonica) mannitol [38]. However, natively, the yeast S. cerevisiae cannot use pentoses, such as xylose, and cannot efficiently ferment all hexoses. Therefore, another major objective of yeast fermentation research is to improve the selection of sugars capable of being fermented by S. cerevisiae for the purpose of industrial fermentation (Fig. 3). In Sect. 2.2 we discuss the currently available substrates for native and engineered S. cerevisiae strains.
2.2 Native and Non-Native Substrate Utilization by Saccharomyces cerevisiae
2.2.1 Glucose
Glucose is the most preferred carbon source for S. cerevisiae [39] and can be fermented more rapidly than any other sugar. To date, no other carbon source has been found to be consumed more rapidly or efficiently than glucose in any wild-type or engineered S. cerevisiae. The major industrial source of glucose is from cornstarch, although wheat is also sometimes used. The first generation of biofuels is based on the hydrolysis of cornstarch and very high gravity (VHG) fermentations have been conducted to decrease the process costs [32, 40, 41]. Several studies have focused on enhancing the fitness of S. cerevisiae in the presence of high concentrations of glucose. For example, Guadalupe-Medina et al. created a GPD1- and GPD2-negative S. cerevisiae that anaerobically produced ethanol at a high yield from glucose [42]. However, this strain became sensitive to high concentrations of ethanol, but the problem was alleviated by employing a laboratory evolution strategy with serial subculturing of the GPD1/GPD2-deleted strain on ethanol [42]. Because glucose fermentations by S. cerevisiae are very rapid and efficient, further improvements for glucose fermentations by engineered S. cerevisiae are likely to focus on improving strain tolerance to harsh fermentation media conditions, especially those found in cellulosic hydrolysates.
2.2.2 Xylose
Harvested terrestrial biomass is processed into a sludge-like product known as a hydrolysate. In terrestrial biomass, hydrolysates contain both C6 and C5 sugars. However, the most widely-used fermenting microorganism, S. cerevisiae, cannot metabolize pentose sugars such as xylose and arabinose which are abundant in cellulosic hydrolysates. Therefore, numerous studies have attempted to construct metabolically engineered S. cerevisiae capable of fermenting pentose as rapidly as glucose [43–45]. Xylose metabolism can be introduced into S. cerevisiae using a bacterial or fungal metabolic route for xylose assimilation [46, 47]. The bacterial pathway uses only one enzyme, xylose isomerase (XI), for converting xylose into xylulose [44, 45]. Xylulose is later phosphorylated by xylulose kinase (XK) into xylulose-5-phosphate (X5P) and then enters the non-oxidative pentose phosphate pathway (PPP) for further metabolism toward pyruvate. Using the XI pathway, an ethanol yield from xylose as high as 0.45 g/g has been achieved [44]. Another study by Lee et al. engineered an S. cerevisiae to harbor a bacterial xylose pathway to express a mutant xylose isomerase (xylA3*) from Piromyces sp. with aldose reductase (GRE3) and PHO13 deletions coupled with overexpression of the S. cerevisiae native xylulokinase (XKS1) and S. stipitis transaldolase (TAL1) [44]. Zhou et al. also overexpressed the Piromyces sp. xylose isomerase gene (XYLA), S. stipitis xylulose kinase (XYL3), and genes of the non-oxidative pentose phosphate pathway [45].
The fungal xylose assimilation pathway consists of two oxidoreductases, NADPH-linked xylose reductase (XR) and NAD-linked xylitol dehydrogenase (XDH) [43]. Several researchers developed platforms for consuming these specific substrates, such as introducing xylose-metabolizing enzymes into S. cerevisiae to produce a rapid and efficient xylose-fermenting strain [47–50]. For example, Kim et al. introduced the fungal pathway by strong and balanced expression of genes from Scheffersomyces stipitis consisting of xylose reductase (XR, encoded by XYL1), xylitol dehydrogenase (XDH, encoded by XYL2), and xylulose kinase (XK, encoded by XYL3) with the addition of the genetic disruption of alkaline phosphatase (PHO13) and acetaldehyde dehydrogenase (ALD6) [43]. This series of genetic manipulations using the fungal XR/XDH/XK pathway resulted in an ethanol yield of 0.35 g/g from xylose [43].
Collectively, these studies have developed numerous xylose-fermenting S. cerevisiae capable of rapid and efficient xylose fermentation. Despite these advances, even the fastest xylose fermentations by engineered yeasts are still slower than the fastest glucose fermentations, and so further studies to improve xylose fermentation rates and yields by S. cerevisiae are ongoing.
2.2.3 Arabinose
Similar to xylose metabolism, different l-arabinose metabolizing pathways have been identified in bacteria [51] and fungi [52, 53]. The bacterial pathway for l-arabinose utilization converts l-arabinose into X5P via l-ribulose-5-phosphate (L5P) using three enzymes (an isomerase, a kinase, and an epimerase). When l-arabinose isomerase (araA), l-ribulokinase (araB), and l-ribulose-5-phosphate 4-epimerase (araD) from Lactobacillus plantarum were expressed in S. cerevisiae, l-arabinose fermentation was observed [51]. The fungal l-arabinose utilization pathway converts l-arabinose into l-arabinitol by aldose reductase (GRE3 from S. cerevisiae or XYL1 from Scheffersomyces stipitis), l-xylulose by l-arabinitol 4-dehydrogenase (LAD from Trichoderma reesei), xylitol by l-xylulose reductase (LXR from T. reesei), d-xylulose by (XDH from S. stipitis), and lastly X5P by xylulokinase (XYL3) [52, 53]. As X5P is a gateway metabolite in the PPP, it can be converted to pyruvate and ethanol. Recently, researchers have used codon optimized bacterial pathways for l-arabinose fermentation in S. cerevisiae because of the inefficient l-arabinose utilization and high byproduct (l-arabinitol) yield of fungal pathways caused by severe redox imbalance [54].
Other than introducing xylose and arabinose pathways into S. cerevisiae, known hexose transporters, as potential xylose and arabinose transporters, have been investigated. Several hexose transporters were proven to be responsible for the uptake of pentose sugars. For instance, Hxt7p, Hxt5p, and Gal2p improve xylose uptake [55] and Gal2p also facilitates the transport of l-arabinose [56]. However, these hexose transporters exhibited very low affinity to pentoses and preferred d-glucose. Therefore, for the improvement of xylose and l-arabinose fermentations, efforts were made to find high-affinity xylose or l-arabinose specific transporters. Heterologous transporters were discovered with higher affinity for xylose over glucose, such as Gxs1p from Candida intermedia [57], Xut3p from S. stipitis [58], and Mgt05196p from Meyerozyma guilliermondii [59]. Nonetheless, it is still challenging to have both the specificity and efficiency of xylose transport, and further evolutionary adaptation and protein engineering are required [59, 60]. Heterologous overexpression of STP2 from Arabidopsis thaliana and ARAT from S. stipitis in S. cerevisiae also led to improved anaerobic l-arabinose fermentation, especially at low l-arabinose concentrations, although these two transporters still are inhibited in the presence of glucose [61]. Recently, Wang et al. have engineered an S. cerevisiae strain capable of producing an ethanol yield of 0.43 g/g from arabinose, one of the highest reported yields to date [62].
2.2.4 Cellobiose
Another major sugar of interest is cellobiose, a β(1,4)-linked dimer of d-glucose, which is readily released from larger cellodextrins from cellulose by cellulases after acidic treatment of terrestrial biomass [63]. However, S. cerevisiae cannot naturally metabolize cellobiose because of the lack of a cellobiose transporter and intracellular β-glucosidase. A high-affinity cellodextrin transporter (cdt-1 or cdt-2) and intracellular β-glucosidase (gh1-1) were identified from the cellulolytic fungus Neurospora crassa [64]. The cellobiose transporters and the intracellular β-glucosidase promote efficient cellobiose fermentation and ethanol production when expressed in S. cerevisiae [64]. The intracellular β-glucosidase can be replaced by cellobiose phosphorylase, which produces glucose and glucose-1-phosphate from cellobiose. Efficient cellobiose fermentation by engineered yeast expressing a cellobiose transporter and a bacterial cellobiose phosphorylase has also been demonstrated [65]. Because cellobiose does not induce glucose inhibition on other carbon sources, simultaneous cofermentation of cellobiose and xylose [66, 67] as well as cellobiose and galactose [68] has been achieved. Simultaneous cofermentation is necessary for efficient and rapid industrial-scale fermentation of hydrolysates.
2.2.5 Alginate and Mannitol
Another type of sustainable non-lignocellulosic biomass is marine biomass, such as macroalgae or seaweed. The most abundant sugars in brown macroalgae are alginate, mannitol, and glucan (presented as laminarin or cellulose). However, industrial microbes are unable to metabolize the alginate, which represents 30–60% of total sugars in brown macroalgae. Alginate is a linear block copolymer of two uronates, β-d-mannuronate (M) and α-l-guluronate (G), arranged in varying sequences [69]. Some microbes can metabolize alginate natively by depolymerization of alginate into oligomers by alginate lyases (Alys). These oligomers are further degraded into unsaturated monomers by oligoalginate lyase (Oal) and the monomers are rearranged spontaneously into 4-deoxy-l-erythro-5-hexoseulose uronic acid (DEH). DEH is then converted into 2-keto-3-deoxyl-glucontate (KDG) by DEH reductase (DehR), and KDG is a common metabolite that can enter into the Entner–Doudoroff (ED) pathway and yield pyruvate and glyceraldehydrate-3-phosphate via KDG kinase (KdgK) and KDG-6-aldolase (Eda).
However, these natural microbes, such as Sphingomonas sp., lack the robustness necessary for industrial fermentation conditions and have limited availability of genetic and metabolic engineering tools. Therefore, researchers have introduced and expressed the genes responsible for the alginate degradation, transport, and metabolism into the well-characterized microorganism Escherichia coli, which is naturally capable of utilizing mannitol and d-glucose. A 36-kb pair DNA fragment from Vibrio splendidus encoding enzymes necessary for alginate degradation, transport, and metabolism was discovered. After introducing the alginate metabolism, the heterologous homoethanol pathway consisting of Zymomonas mobilis pyruvate decarboxylase (pdc) and alcohol dehydrogenase B (adhB) were also introduced for efficient ethanol production [70], which later demonstrated the feasibility of utilizing macroalgae as a microbial host for ethanol production.
Although engineered E. coli provided the proof of concept for metabolizing alginate, mannitol, and d-glucose, S. cerevisiae is a more amenable host for industrial-scale ethanol production. Therefore, Enquist-Newman et al. attempted to re-engineer the alginate and mannitol catabolic pathways into S. cerevisiae [71]. They discovered an alginate monomer (DEHU) transporter from the alginolytic eukaryote Asteromyces cruciatus. Through the genome integration and overexpression of this transporter and with the necessary bacterial alginate degradation genes and essential genes for mannitol consumption, including an NAD+-dependent mannitol-2-dehydrogenase (M2DH) and a mannitol transporter, the engineered S. cerevisiae was able to metabolize DEHU and mannitol [71]. As a result, the engineered S. cerevisiae strain produced ethanol from mannitol and DEHU at 83% of the maximum theoretical yield.
2.2.6 Galactose
S. cerevisiae are naturally capable of fermenting galactose, a C6 monosaccharide, into ethanol through the Leloir pathway. In the Leloir pathway, galactose is converted to UDP-glucose and then glucose-1-phosphate. Phosphoglucomutase converts glucose 1-phosphate to glucose 6-phosphate. Whereas the rest of the metabolic pathway is identical, the ethanol yield and productivity from galactose by S. cerevisiae is significantly lower than from glucose [72]. Through overexpression of a truncated TUP1 gene, which codes for a general transcription repressor, Lee et al. were able to improve galactose consumption rate and ethanol productivity by 250% compared to a control S. cerevisiae [72]. By combining enhanced galactose metabolism with a heterologous cellobiose pathway, an engineered S. cerevisiae could be employed for fermenting red seaweed hydrolysates. The major components of red seaweed (Gelidium amansii), cellulose and galactan, can be hydrolyzed to produce a mixture of cellobiose and galactose [36, 68]. Cellobiose and galactose can be cofermented by engineered yeast [68] because the two sugars are transported with high affinity by independent transporters (CDT-1 and Gal2). Recently, one research group has focused on using high concentrations of galactose as an adaptation pressure on yeast to improve galactose consumption rates and ethanol productivity [73, 74]. Further improvements for producing ethanol from galactose and red seaweed are necessary for industrial-scale ethanol production, especially as demand for second-generation biofuels continues to grow.
2.2.7 Acetate
Acetate is one of the major inhibitors present in lignocellulosic hydrolysates which can hamper S. cerevisiae fermentation capabilities. In addition, acetate is also produced as a major component from the pyrolysis of lignin [75, 76]. Recently, an interesting solution was developed to convert acetate from a fermentation component or inhibitor into a valuable product. By coupling the consumption of acetate and xylose, the redox imbalance of xylose fermentation by the heterologous xylose reductase (XR), xylitol dehydrogenase (XDH), and xylulokinase (XK) pathway can be alleviated and the inhibitor (acetate) can be detoxified [77]. As a major result, the entire bioethanol fermentation process was improved compared to the control, increasing the ethanol yield by 6% (to 0.414 g/g) and reducing byproduct formation by 11% [77]. This process was further advanced by generating an engineered S. cerevisiae which expresses a cellobiose-utilizing pathway in addition to the aforementioned acetate and xylose pathways, allowing for efficient fermentation of multiple lignocellulosic sugars (xylose and cellobiose) and fermentation inhibitors (acetate) [78]. Finally, a peak ethanol yield of 0.463 g ethanol/g xylose was achieved by an XR/XDH-expressing S. cerevisiae through upregulation of acetylating acetaldehyde dehydrogenase (AADH) and acetyl-CoA synthetase (ACS) [79]. Compared to the control strain, the engineered strain was able to produce 18.4% more ethanol, 41.3% less glycerol, and consume 4.1 g/L of acetate from a cellulosic hydrolysate [79]. Collectively, these acetate-utilization studies are a significant breakthrough for the in situ detoxification of acetate by S. cerevisiae for ethanol production. Additional improvements could convert this fermentation into an industrial-scale ready process.
2.2.8 Lactose
Lactose is a disaccharide consisting of the monomers glucose and galactose. The primary source of lactose is from milk or fermented dairy products. Annually, millions of tons of lactose are produced by the dairy industry. As a result of the acid whey fermentation process, a significant amount of lactose is trapped in the harsh and acidic acid whey slurry. Many studies have been conducted to find efficient uses for this trapped lactose.
Several studies have attempted to create a lactose-consuming S. cerevisiae by introducing LAC4 and LAC12 from Kluyveromyces marxianus and Kluyveromyces lactis into S. cerevisiae [80–84]. These studies resulted in the development of engineered S. cerevisiae capable of fermenting lactose. By expressing the LAC4 and LAC12 genes into the MIG1 and NTH1 gene-encoding regions in S. cerevisiae, respectively, Zou et al. engineered a strain capable of producing 63.3 g/L of ethanol from approximately 150 g/L lactose in 120 h from concentrated cheese whey [84]. By disrupting the function of the MIG1 and NTH1 genes, the engineered strain had highly reduced glucose repression. Although Kluyveromyces spp. are yeasts which can natively ferment lactose, their genetics are not as well-understood as that of S. cerevisiae, suggesting that improvements of S. cerevisiae for lactose fermentation may be ideal.
3 Biofuel Production by Engineered or Evolved Yeast
S. cerevisiae offers many advantages for producing sustainable and economically viable biofuels from renewable feedstocks. It has been widely used as an important eukaryotic model for fundamental molecular biology research with numerous synthetic biology tools developed as compared to most other microorganisms, perhaps second only to E. coli. Recently developed yeast engineering tools include the use of zinc-finger nucleases [85], yeast oligo-mediated genome engineering [21], and most notably the clustered regularly-interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas9) system) [22]. However, S. cerevisiae is considered to be more robust than E. coli, with S. cerevisiae possessing a higher tolerance to low pH/high acid conditions, resulting in preference for the eukaryote for fermentation of biomass hydrolysate. S. cerevisiae has been used extensively as a platform cell factory for first-generation, industrial-scale bioethanol production [86]. Because of its unique robustness toward harsh fermentation conditions and the substantial availability of yeast engineering tools, introducing new metabolic engineering pathways into S. cerevisiae has been used for producing alternative products beyond bioethanol (Fig. 4 and Table 1).
3.1 Biofuel Production by S. cerevisiae
3.1.1 Ethanol
First-generation biofuel production focused almost entirely on producing bioethanol from corn or sugarcane juice. Although many research directions were investigated to improve bioethanol production, one major direction focused on glycerol, a common byproduct of ethanol fermentations. During anaerobic yeast fermentations, the biosynthesis of proteins, nucleic acids, and lipids from biomass production generate excess cytosolic reduced redox cofactors such as NADH. Formation of glycerol serves as an essential electron sink for oxidizing NADH into NAD+ in the cytosol. Tremendous research efforts focused on minimizing the formation of the unwanted glycerol byproduct generated during the bioethanol production process because carbon directed toward glycerol reduced carbon availability for ethanol synthesis. Two structural genes encoding cytosolic NADH-dependent glycerol-3-phosphate dehydrogenases, GPD1 and GPD2, play important roles in redox balance and osmoregulation. These genes are also both induced under high osmotic conditions and during anaerobic fermentation. Glycerol formation can be reduced by deleting one or both genes [87]. However, yeast cells with this double deletion of GPD1 and GPD2 become unable to grow anaerobically because of the lack of alternative pathways to oxidize NADH. The single deletion of GPD2 showed improved ethanol yields by decreasing glycerol production, but the deletion also hindered cell growth and ethanol productivity [88]. Reduced glycerol production also increased the osmosensitivity and diminished the general robustness of the engineered yeast [89].
Other studies have focused on the metabolic engineering of the cellular redox metabolism. An ammonium assimilation pathway that consumes NADH and ATP was utilized. Deletion of the NADPH-dependent glutamate dehydrogenase gene GDH1 and respectively co-overexpression of the glutamate synthase gene GLT1 and the glutamine synthetase gene GLN1 showed a significant reduction in glycerol by 38% and improved ethanol yield by 10% [90]. Another alternative way of redox engineering would be the reduction of the surplus cytosolic NADH with lower ATP production by replacing the natural glyceraldehyde-3-phosphate dehydrogenase with a non-phosphorylating, NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase (gapN) from Streptococcus sp. mutants or Bacillus cereus [91, 92]. One interesting demonstration of cofactor metabolism is that overexpression of E. coli mhpF was able to restore the anaerobic growth of a GPD1- and GPD2-deleted mutant under the presence of acetate by re-oxidizing the NADH through the reduction of acetic acid to ethanol [93]. By combining these genetic modifications, overexpression of the NAD+-dependent fumarate reductase frdA or NAD+-dependent acetaldehyde dehydrogenase mhpF from E. coli with gapN can improve the ethanol yield to above 97% of the maximum theoretical yield compared to wild-type yeast [94]. Furthermore, gapN expression with the combination of TPS1 and TPS2 overexpression showed reduced glycerol production and improved ethanol yield [92]. By blocking the export of glycerol through deletion of FPS1 encoding a glycerol facilitator, yet another method to reduce glycerol production and improve ethanol yield was uncovered [95].
Promoter engineering has been used as an alternative approach to modulate the expression of GPD1 and GPD2. For example, S. cerevisiae mutants with the lower-strength TEF1 promoter replacing the native GPD1 and/or GPD2 promoters produced less glycerol and more ethanol without reducing the robustness of the host strain toward osmotic stress [96]. With the FPS1- and GPD2-deleted yeast strain background (KAM15 strain), the mutants with 3′ truncation of the GPD1 promoter by 20-, 60-, or 80-bp displayed varied expression strength of GPD1 and had an unaffected osmotic response. The glycerol production by the engineered yeast was also reduced by 16% and 31% in mutants with 60- and 80-bp truncated promoters, respectively, in high-gravity (VHG) fermentations. The ethanol yield reached 0.499 g/g in the mutant with an 80-bp truncated promoter [40].
3.1.2 1-Butanol
Higher-chain alcohols provide higher energy density and are considered as potential next-generation gasoline substitutes. One of the primary target alcohols is 1-butanol. Although 1-butanol was traditionally produced from Clostridium species through the acetone-butanol-ethanol (ABE) fermentation process, or by engineered E. coli with a titer up to 30 g/L [97], there are several advantages of using S. cerevisiae for 1-butanol production. In addition to the general robustness of S. cerevisiae toward fermentation inhibitors and low pH, S. cerevisiae also does not have phage contamination issues and has better resistance to high 1-butanol concentrations. However, only a low concentration of 1-butanol was produced from the native 1-butanol metabolic pathway in S. cerevisiae, which prompted several labs to look for heterologous pathways to improve 1-butanol production.
Steen et al. introduced and expressed in S. cerevisiae several isozymes from different organisms to create a biosynthetic 1-butanol pathway with a peak 1-butanol titer of 2.5 mg/L [98]. This pathway consisted of converting acetyl-CoA into acetoacetyl-CoA, which was reduced to 3-hydroxybutyryl-CoA, and later crotonyl-CoA. Butyryl-CoA is the reduced form of crotonyl-CoA which is later further reduced into butyraldehyde and finally reduced into 1-butanol. This pathway consisted of overexpression of a thiolase (ERG10) from S. cerevisiae, an NADH-dependent 3-hydroxybutyryl-CoA dehydrogenase (hbd) and crotonase (crt) from Clostridium beijerinckii, an NADH-dependent crotonyl-CoA reductase (ccr) from Streptomyces collinus and butanol dehydrogenase (adhe2) from C. beijerinckii [98].
Krivoruchko et al. initially increased 1-butanol titers up to 6.6 mg/L by engineering yeast with higher flux toward cytosolic acetyl-CoA, which is the precursor for 1-butanol biosynthesis in addition to the overexpression of the heterologous enzymes for the 1-butanol biosynthetic pathway as follow [99]. First, NADH-dependent crotonyl-CoA-specific trans-enoyl-CoA reductase (Ter) from Treponema denticola replaced the ccr to avoid the reverse oxidation of butyryl-CoA to crotonyl-CoA. Second, to increase the cytosolic acetyl-CoA supply, a pyruvate dehydrogenase (PDH) bypass was created by overexpression of endogenous alcohol dehydrogenase (ADH2), NADP-dependent aldehyde dehydrogenase (ALD6), codon-optimized acetyl-CoA synthetase (ACS2) from Salmonella enterica, and acetyl-CoA acetyltransferase (ERG10). Lastly, deletion of malate synthase (MLS1) or citrate synthase (CIT2) reduced the drainage of acetyl-CoA through the glyoxylate pathway, and the 1-butanol titer increased to 16.3 mg/L [99].
Therefore, intracellular availability of cytosolic acetyl-CoA is considered an important factor for 1-butanol production in yeast. NADH availability could also be a strong driving force toward 1-butanol production. Therefore, NADH-dependent alcohol dehydrogenase (ADH) and glycerol-3-phosphate dehydrogenase (GPD) can be deleted to increase the NADH availability and reduce the unwanted byproducts such as ethanol and glycerol. Lian et al. produced up to 120 mg/L 1-butanol by inactivating ADH and GPD, introducing the butanol biosynthesis pathway genes, and, most importantly, introducing a PDH-bypass pathway, cytosolic localized PDH, and ATP-dependent citrate lyase (ACL) [100].
Despite the limited accumulation of 1-butanol from the native S. cerevisiae pathway, some researchers have focused on improving the native pathway by focusing on threonine catabolism. Si et al. utilized genes from leucine biosynthesis (LEU1, LEU2, LEU4, and LEU9), together with threonine deaminase genes (ILV1/CHA1), 2-keto acid decarboxylases (KDCs) from Lactococcus lactis, and alcohol dehydrogenases (ADHs) from S. cerevisiae [101]. The pathway consists of many steps, starting with l-threonine to 2-ketobutyrate to 2-ketovalerate, and so forth, eventually ending at 1-butanol. Deletion of ADH allowed the engineered S. cerevisiae to produce more than 120 mg/L of 1-butanol from glucose in a complex yeast-peptone medium. By amplifying the leucine biosynthesis pathway via overexpression of several key genes and eliminating the competing pathways, the highest reported 1-butanol titer of 242.8 mg/L in S. cerevisiae with ADH1- and ILV2-deletions was achieved [101].
3.1.3 Isobutanol
Isobutanol is another example of a target alcohol which has a higher energy density than ethanol. The isobutanol biosynthesis pathway is closely linked to the biosynthesis of branched-chain amino acids via the Ehrlich pathway. 2-Ketoisovalerate (KIV), an intermediate of valine biosynthesis, is decarboxylated to isobutyraldehyde by 2-ketoacid decarboxylase (KDC) and later reduced into isobutanol by alcohol dehydrogenase (ADH). However, the protein synthesis of KIV occurs in the yeast mitochondria whereas the other two enzymes, Kdc and Adh, are found in the yeast cytosol. For isobutanol synthesis in S. cerevisiae, pyruvate must transfer into mitochondria and then KIV must be transported into the cytosol.
The first report for isobutanol overproduction in yeast utilized simultaneous overexpression of endogenous genes (ILV2, ILV3, and ILV5) of the mitochondrial valine biosynthesis pathway. The resulting strain produced isobutanol with a yield up to 0.97 mg isobutanol/g glucose in minimal medium [102]. Additional overexpression of the cytosolic branched-chain amino acid aminotransferase (BAT2) increased the isobutanol yield up to 3.86 mg/g glucose [102]. Finally, a yield of 4.12 mg/g glucose was achieved by the engineered yeast in an aerobic condition with complex yeast-peptone medium [102]. Avalos et al. demonstrated that locating the complete isobutanol pathway into the mitochondria resulted in substantial increases in isobutanol as compared with the native pathway which is split between the cytosol and the mitochondria. KDCs and ADHs were overexpressed in the cytosol or imported into mitochondria by fusing them with an N-terminal targeting signal, and the isobutanol yield reached up to 6.40 mg/g glucose with a titer up to 0.635 g/L [103]. This study suggested that the availability of the KIV intermediate and the increased local enzyme concentration would be beneficial for isobutanol production. Another research group, Yuan and Ching, developed a similar approach with a δ-integration system to assemble the genes into the yeast chromosomes with the resulting isobutanol yield up to 15 mg/g glucose [104].
The opposite strategy is to relocate the pathway into the cytosol. By re-localization and codon-optimization of the mitochondrial valine synthesis enzymes together, along with the overexpression of decarboxylase (ARO10) and alcohol dehydrogenase (ADH2) genes, isobutanol production was improved to the highest titer of 0.63 g/L and a yield of approximately 15 mg/g glucose [105]. Isobutanol production was further improved in engineered S. cerevisiae by two strategies. First, the elimination of competing pathways by deletion of a pyruvate dehydrogenase complex component (LPD1) to avoid competing with acetyl-CoA biosynthesis in the mitochondria. Second, resolving cofactor imbalance by the implementation of the transhydrogenase-like shunt, which pyruvate cyclically converted into oxaloacetate, malate, and back to pyruvate causing simultaneous conversion of NADH to NADPH. The final isobutanol titer reached 1.62 g/L and a yield of 16 mg/g glucose [106]. However, even this heightened result is still considerably below that of engineered E. coli, reported to generate isobutanol titers up to several grams per liter [107]. These results suggest that considerable improvements are necessary before yeast-based isobutanol production can be competitive on an industrial scale.
3.1.4 Fatty Acids
Fatty acids (FAs) and lipids are also valuable chemicals for numerous industrial applications. Lipids are condensed from a glycerol-3-phosphate backbone with the completed FA synthesized from acetyl-CoA. Fatty acid ethyl esters (FAEEs) can be used for diesel or jet fuel production. FAEEs can be formed by esterification of fatty acyl-CoAs and ethanol. Kalscheuer et al. first studied FAEE production in yeast [108] by heterologous expression of an unspecific bacterial acyltransferase, a wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase (WS/DGAT), from Acinetobacter calcoaceticus ADP1. Later, Shi et al. screened five different wax ester synthases in S. cerevisiae and found the wax ester synthase from Marinobacter hydrocarbonoclasticus performed best with the highest titer of FAEE at 6.3 mg/L [109]. Overexpression of acetyl-coA carboxylase (ACC1) led to an increase of FAEE titer to 8.2 mg/L [109]. de Jong et al. continued the study by increasing the acyl-CoA synthesis which later enhanced the production of FAEE by increasing the NADPH and acetyl-CoA pools in two ways [110]. First, overexpression of alcohol dehydrogenase (ADH2), acetaldehyde dehydrogenase (ALD6), and a heterologous acetyl-CoA synthase variant from Salmonella enterica (acs SE L641P) was conducted to re-channel the carbon flow for acetyl-CoA with the ethanol degradation pathway. Wax ester synthase from M. hydrocarbonoclasticus was also overexpressed. Second, a phosphoketolase pathway was established by overexpression of xpkA and ack from Aspergillus nidulans for the conversion of xylulose-5-phosphate to acetyl-phosphate and glyceraldehyde-3-phosphate and acetyl phosphate to acetate. The resulting engineered S. cerevisiae strain proved to have a 1.7-fold improvement for FAEE production compared to the control strain, with 5.1 mg/g dry cell weight [110].
In the same year, Valle-Rodrigez et al. eliminated the non-essential fatty acid utilization pathway such as steryl esters (SEs) and triacylglycerols (TAGs) by deletion of DGA1, LRO1, ARE1, and ARE2 [111]. The researchers also deleted POX1 to avoid degradation of FAs and overexpressed wax ester synthase (WS) from M. hydrocarbonoclasticus DSM 8798 which generated a final FAEE titer of up to 17.2 mg/L [111]. Recently, Eriksen et al. investigated the heterologous expression of Type-I fatty acid synthase (FAS) from Brevibacterium ammoniagenes coupled with WS/DGAT [112]. They found the strain harboring the orthologous FAS yielded a 6.3-fold increased FAEE titer compared to strains without FAS. The FAEE titer was 10.498 mg/g DCW with the overexpression of Type-I fatty acid synthase (bafas and ppt1) from Brevibacterium ammoniagenes, FAA1 from S. cerevisiae, and wax ester synthase from M. hydrocarbonoclasticus [112]. However, additional studies and demonstrations must be conducted for further improvement of the titers for FAEE, because the above-mentioned titers from engineered S. cerevisiae are still relatively low for industrial applications.
4 Chemical Production by Engineered or Evolved Yeast
There has been an intensive effort for the engineering of S. cerevisiae to produce non-fuel, value-added chemicals. Historically, S. cerevisiae has been used for ethanol production by the food or fuel industries, but scientific advances for the purpose of ethanol production by yeast can often easily be applied to non-fuel production. As mentioned in previous sections of this review, S. cerevisiae has GRAS status and their genetic system has been studied heavily. Thus, many genetic tools are available [21, 22, 85] which ease the engineering of this host organism to produce nonconventional target products. These products include food additives, pharmaceuticals, advanced biofuels, and valuable chemicals for industrial applications.
Natively, S. cerevisiae produces numerous minor and major intermediates and metabolites, especially those throughout the glycolytic pathway, the pentose phosphate pathway, and the tricarboxylic acid pathway [113]. However, to accumulate a significant concentration of these intermediates (or other, non-native compounds) for industrial purposes, considerable engineering or evolution of S. cerevisiae is often necessary. Methods, such as the Design, Build, Test, and Learn approach (Fig. 1) or tools such as CRISPR/Cas9 [22] have been largely applied for the purpose of producing ethanol by yeast fermentations, but can be and have been easily re-tooled for constructing yeast capable of producing many other chemicals. These chemicals cover many broad categories including isoprenoids, fatty acids, organic acids, rare sugars, sugar alcohols, and others. A recent tour de force of S. cerevisiae engineering came from Galanie et al., in which the group required 23 enzymes from bacteria, mammals, plants, and yeast to produce a tiny amount of opioids, albeit at roughly five orders of magnitude below what would be necessary for industrial scale-up [114]. However, this demonstrates a future for yeast biotechnology in which a single biosynthetic pathway can create downstream products that may otherwise take multiple chemical catalysis steps (Fig. 5 and Table 2).
4.1 Chemical Production by S. cerevisiae
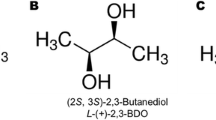
4.1.1 2,3-Butanediol
2,3-Butanediol (2,3-BD) is an increasingly popular target chemical because of its wide applications for synthesizing diverse products such as pharmaceuticals, cosmetics, and industrial solvents. As 2,3-BD is mostly produced by pathogenic bacteria, it is difficult to apply the bacteria to industrial fermentations. S. cerevisiae can produce 2,3-BD naturally, but at a very low concentration, because of ethanol serving as the major fermentative end product. Therefore, researchers have engineered S. cerevisiae to generate a higher titer of 2,3-BD by the elimination of ethanol production through the disruption of alcohol dehydrogenases (ADH1, ADH3, and ADH5). Ng et al. achieved a titer of 2.29 g 2,3-BD/L with a yield of 0.113 g/g glucose [115]. Kim et al. further eliminated the competing pathways by deleting all three pyruvate decarboxylase genes (PDC1, PDC5, and PDC6) and generated a Pdc-deficient mutant to improve the 2,3-BD titer [116]. However, Pdc-deficient mutants had defects such as slow growth, and they required acetate or ethanol supplementation as a carbon source. The Pdc-deficient mutants also suffered from redox imbalance because of glucose repression. The researchers identified point mutation A81P in the transcription regulator Mth1 involved in glucose sensing, which is necessary for glucose tolerance. They also introduced a bacterial 2,3-BD pathway by converting pyruvate into α-acetolactate and then acetoin, respectively, by acetolactate synthase (alsS) and acetolactate decarboxylase (alsD), and then acetoin is reduced into 2,3-BD by butanediol dehydrogenase (BDH1) from Bacillus subtilis. Finally, the engineered S. cerevisiae produced a titer up to 96.2 g/L under a fed-batch fermentation with a yield of 0.28 g/g glucose [116].
Recently, Kim et al. attempted to minimize the glycerol byproduct formation by decreasing the intracellular NADH/NAD+ from the expression of NADH oxidase (noxE) from L. lactis, and the resulting engineered yeast strain was able to produce 2,3-BD with a yield of 0.359 g/g glucose [117]. With a similar approach, Kim and Hahn tried to minimize glycerol production in engineered S. cerevisiae with the additional deletion of glycerol-3-phosphate dehydrogenase (GPD1 and GPD2), creating a strain which could produce a 2,3-BD titer of up to 72.9 g/L in a fed-batch fermentation and with a yield of up to 0.41 g/g glucose [118].
4.1.2 Isoprenoids
Isoprenoids, also known as terpenes, are a diverse group of chemical compounds typically utilized as medicines, cosmetics, nutritional supplements, food additives, or even as a potential future biofuels [119]. S. cerevisiae harbor natural metabolic pathways to produce certain isoprenoids, although yields and productivities are very poor [120]. Despite the poor natural production, isoprenoids are of great interest because of their diverse structures and wide range of potential uses. Monoterpenes (C10) and sesquiterpenes (C15) are two of the main candidates for jet fuel and biodiesel alternatives because of their low freezing temperature and high ignition stability properties. To produce isoprenoids, acetyl-CoA production is of a high importance because all isoprenoids share the mevalonate metabolic pathway starting from acetyl-CoA [121–123]. Either the bacterial 1-deoxyl-d-xylulose 5-phosphate (DXP) pathway or the eukaryote/archaea mevalonate (MVA) pathway is essential for the biosynthesis of isoprenoids. Both pathways end with the formation of five-carbon monomers dimethylallyl pyrophosphate (DMAPP) and isopentenyl pyrophosphate (IPP). DMAPP and IPP are then condensed and modified by prenyltransferases to form isoprenoid precursors such as geranyl pyrophosphate (GPP, C10) and farnesyl pyrophosphate (FPP, C15) [124].
Monoterpenes (C10) are derived from GPP by monoterpene synthases. Fischer et al. is the first group able to produce geraniol, a monoterpene and alcohol, with a titer of up to 5 mg/L in S. cerevisiae by a mutation of ERG20 (farnesyl pyrophosphate synthase) and the overexpression of heterologous geraniol synthase (monoterpene synthases) from Ocimum basilicum [122]. To improve the monoterpene biosynthesis, Ignea et al. used the yeast sterol biosynthesis pathway genes HMG2, ERG20, and IDI1 and co-expression of two terpene synthase enzymes (cineole synthase) from Salvia fruticosa and Salvia pomifera. The final titer of cineole was up to 1 g/L [125].
Sesquiterpenes (C15) are another isoprenoid-derived potential fuel source which has recently gained interest for several industrial applications. Bisabolene, a precursor of bisabolane, was produced at a titer of over 900 mg/L in engineered S. cerevisiae by Peralta-Yahya et al. [126]. The yeast was first engineered by overexpression of acetyl-CoA acetyltransferase (ERG10), isoprenyl diphosphate isomerase (IDI1), and farnesyl pyrophosphate synthase (ERG20), truncated HMG-CoA reductase (tHMGR), and the transcriptional regulator of the sterol pathway (Upc2-1). Then researchers examined six different bisabolene synthases isolated from Arabidopsis thaliana, Picea abies, Pseudotsuga menziesii, and Abies grandis. Finally they developed the highest titer with the codon-optimized bisabolene synthase (BIS) from A. grandis [126]. Recently, Özaydın et al. screened the S. cerevisiae deletion collection for carotenoid production and constructed a strain producing the highest titer of up to 5.2 g/L of bisabolene through double deletion of YJL064W and YPL062W [127].
Several diterpenes (C20) have also been produced by engineered yeast. In 2008, a titer of 8.7 mg/L of taxadiene was achieved from engineered S. cerevisiae [128]. This feat was achieved through two general metabolic modifications: (1) coexpression of a codon-optimized Taxus chinensis taxadiene synthase and a Sulfolobus acidocaldarius geranylgeranyl diphosphate synthase and (2) expression of a truncated 3-hydroxyl-3-methylglutaryl-CoA reductase isoenzyme and a mutant regulatory protein UPC2-1 allowing for steroid uptake in anerobic conditions. In 2012, miltiradiene, another diterpene, was overproduced through metabolic engineering of S. cerevisiae. Zhou et al. achieved a peak titer of miltiradiene of 365 mg/L [129] and Dai et al. obtained 488 mg/L through a fed-batch fermentation [130]. The 488 mg/L titer was achieved through multiple S. cerevisiae metabolic engineering and fermentation technology steps: (1) overexpression of a mutated global regulatory factor (upc2.1) and a truncated 3-hydroxyl-3-methylglutaryl-CoA reductase (tHMGR), (2) copalyl diphosphate synthase was first expressed, (3) overexpression of a fusion gene of farnesyl diphosphate synthase (ERG20) and an endogenous geranylgeranyl diphosphate (BTS1) together with a geranylgeranyl diphosphate synthase from Sulfolobus acidocaldarius (SaGGPS), and (4) use of a fed-batch fermentation [130].
Artemisinin is a sesquiterpene lactone which has received notoriety as an antimalarial drug following its discovery by You-You Tu in the 1970s [131, 132]. Unfortunately, the natural isolation and industrial production process for artemisinin is not always reliable, and shortages of this vital drug have been reported [133]. Production of artemisinin through a reliable and sustainable microbial cell factory could be a viable alternative. Several labs have worked to construct such a process. An important precursor for artemisinin production, amorpha-4,11-diene, was produced by Lindahl et al. in 2006 [134]. This result was achieved by subcloning the amorpha-4,11-diene synthase from Artemisia annua into a galactose-inducible, high-copy number pYeDP60 plasmid and subsequent transformation of the plasmid into an S. cerevisiae strain. Although further optimizations are needed before industrial-scale applications, the final titer, 600 μg/L, served as an important step toward microbial production of artemisinin.
Within a year of the report of amorpha-4,11-diene the process of producing artemisinic acid from engineered yeast was published. Artemisinic acid serves as the immediate precursor of artemisinin and can undergo further chemical synthesis to produce artemisinin. In their report, Ro et al. achieved a peak titer of ~100 mg/L of artemisinic acid [135]. A multitude of engineering steps were necessary to achieve this production in an engineered S. cerevisiae, broadly including increasing farnesyl pyrophosphate (FPP) production and reducing its use for sterols, expressing the amorphadiene synthase gene from A. annua into the improved FPP-producing strain, and cloning a novel cytochrome P450 to provide a three-step oxidation pathway from amorphadiene to artemisinic acid.
More recently, significant boosts in the production of both amorpha-4,11-diene and artemisinic acid from engineered S. cerevisiae have been reported. Lenihan et al. produced a titer of 2.5 g/L of artemisinic acid from an engineered S. cerevisiae by using a defined medium containing galactose as a carbon source and inducer in a fed-batch process which utilized a precise agitation and feed pump rate [5]. A Pmet3 promoter was used to control ERG9, which improved precursor availability for artemisinic acid synthesis by limiting sterol synthesis. Later, Westfall et al. hypothesized that high titers of artemisinic acid may be unachievable without improvement to the production of necessary precursors [136], such as the previously discussed amorpha-4,11-diene. Through overexpression of every mevalonate pathway enzyme through ERG20 and fermentation optimization resulted in a considerably titer of 40 g/L amorpha-4,11-diene [136].
4.1.3 Organic Acids
Organic acids are widely used for many applications including usage as food additives. However, organic acids also serve as building blocks of many larger polymers by undergoing several steps of chemical catalysis. For example, lactic acid is produced by engineered S. cerevisiae by introducing lactate dehydrogenase (ldh). Through catalysis, polylactic acid (also known as polylactide; PLA) can be produced [137]. PLA is a renewable and biodegradable polyester used for many purposes including as a filament for 3D printing, for producing medical screws/implants, and for producing plastic dinnerware. Numerous studies have been conducted for producing lactic acid from engineered S. cerevisiae from a variety of feedstocks including glucose [138, 139], xylose [140], and cellobiose [141]. Currently, no study using engineered yeast has been able to achieve the theoretical maximum of lactic acid production from glucose, xylose, cellobiose, or a mixture of these carbon sources, so work is ongoing to improve these fermentation processes. Of the studies which have generated lactic acid-producing S. cerevisiae, a variety of ldh sources have been used, including bovine materials [142, 143], Rhizopus oryzae [140, 141, 144], Bifidobacterium longum [142], and Lactobacillus plantarum [145, 146]. Moving forward, expression of ldh from yet-unstudied sources into S. cerevisiae may prove useful for producing specific ratios of l- or d-lactic acid, which can be beneficial for specific industrial applications.
Because itaconic acid has many industrial uses, including serving as a copolymer for producing plastics and rubbers [147], this compound is another interesting organic acid which has recently been produced at a laboratory-scale in engineered S. cerevisiae. To achieve a peak titer of 168 mg/L of itaconic acid from S. cerevisiae, several metabolic engineering steps were implemented [148]. First, the cis-aconitic acid decarboxylase encoding gene (CAD) from Aspergillus terreus was expressed in an S. cerevisiae strain under the control of a strong “Enhanced” GPD promoter. Second, several gene targets including ADE3, BNA2, and TES1 were identified by a genome-wide stoichiometric model, deleted, and assessed for itaconic acid production improvements. Finally, the triple deletion strain expressing the A. terreus CAD was grown in optimized fermentation conditions including a high cell density to provide the peak titer of 168 mg/L itaconic acid. However, scale-up to a cost-effective and efficient industrial-scale process require further optimization, as a titer of more than 80 g/L of itaconic acid is considered necessary [148].
As with itaconic acid, muconic acid is another platform chemical which can act as a precursor for the production of many useful products, including various renewable plastics [149]. The first reported instance of muconic acid production by engineered S. cerevisiae was in 2012, resulting in a peak titer of approximately 1.56 mg/L muconic acid [150]. However, by 2013, several metabolic engineering improvements allowed for production of 141 mg/L muconic acid from an engineered S. cerevisiae [151]. Several metabolic engineering steps were needed to produce this result. First, Candida albicans catechol 1,2-dioxygenase, Enterobacter cloacae protocatechuic acid decarboxylase, and Podospora anserine dehydroshikimate dehydratase were expressed in an S. cerevisiae strain. Then ARO3 was deleted and a feedback-resistant mutant ARO4 was expressed to reduce shikimate pathway feedback inhibition. Next, ZWF1 was deleted and TKL1 was overexpressed to increase precursor flux into the target pathway. Finally, several heterologous enzyme levels were balanced, resulting in the final titer of 141 mg/L muconic acid [151].
Succinic acid is a value-added organic acid which can be overproduced by engineered yeast [152–154]. Similar to lactic acid, succinic acid can be used as a precursor to several polyesters [155]. Furthermore, succinic acid is designated as GRAS by the U.S. Food and Drug Administration, which has allowed its use in the food industry as an acidity regulator. As an intermediate of the citric acid cycle (or tricarboxylic acid cycle), yeast natively produces succinic acid if provided with an aerobic environment, but overproduction of succinic acid requires multiple genetic perturbations. For example, Otero et al. constructed an engineered S. cerevisiae with deletions of SDH3, SER3, and SER33 to reduce primary succinate-consuming reactions and to interrupt glycolysis-derived serine [154]. The resulting engineered yeast displayed a 30-fold improvement in succinic acid titer and a 43-fold improvement in succinic acid yield as compared to the control strain.
Beyond succinic acid, glycolic acid, a C2 hydroxy acid, has gained attention in recent years. The global glycolic acid production in 2011 was approximately 40,000,000 kg with this expected to more than double by 2018 [156]. Glycolic acid is often used as a building block of a polyglycolate. The polyglycolate polymer is used as a packaging material because of its high gas permeability and mechanical strength. However, most glycolic acid is produced in a chemical process which relies on non-renewable fossil resources [156]. As an alternative, a biological route for the production of glycolic acid exists which involves converting glyoxylate through glyoxylate reductase into glycolic acid. To overproduce glycolic acid successfully, efficient glyoxylate reductase activity in an engineered S. cerevisiae is required. A further improvement, up to approximately 1 g/L glycolic acid, can be achieved by deletions of the malate synthase (MLS1) and the cytosolic form of isocitrate dehydronase (IDP2) genes [156]. As the current generation of organic acids produced by S. cerevisiae continues to improve and develop, it is likely that new, rare, or hard-to-obtain organic acids can be produced in laboratories by engineered S. cerevisiae strains.
4.1.4 Rare Sugars, Sugar Alcohols, and Antioxidants
Sugars such as l-ribose, d-allose, d-tagatose, and d-psicose are classified as rare sugars. As the name implies, these sugars are rarely found in nature, but they have beneficial health properties. l-Ribose, for example, is considered a very important intermediate to produce chemicals for pharmaceutical and food products [157, 158]. Although d-ribose is very common in nature, l-ribose is not found in nature based on current knowledge. The driving demand for l-ribose production is its potential as a building block for l-nucleoside-based pharmaceutical compounds. l-Nucleoside-based compounds or analogs play an important role in treating viral infections and cancers [159]. Currently, research regarding rare sugar production by engineered yeast is very limited.
Sugar alcohols such as erythritol, xylitol, or sorbitol have a high demand in the food industry because of their sweetening properties without causing dental caries [160]. Although generally difficult, one positive aspect of sugar alcohol production is that, in general, sugar alcohols are not fermentable by S. cerevisiae, which limits reuptake by engineered yeast designed to overproduce target sugar alcohols. The interest in producing sugar alcohols dates back more than 50 years, with at least one study investigating d-arabitol production in Saccharomyces spp. [161]. More recently, a minute titer of 44 μg/mL mannitol was produced by expression of multiple copies of the E. coli mannitol-1-phosphate dehydrogenase gene (mtlD) into S. cerevisiae [162]. This titer was later improved upon by Costenoble et al. by producing a titer of nearly 400 mg/L of mannitol in an engineered S. cerevisiae in anaerobic conditions [163]. Primarily, this was achieved by expression of the E. coli mtlD into an S. cerevisiae strain and deletion of GPD1 and GPD2 followed by an oxygen-sparged fermentation which was switched to nitrogen-sparging during the exponential growth phase.
As one primary example of a well-known sugar alcohol, xylitol shares similar sweetening power with sucrose, but it does not contribute to dental caries and has a cooling effect when eaten. A chemical hydrogenation process to produce xylitol has existed for decades [164] but, more recently, several groups have produced high xylitol titers and yields from biological, engineered yeast systems [165–167]. Oh et al. were able to produce xylitol rapidly and efficiently using an engineered S. cerevisiae expressing xylose reductase (XYL1), a cellodextrin transporter (cdt-1), and an intracellular β-glucosidase (gh1-1) via simultaneous utilization of xylose and cellobiose [166]. As a result, the engineered S. cerevisiae was able to produce xylitol at the maximum theoretical yield by co-utilization of xylose and cellobiose.
Because antioxidants have been considered potentially beneficial as supplements to the human diet, there has been increased interest in efficiently producing these compounds from a consistently obtainable source rather than depending on extraction from seasonally-available produce. Resveratrol is one of these compounds of interest, as it is a common component of grape skins and wines made from these skins [168]. Many studies discussing the engineering of S. cerevisiae and other microbes for the microbe-based production of resveratrol have been published in recent years [169–172]. In one example, an engineered S. cerevisiae expressing a codon-optimized bacterial tyrosine ammonia lyase and an E. coli high-capacity, low-affinity arabinose transporter (araE) were able to produce a peak of 3.44 mg/L at 48 h in a laboratory-scale grape juice fermentation [172]. This result is an important step from an industrial standpoint, as it represented a method to increase the resveratrol concentration in white wine, which in most cases has a significantly lower resveratrol concentration than red wine.
As with resveratrol production, glutathione is another antioxidant which has been extensively studied for production by engineered S. cerevisiae [173–178]. Microbe-based production of glutathione is currently the primary industrial process for glutathione synthesis, although it can also be produced by chemical synthesis [179]. Because the microbe-based process is the major method of industrial-scale production, many varied processes to improve the titer, yield, and productivity have been explored. Recently, a titer of 320 mg/L of glutathione was achieved by a laboratory-evolved S. cerevisiae strain in an acrolein-containing medium [178]. Acrolein is an aldehyde which is toxic to yeast cells [180], although glutathione has been shown to act as a defense against acrolein toxicity, suggesting that cells which have increased resistance to acrolein may be overproducing glutathione [181]. Based on this knowledge, several S. cerevisiae strains were evolved over 250 generations on increasing concentrations of acrolein. Finally, S. cerevisiae strain A4-19 was isolated, which displayed glucose consumption rates, growth rates, and ethanol production rates similar to the parental A4 strain, yet had increased acrolein resistance and a glutathione titer of 320 mg/L, approximately twofold larger than the parental strain [178].
5 Current Scope and Future Outlook of Industrial Fuel and Chemical Production by Yeast
As discussed in Sects. 3 and 4, many advances have been made in recent years in yeast metabolic engineering and synthetic biology for the purpose of biofuel and renewable chemical production. Collectively, these new technologies have resulted in S. cerevisiae strains capable of fermenting a variety of substrates, such as xylose and cellobiose, with improved target product yields and productivities. Only a fraction of these laboratory developments have seen implementation at an industrial scale because of prohibitive costs, difficulty in scale-up, or low yields and productivities. For industrial-scale biofuel production, S. cerevisiae is the primary yeast species seeing usage, although lab-scale biofuel production by non-S. cerevisiae yeast, such as Yarrowia lipolytica and Schizosaccharomyces pombe, has seen growth in recent years [182, 183]. However, several non-S. cerevisiae microbes are used for industrial chemical production because of the wide range of target chemicals produced by the biobased chemical industry. Although S. cerevisiae is extremely hardy and can be easily engineered, there are instances where other microbes are preferred for a target product. Perhaps the most notable example is the use of engineered E. coli for the production of recombinant insulin [184], and over 150 recombinant therapeutics have been approved by the European Medicines Agency [185]. However, only approximately one-third of approved therapeutics utilize engineered E. coli, with S. cerevisiae and other yeasts also accounting for a significant portion of industrial therapeutics, fuels, and chemicals [185].
At the industrial scale, ethanol is the major biofuel target, especially by engineered S. cerevisiae [186, 187]. Ethanol is commonly used as a fuel additive for the creation of gasoline-ethanol blends. The use of ethanol blends in the United States has grown from less than 5 vol% to over 10 vol% in the past decade [188]. This growth is at least partially attributed to the United States Environmental Protection Agency’s Renewable Fuel Standard, which requires up to 17.4 billion gallons of renewable fuel production by 2016, of which 0.21 billion gallons must be cellulosic biofuel [189]. The total production requirement for renewable fuels can increase to 36 billion gallons by 2020 [190].
To achieve the renewable fuel standards set by the United States and other governments, industrial fuel producers have used S. cerevisiae as their platform microbial strain of choice. As of 2014, approximately 23.8 billion gallons of ethanol are produced on an annual basis worldwide, almost entirely from fermentation by S. cerevisiae [28]. The United States and Brazil are responsible for the vast majority of global bioethanol production, annually producing 14.3 billion gallons and 6.2 billion gallons, respectively [28]. In the United States, corn serves as the primary feedstock, whereas in Brazil, sugarcane is the major feedstock for the purpose of bioethanol production [191, 192].
As the two major bioethanol-producing countries, both nations have considerable motivation for the success of their respective ethanol industries. In Brazil, ethanol serves as a transportation fuel at nearly a 1:1 ratio with gasoline [193]. In the United States, roughly 40% of corn produced is used for the purpose of producing ethanol [194]. Both nations provide protection to their bioethanol industries in the form of tax breaks, subsidies, or increased tariffs toward imported ethanol. Moving forward, it is expected that these economic benefits are likely to shift away from first-generation biofuels (using corn and sugarcane juice as the feedstock) toward second-generation biofuels (using corn stover, switchgrass, and miscanthus). As government and environmental protection groups provide further incentives for renewable biofuel production by engineered yeast, scientific advances developed for producing fuel can be modified and applied to the production of non-fuel chemicals by engineered yeast. However, despite legislation in the United States and elsewhere to encourage biofuel production, no equivalent guidelines exist to provide incentive specifically for the purpose of biobased, non-fuel chemical production. A global effort to limit average global Earth surface temperatures to increasing by no more than 2°C relative to temperatures in the late nineteenth century by reducing greenhouse gas emissions has provided a minor incentive for renewable chemical production [195]. The influence this legislation has on biobased chemicals is small because of less than 10% of total fossil fuels being employed for chemical catalysis, with the vast majority going toward the energy and transportation fuel industries [196, 197].
Since early 2014, global oil prices have fallen rapidly and dramatically [198]. Unsurprisingly, as fossil fuel costs decrease, the economic production of biofuels and renewable chemicals becomes increasingly less viable. Not only are second-generation (lignocellulosic) biofuels at economic risk, but even the currently more cost-effective first-generation biofuels become difficult to produce in a cost-effective manner. Roboredo et al. suggest that “huge state subsidies” would be needed to maintain viable biofuel production amidst the crashing oil prices [199]. Although the short-term outlook on biofuel and renewable chemical production is uncertain, it is anticipated that the continuing volatility of oil prices is likely to encourage further research for efficient, economical, and renewable biofuel and renewable chemical production.
Although there are many companies which produce renewable fuels or fuel additives, there also exist many companies worldwide which employ microbial fermentation for the production of non-fuel, renewable chemicals. In many cases, the exact specifications of the species of microbe used or the precise metabolic pathway engineering protocol are not entirely disclosed. However, some of the more notable companies using a yeast-based fermentation platform include DSM, Verdezyne, BioAmber, Amyris, and NatureWorks, which produce, respectively, succinic acid [200], adipic acid [201], 1,4-butanediol [202], farnesene [203], and lactic acid [204].
6 Conclusion
Equipped with rapid advances in metabolic engineering, synthetic biology, and genomics, the production of fuels and non-fuel chemicals by engineered S. cerevisiae has developed tremendously. Several of these advances have transitioned to industrial-scale fermentation processes, allowing for the sustainable production of many valuable chemicals from renewable biomass. Despite these advances and growing numbers of industrial examples, many barriers still exist, which can hinder the further adoption of S. cerevisiae industrial fermentations.
Currently, global oil prices have reached the lowest levels in approximately a decade [199]. Low oil prices are a major detriment not only to the cost-effective production of renewable fuels and chemicals but also to consumer and government sentiment regarding the short-term importance of developing a renewable chemical industry infrastructure. Furthermore, reduced oil prices significantly lower the cost of petroleum-based chemicals, which places additional pressure on renewable, fermentation-based biochemical production. Despite these pressures, many industrial biobased processes, such as succinic acid production (from E. coli) [205] and bioethanol production (from S. cerevisiae) [186, 187], are still considered to be feasible or even preferential to petrochemical production.
Moving forward, newer and more complex industrial-scale fuels and chemicals can be produced by engineered S. cerevisiae as volatile oil prices and depletion of finite fossil fuels encourage investment in biobased alternatives. Nearly all industrial-scale S. cerevisiae fermentations start as laboratory-scale studies following the “Design, Build, Test, and Learn” cycle (Fig. 1), but simpler single-step metabolic pathways, such as producing lactic acid by a heterologous lactate dehydrogenase [140], can give way to complex, multi-step pathways, such as producing opioids [114]. Collectively, the impact of engineered S. cerevisiae on the biobased fuel and chemical industries is likely to expand in the near future.
References
Hubbert MK (1956) Nuclear energy and the fossil fuel. Drilling Prod Pract 36
Nigam PS, Singh A (2011) Production of liquid biofuels from renewable resources. Prog Energy Combust Sci 37:52–68
Escobar JC, Lora ES, Venturini OJ, Yáñez EE, Castillo EF, Almazan O (2009) Biofuels: environment, technology and food security. Renew Sustain Energy Rev 13:1275–1287
Zeng A, Biebl H (2002) Bulk chemicals from biotechnology: the case of 1,3-propanediol production and the new trends. In: Tools and applications of biochemical engineering science. Springer, Berlin, pp 239–259
Lenihan JR, Tsuruta H, Diola D, Renninger NS, Regentin R (2008) Developing an industrial artemisinic acid fermentation process to support the cost-effective production of antimalarial artemisinin-based combination therapies. Biotechnol Prog 24:1026–1032
Martinez FAC, Balciunas EM, Salgado JM, González JMD, Converti A, de Souza Oliveira RP (2013) Lactic acid properties, applications and production: a review. Trends Food Sci Technol 30:70–83
Berg P, Boland A (2014) Analysis of ultimate fossil fuel reserves and associated CO2 emissions in IPCC scenarios. Nat Resour Res 23:141–158
Shafiee S, Topal E (2009) When will fossil fuel reserves be diminished? Energy Policy 37:181–189
Keranen KM, Weingarten M, Abers GA, Bekins BA, Ge S (2014) Induced earthquakes. Sharp increase in central Oklahoma seismicity since 2008 induced by massive wastewater injection. Science 345:448–451
Throupe R, Simons R, Mao X (2013) A review of hydro “fracking” and its potential effects on real estate. J Real Estate Lit 21:205–232
Jackson RB, Vengosh A, Carey JW, Davies RJ, Darrah TH, O’Sullivan F, Pétron G (2014) The environmental costs and benefits of fracking. Annu Rev Env Resour 39:327–362
Somerville RC, Hassol SJ (2011) The science of climate change. Phys Today 64:48
Hoegh-Guldberg O, Bruno JF (2010) The impact of climate change on the world’s marine ecosystems. Science 328:1523–1528
Friedlingstein P, Andrew R, Rogelj J, Peters G, Canadell J, Knutti R, Luderer G, Raupach M, Schaeffer M, van Vuuren D (2014) Persistent growth of CO2 emissions and implications for reaching climate targets. Nat Geosci 7:709–715
Montzka SA, Dlugokencky EJ, Butler JH (2011) Non-CO2 greenhouse gases and climate change. Nature 476:43–50
Stephanopoulos G (2012) Synthetic biology and metabolic engineering. ACS Synth Biol 1:514–525
Jang Y, Kim B, Shin JH, Choi YJ, Choi S, Song CW, Lee J, Park HG, Lee SY (2012) Bio-based production of C2–C6 platform chemicals. Biotechnol Bioeng 109:2437–2459
Ferreira I, Pinho O, Vieira E, Tavarela J (2010) Brewer’s Saccharomyces yeast biomass: characteristics and potential applications. Trends Food Sci Technol 21:77–84
Krivoruchko A, Siewers V, Nielsen J (2011) Opportunities for yeast metabolic engineering: lessons from synthetic biology. Biotechnol J 6:262–276
Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED (2012) Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705
DiCarlo JE, Conley AJ, Penttilä M, Jäntti J, Wang HH, Church GM (2013) Yeast oligo-mediated genome engineering (YOGE). ACS Synth Biol 2:741–749
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343
Abbott DA, Zelle RM, Pronk JT, Van Maris AJ (2009) Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res 9:1123–1136
Benjaphokee S, Hasegawa D, Yokota D, Asvarak T, Auesukaree C, Sugiyama M, Kaneko Y, Boonchird C, Harashima S (2012) Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. New Biotechnol 29:379–386
Crook NC, Schmitz AC, Alper HS (2013) Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synth Biol 3:307–313
Paddon CJ, Keasling JD (2014) Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol 12:355–367
Panagiotopoulos I, Bakker R, de Vrije T, Claassen P, Koukios E (2013) Integration of first and second generation biofuels: fermentative hydrogen production from wheat grain and straw. Bioresour Technol 128:345–350
Renewable Fuels Association (2014) World Fuel Ethanol Production. In: http://ethanolrfa.org/pages/World-Fuel-Ethanol-Production. Accessed 21 Sept 2015
Scott F, Conejeros R, Aroca G (2013) Attainable region analysis for continuous production of second generation bioethanol. Biotechnol Biofuels 6:171-6834-6-171
Wang P, Dudareva N, Morgan JA, Chapple C (2015) Genetic manipulation of lignocellulosic biomass for bioenergy. Curr Opin Chem Biol 29:32–39
Parreiras LS, Breuer RJ, Narasimhan RA, Higbee AJ, La Reau A, Tremaine M, Qin L, Willis LB, Bice BD, Bonfert BL (2014) Engineering and two-stage evolution of a lignocellulosic hydrolysate-tolerant Saccharomyces cerevisiae strain for anaerobic fermentation of xylose from AFEX pretreated corn stover. PLoS One 9:e107499
Pereira FB, Guimarães PM, Teixeira JA, Domingues L (2010) Optimization of low-cost medium for very high gravity ethanol fermentations by Saccharomyces cerevisiae using statistical experimental designs. Bioresour Technol 101:7856–7863
Macrelli S, Galbe M, Wallberg O (2014) Effects of production and market factors on ethanol profitability for an integrated first and second generation ethanol plant using the whole sugarcane as feedstock. Biotechnol Biofuels 7:26–41
Mullet J, Morishige D, McCormick R, Truong S, Hilley J, McKinley B, Anderson R, Olson SN, Rooney W (2014) Energy sorghum—a genetic model for the design of C4 grass bioenergy crops. J Exp Bot 65:3479–3489
Behera S, Singh R, Arora R, Sharma NK, Shukla M, Kumar S (2015) Scope of algae as third generation biofuels. Front Bioeng Biotechnol 2:90
Wei N, Quarterman J, Jin Y (2013) Marine macroalgae: an untapped resource for producing fuels and chemicals. Trends Biotechnol 31:70–77
Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60:165–182
Kim N, Li H, Jung K, Chang HN, Lee PC (2011) Ethanol production from marine algal hydrolysates using Escherichia coli KO11. Bioresour Technol 102:7466–7469
Busti S, Coccetti P, Alberghina L, Vanoni M (2010) Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae. Sensors 10:6195–6240
Ding WT, Zhang GC, Liu JJ (2013) 3′ Truncation of the GPD1 promoter in Saccharomyces cerevisiae for improved ethanol yield and productivity. Appl Environ Microbiol 79:3273–3281
Puligundla P, Smogrovicova D, Obulam VSR, Ko S (2011) Very high gravity (VHG) ethanolic brewing and fermentation: a research update. J Ind Microbiol Biotechnol 38:1133–1144
Guadalupe-Medina V, Metz B, Oud B, Der Graaf CM, Mans R, Pronk JT, Maris AJ (2014) Evolutionary engineering of a glycerol-3-phosphate dehydrogenase-negative, acetate-reducing Saccharomyces cerevisiae strain enables anaerobic growth at high glucose concentrations. Microb Biotechnol 7:44–53
Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin Y (2013) Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8, e57048
Lee SM, Jellison T, Alper HS (2014) Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels 7:122-014-0122-x. eCollection 2014
Zhou H, Cheng J, Wang BL, Fink GR, Stephanopoulos G (2012) Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 14:611–622
Sarthy AV, McConaughy BL, Lobo Z, Sundstrom JA, Furlong CE, Hall BD (1987) Expression of the Escherichia coli xylose isomerase gene in Saccharomyces cerevisiae. Appl Environ Microbiol 53:1996–2000
Kötter P, Amore R, Hollenberg CP, Ciriacy M (1990) Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet 18:493–500
Walfridsson M, Hallborn J, Penttila M, Keranen S, Hahn-Hagerdal B (1995) Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol 61:4184–4190
Ho NW, Chen Z, Brainard AP (1998) Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859
Chu BC, Lee H (2007) Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv 25:425–441
Wisselink HW, Toirkens MJ, del Rosario Franco Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJ (2007) Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of L-arabinose. Appl Environ Microbiol 73:4881–4891
Richard P, Londesborough J, Putkonen M, Kalkkinen N, Penttila M (2001) Cloning and expression of a fungal L-arabinitol 4-dehydrogenase gene. J Biol Chem 276:40631–40637
Richard P, Putkonen M, Väänänen R, Londesborough J, Penttilä M (2002) The missing link in the fungal L-arabinose catabolic pathway, identification of the L-xylulose reductase gene. Biochemistry (NY) 41:6432–6437
Bettiga M, Bengtsson O, Hahn-Hagerdal B, Gorwa-Grauslund MF (2009) Arabinose and xylose fermentation by recombinant Saccharomyces cerevisiae expressing a fungal pentose utilization pathway. Microb Cell Fact 8:40-2859-8-40
Hamacher T, Becker J, Gardonyi M, Hahn-Hagerdal B, Boles E (2002) Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783–2788
Becker J, Boles E (2003) A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Appl Environ Microbiol 69:4144–4150
Leandro M, Gonçalves P, Spencer-Martins I (2006) Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H symporter. Biochem J 395:543–549
Young EM, Comer AD, Huang H, Alper HS (2012) A molecular transporter engineering approach to improving xylose catabolism in Saccharomyces cerevisiae. Metab Eng 14:401–411
Wang C, Bao X, Li Y, Jiao C, Hou J, Zhang Q, Zhang W, Liu W, Shen Y (2015) Cloning and characterization of heterologous transporters in Saccharomyces cerevisiae and identification of important amino acids for xylose utilization. Metab Eng 30:79–88
Young EM, Tong A, Bui H, Spofford C, Alper HS (2014) Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc Natl Acad Sci U S A 111:131–136
Subtil T, Boles E (2011) Improving L-arabinose utilization of pentose fermenting Saccharomyces cerevisiae cells by heterologous expression of L-arabinose transporting sugar transporters. Biotechnol Biofuels 4:38-6834-4-38
Wang C, Shen Y, Zhang Y, Suo F, Hou J, Bao X (2013) Improvement of L-arabinose fermentation by modifying the metabolic pathway and transport in Saccharomyces cerevisiae. BioMed Res Int 2013:1–9
Hamelinck CN, Van Hooijdonk G, Faaij AP (2005) Ethanol from lignocellulosic biomass: techno-economic performance in short-, middle- and long-term. Biomass Bioenergy 28:384–410
Galazka JM, Tian C, Beeson WT, Martinez B, Glass NL, Cate JH (2010) Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86
Ha S, Galazka JM, Oh EJ, Kordić V, Kim H, Jin Y, Cate JH (2013) Energetic benefits and rapid cellobiose fermentation by Saccharomyces cerevisiae expressing cellobiose phosphorylase and mutant cellodextrin transporters. Metab Eng 15:134–143
Ha SJ, Galazka JM, Kim SR, Choi JH, Yang X, Seo JH, Glass NL, Cate JH, Jin YS (2011) Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci U S A 108:504–509
Li S, Du J, Sun J, Galazka JM, Glass NL, Cate JH, Yang X, Zhao H (2010) Overcoming glucose repression in mixed sugar fermentation by co-expressing a cellobiose transporter and a β-glucosidase in Saccharomyces cerevisiae. Mol BioSyst 6:2129–2132
Ha SJ, Wei Q, Kim SR, Galazka JM, Cate JH, Jin YS (2011) Cofermentation of cellobiose and galactose by an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol 77:5822–5825
Pawar SN, Edgar KJ (2012) Alginate derivatization: a review of chemistry, properties and applications. Biomaterials 33:3279–3305
Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y (2012) An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313
Enquist-Newman M, Faust AME, Bravo DD, Santos CNS, Raisner RM, Hanel A, Sarvabhowman P, Le C, Regitsky DD, Cooper SR (2014) Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 505:239–243
Lee K, Hong M, Jung S, Ha S, Yu BJ, Koo HM, Park SM, Seo J, Kweon D, Park JC (2011) Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol Bioeng 108:621–631
Ra CH, Kim YJ, Lee SY, Jeong G, Kim S (2015) Effects of galactose adaptation in yeast for ethanol fermentation from red seaweed, Gracilaria verrucosa. Bioprocess Biosyst Eng :1–8
Kim H, Ra CH, Kim S (2013) Ethanol production from seaweed (Undaria pinnatifida) using yeast acclimated to specific sugars. Biotechnol Bioprocess Eng 18:533–537
Patwardhan PR, Brown RC, Shanks BH (2011) Understanding the fast pyrolysis of lignin. ChemSusChem 4:1629–1636
Wang K, Kim KH, Brown RC (2014) Catalytic pyrolysis of individual components of lignocellulosic biomass. Green Chem 16:727–735
Wei N, Quarterman J, Kim SR, Cate JH, Jin Y (2013) Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat Commun 4:2580
Wei N, Oh EJ, Million G, Cate JH, Jin Y (2015) Simultaneous utilization of cellobiose, xylose, and acetic acid from lignocellulosic biomass for biofuel production by an engineered yeast platform. ACS Synth Biol 4:707–713
Zhang G, Kong II, Wei N, Peng D, Turner TL, Sung BH, Sohn J, Jin Y (2016) Optimization of an acetate reduction pathway for producing cellulosic ethanol by engineered yeast. Biotechnol Bioeng 113:2587–2596
Domingues L, Lima N, Teixeira JA (2001) Alcohol production from cheese whey permeate using genetically modified flocculent yeast cells. Biotechnol Bioeng 72:507–514
Guimaraes PM, Francois J, Parrou JL, Teixeira JA, Domingues L (2008) Adaptive evolution of a lactose-consuming Saccharomyces cerevisiae recombinant. Appl Environ Microbiol 74:1748–1756
Guimarães PM, Teixeira JA, Domingues L (2008) Fermentation of high concentrations of lactose to ethanol by engineered flocculent Saccharomyces cerevisiae. Biotechnol Lett 30:1953–1958
Sreekrishna K, Dickson RC (1985) Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc Natl Acad Sci U S A 82:7909–7913
Zou J, Guo X, Shen T, Dong J, Zhang C, Xiao D (2013) Construction of lactose-consuming Saccharomyces cerevisiae for lactose fermentation into ethanol fuel. J Ind Microbiol Biotechnol 40:353–363
Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC (2011) Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8:74–79
Mosier NS, Ileleji KE (2014) How fuel ethanol is made from corn. Bioenerg Biomass Biofuel 379–384
Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L (1997) The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J 16:2179–2187
Valadi H, Larsson C, Gustafsson L (1998) Improved ethanol production by glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae. Appl Microbiol Biotechnol 50:434–439
Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372
Nissen TL, Kielland-Brandt MC, Nielsen J, Villadsen J (2000) Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab Eng 2:69–77
Bro C, Regenberg B, Förster J, Nielsen J (2006) In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng 8:102–111
Guo Z, Zhang L, Ding Z, Shi G (2011) Minimization of glycerol synthesis in industrial ethanol yeast without influencing its fermentation performance. Metab Eng 13:49–59
Guadalupe Medina V, Almering MJ, van Maris AJ, Pronk JT (2010) Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol 76:190–195
Zhang L, Tang Y, Guo Z, Ding Z, Shi G (2011) Improving the ethanol yield by reducing glycerol formation using cofactor regulation in Saccharomyces cerevisiae. Biotechnol Lett 33:1375–1380
Zhang A, Kong Q, Cao L, Chen X (2007) Effect of FPS1 deletion on the fermentation properties of Saccharomyces cerevisiae. Lett Appl Microbiol 44:212–217
Hubmann G, Guillouet S, Nevoigt E (2011) Gpd1 and Gpd2 fine-tuning for sustainable reduction of glycerol formation in Saccharomyces cerevisiae. Appl Environ Microbiol 77:5857–5867
Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol 77:2905–2915
Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD (2008) Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Fact 7:36
Krivoruchko A, Serrano-Amatriain C, Chen Y, Siewers V, Nielsen J (2013) Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism. J Ind Microbiol Biotechnol 40:1051–1056
Lian J, Si T, Nair NU, Zhao H (2014) Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng 24:139–149
Si T, Luo Y, Xiao H, Zhao H (2014) Utilizing an endogenous pathway for 1-butanol production in Saccharomyces cerevisiae. Metab Eng 22:60–68
Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K (2011) Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels 4:2089–2090
Avalos JL, Fink GR, Stephanopoulos G (2013) Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol 31:335–341
Yuan J, Ching CB (2014) Combinatorial assembly of large biochemical pathways into yeast chromosomes for improved production of value-added compounds. ACS Synth Biol 4:23–31
Brat D, Weber C, Lorenzen W, Bode HB, Boles E (2012) Cytosolic re-localization and optimization of valine synthesis and catabolism enables increased isobutanol production with the yeast Saccharomyces cerevisiae. Biotechnol Biofuels 5:65
Matsuda F, Ishii J, Kondo T, Ida K, Tezuka H, Kondo A (2013) Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance. Microb Cell Fact 12:119
Huo Y, Cho KM, Rivera JGL, Monte E, Shen CR, Yan Y, Liao JC (2011) Conversion of proteins into biofuels by engineering nitrogen flux. Nat Biotechnol 29:346–351
Kalscheuer R, Luftmann H, Steinbuchel A (2004) Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Appl Environ Microbiol 70:7119–7125
Shi S, Valle-Rodríguez JO, Khoomrung S, Siewers V, Nielsen J (2012) Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol Biofuels 5:10.1186
de Jong BW, Shi S, Siewers V, Nielsen J (2014) Improved production of fatty acid ethyl esters in Saccharomyces cerevisiae through up-regulation of the ethanol degradation pathway and expression of the heterologous phosphoketolase pathway. Microb Cell Fact 13:39
Valle-Rodríguez JO, Shi S, Siewers V, Nielsen J (2014) Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid ethyl esters, an advanced biofuel, by eliminating non-essential fatty acid utilization pathways. Appl Energy 115:226–232
Eriksen DT, HamediRad M, Yuan Y, Zhao H (2015) Orthogonal fatty acid biosynthetic pathway improves fatty acid ethyl ester production in Saccharomyces cerevisiae. ACS Synth Biol 4:808–814
Costenoble R, Picotti P, Reiter L, Stallmach R, Heinemann M, Sauer U, Aebersold R (2011) Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol Syst Biol 7:464
Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD (2015) Complete biosynthesis of opioids in yeast. Science 349:1095–1100
Ng CY, Jung M, Lee J, Oh M (2012) Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb Cell Fact 11:68
Kim S, Seo S, Jin Y, Seo J (2013) Production of 2,3-butanediol by engineered Saccharomyces cerevisiae. Bioresour Technol 146:274–281
Kim J, Seo S, Zhang G, Jin Y, Seo J (2015) Expression of Lactococcus lactis NADH oxidase increases 2,3-butanediol production in Pdc-deficient Saccharomyces cerevisiae. Bioresour Technol 191:512–519
Kim S, Hahn J (2015) Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng 31:94–101
George KW, Alonso-Gutierrez J, Keasling JD, Lee TS (2015) Isoprenoid drugs, biofuels, and chemicals—artemisinin, farnesene, and beyond. In: Biotechnology of isoprenoids. Springer, Cham, pp 355–389
Tippmann S, Chen Y, Siewers V, Nielsen J (2013) From flavors and pharmaceuticals to advanced biofuels: production of isoprenoids in Saccharomyces cerevisiae. Biotechnol J 8:1435–1444
Brennan TC, Turner CD, Krömer JO, Nielsen LK (2012) Alleviating monoterpene toxicity using a two‐phase extractive fermentation for the bioproduction of jet fuel mixtures in Saccharomyces cerevisiae. Biotechnol Bioeng 109:2513–2522
Fischer MJ, Meyer S, Claudel P, Bergdoll M, Karst F (2011) Metabolic engineering of monoterpene synthesis in yeast. Biotechnol Bioeng 108:1883–1892
Albertsen L, Chen Y, Bach LS, Rattleff S, Maury J, Brix S, Nielsen J, Mortensen UH (2011) Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl Environ Microbiol 77:1033–1040
d’Espaux L, Mendez-Perez D, Li R, Keasling JD (2015) Synthetic biology for microbial production of lipid-based biofuels. Curr Opin Chem Biol 29:58–65
Ignea C, Cvetkovic I, Loupassaki S, Kefalas P, Johnson CB, Kampranis SC, Makris AM (2011) Improving yeast strains using recyclable integration cassettes, for the production of plant terpenoids. Microb Cell Fact 10:1–18
Peralta-Yahya PP, Ouellet M, Chan R, Mukhopadhyay A, Keasling JD, Lee TS (2011) Identification and microbial production of a terpene-based advanced biofuel. Nat Commun 2:483
Özaydın B, Burd H, Lee TS, Keasling JD (2013) Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab Eng 15:174–183
Engels B, Dahm P, Jennewein S (2008) Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab Eng 10:201–206
Zhou YJ, Gao W, Rong Q, Jin G, Chu H, Liu W, Yang W, Zhu Z, Li G, Zhu G (2012) Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc 134:3234–3241
Dai Z, Liu Y, Huang L, Zhang X (2012) Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol Bioeng 109:2845–2853
White NJ (1997) Assessment of the pharmacodynamic properties of antimalarial drugs in vivo. Antimicrob Agents Chemother 41:1413–1422
Tu Y (2011) The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med 17:1217–1220
Kindermans J, Pilloy J, Olliaro P, Gomes M (2007) Ensuring sustained ACT production and reliable artemisinin supply. Malar J 6:125
Lindahl A, Olsson ME, Mercke P, Tollbom Ö, Schelin J, Brodelius M, Brodelius PE (2006) Production of the artemisinin precursor amorpha-4,11-diene by engineered Saccharomyces cerevisiae. Biotechnol Lett 28:571–580
Ro D, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943
Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, Fickes S, Diola D, Benjamin KR, Keasling JD, Leavell MD, McPhee DJ, Renninger NS, Newman JD, Paddon CJ (2012) Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci U S A 109:E111–E118
Auras RA, Lim L, Selke SE, Tsuji H (2011) Poly(lactic acid): synthesis, structures, properties, processing, and applications. Wiley
John RP, Nampoothiri KM, Pandey A (2007) Fermentative production of lactic acid from biomass: an overview on process developments and future perspectives. Appl Microbiol Biotechnol 74:524–534
Ishida N, Saitoh S, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, Takahashi H (2006) The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Biosci Biotechnol Biochem 70:1148–1153
Turner TL, Zhang G, Kim SR, Subramaniam V, Steffen D, Skory CD, Jang JY, Yu BJ, Jin Y (2015) Lactic acid production from xylose by engineered Saccharomyces cerevisiae without PDC or ADH deletion. Appl Microbiol Biotechnol 99(19):8023–8033
Turner TL, Zhang G, Oh EJ, Subramaniam V, Adiputra A, Subramaniam V, Skory CD, Jang JY, Yu BJ, Park I (2015) Lactic acid production from cellobiose and xylose by engineered Saccharomyces cerevisiae. Biotechnol Bioeng 113:1075–1083
Ishida N, Saitoh S, Tokuhiro K, Nagamori E, Matsuyama T, Kitamoto K, Takahashi H (2005) Efficient production of L-lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated L-lactate dehydrogenase gene. Appl Environ Microbiol 71:1964–1970
Saitoh S, Ishida N, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, Takahashi H (2005) Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl Environ Microbiol 71:2789–2792
Skory CD (2003) Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J Ind Microbiol Biotechnol 30:22–27
Colombié S, Dequin S, Sablayrolles JM (2003) Control of lactate production by Saccharomyces cerevisiae expressing a bacterial LDH gene. Enzyme Microb Technol 33:38–46
Colombié S, Sablayrolles J (2004) Nicotinic acid controls lactate production by K1-LDH: a Saccharomyces cerevisiae strain expressing a bacterial LDH gene. J Ind Microbiol Biotechnol 31:209–215
Tate BE (1967) Polymerization of itaconic acid and derivatives. In: Fortschritte der Hochpolymeren-Forschung. Springer, Berlin, pp 214–232
Blazeck J, Miller J, Pan A, Gengler J, Holden C, Jamoussi M, Alper HS (2014) Metabolic engineering of Saccharomyces cerevisiae for itaconic acid production. Appl Microbiol Biotechnol 98:8155–8164
Xie N, Liang H, Huang R, Xu P (2014) Biotechnological production of muconic acid: current status and future prospects. Biotechnol Adv 32:615–622
Weber C, Bruckner C, Weinreb S, Lehr C, Essl C, Boles E (2012) Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl Environ Microbiol 78:8421–8430
Curran KA, Leavitt JM, Karim AS, Alper HS (2013) Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab Eng 15:55–66
Yan D, Wang C, Zhou J, Liu Y, Yang M, Xing J (2014) Construction of reductive pathway in Saccharomyces cerevisiae for effective succinic acid fermentation at low pH value. Bioresour Technol 156:232–239
Agren R, Otero JM, Nielsen J (2013) Genome-scale modeling enables metabolic engineering of Saccharomyces cerevisiae for succinic acid production. J Ind Microbiol Biotechnol 40:735–747
Otero JM, Cimini D, Patil KR, Poulsen SG, Olsson L, Nielsen J (2013) Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS One 8, e54144
Goerz O, Ritter H (2013) Polymers with shape memory effect from renewable resources: crosslinking of polyesters based on isosorbide, itaconic acid and succinic acid. Polym Int 62:709–712
Koivistoinen OM, Kuivanen J, Barth D, Turkia H, Pitkänen J, Penttilä M, Richard P (2013) Glycolic acid production in the engineered yeasts Saccharomyces cerevisiae and Kluyveromyces lactis. Microb Cell Fact 12:1
Beerens K, Desmet T, Soetaert W (2012) Enzymes for the biocatalytic production of rare sugars. J Ind Microbiol Biotechnol 39:823–834
Hu C, Li L, Zheng Y, Rui L, Hu C (2011) Perspectives of biotechnological production of L-ribose and its purification. Appl Microbiol Biotechnol 92:449–455
Okano K (2009) Synthesis and pharmaceutical application of L-ribose. Tetrahedron 65:1937–1949
Milgrom P, Ly KA (2012) The role of sugar alcohols, xylitol, and chewing gum in preventing dental diseases. Comprehensive preventive dentistry. Wiley, West Sussex, pp 146–158
Ingram JM, Wood WA (1965) Enzymatic basis for D-arbitol production by Saccharomyces rouxii. J Bacteriol 89:1186–1194
Chaturvedi V, Bartiss A, Wong B (1997) Expression of bacterial mtlD in Saccharomyces cerevisiae results in mannitol synthesis and protects a glycerol-defective mutant from high-salt and oxidative stress. J Bacteriol 179:157–162
Costenoble R, Adler L, Niklasson C, Liden G (2003) Engineering of the metabolism of Saccharomyces cerevisiae for anaerobic production of mannitol. FEMS Yeast Res 3:17–25
Wisniak J, Hershkowitz M, Leibowitz R, Stein S (1974) Hydrogenation of xylose to xylitol. Ind Eng Chem Prod Res Dev 13:75–79
Oh E, Bae Y, Kim K, Park Y, Seo J (2012) Effects of overexpression of acetaldehyde dehydrogenase 6 and acetyl-CoA synthetase 1 on xylitol production in recombinant Saccharomyces cerevisiae. Biocatal Agric Biotechnol 1:15–19
Oh EJ, Ha S, Rin Kim S, Lee W, Galazka JM, Cate JH, Jin Y (2013) Enhanced xylitol production through simultaneous co-utilization of cellobiose and xylose by engineered Saccharomyces cerevisiae. Metab Eng 15:226–234
Jo J, Oh S, Lee H, Park Y, Seo J (2015) Dual utilization of NADPH and NADH cofactors enhances xylitol production in engineered Saccharomyces cerevisiae. Biotechnol J 10:1935–1943
Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444:337–342
Becker JV, Armstrong GO, van der Merwe MJ, Lambrechts MG, Vivier MA, Pretorius IS (2003) Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res 4:79–85
Beekwilder J, Wolswinkel R, Jonker H, Hall R, de Vos CH, Bovy A (2006) Production of resveratrol in recombinant microorganisms. Appl Environ Microbiol 72:5670–5672
Sydor T, Schaffer S, Boles E (2010) Considerable increase in resveratrol production by recombinant industrial yeast strains with use of rich medium. Appl Environ Microbiol 76:3361–3363
Wang Y, Halls C, Zhang J, Matsuno M, Zhang Y, Yu O (2011) Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab Eng 13:455–463
Hara KY, Kiriyama K, Inagaki A, Nakayama H, Kondo A (2012) Improvement of glutathione production by metabolic engineering the sulfate assimilation pathway of Saccharomyces cerevisiae. Appl Microbiol Biotechnol 94:1313–1319
Taskin M (2013) A new strategy for improved glutathione production from Saccharomyces cerevisiae: use of cysteine‐and glycine‐rich chicken feather protein hydrolysate as a new cheap substrate. J Sci Food Agric 93:535–541
Zhao Y, Bian X, You X, Shao F, Xiang X, Deng X, Zhao G, Xu J (2013) Nystatin‐enhanced glutathione production by Saccharomyces cerevisiae depends on γ‐glutamylcysteine synthase activity and K. Eng Life Sci 13:156–162
Mezzetti F, De Vero L, Giudici P (2014) Evolved Saccharomyces cerevisiae wine strains with enhanced glutathione production obtained by an evolution-based strategy. FEMS Yeast Res 14:977–987
Oraby MM, Allababidy M, Ramadan E (2014) Enhancement of antioxidant glutathione production by Saccharomyces cerevisiae growing under stressful condition. Int J 5:160–165
Patzschke A, Steiger MG, Holz C, Lang C, Mattanovich D, Sauer M (2015) Enhanced glutathione production by evolutionary engineering of Saccharomyces cerevisiae strains. Biotechnol J 10:1719–1726
Li Y, Wei G, Chen J (2004) Glutathione: a review on biotechnological production. Appl Microbiol Biotechnol 66:233–242
Kwolek-Mirek M, Bednarska S, Bartosz G, Biliński T (2009) Acrolein toxicity involves oxidative stress caused by glutathione depletion in the yeast Saccharomyces cerevisiae. Cell Biol Toxicol 25:363–378
Vollenweider S, Evers S, Zurbriggen K, Lacroix C (2010) Unraveling the hydroxypropionaldehyde (HPA) system: an active antimicrobial agent against human pathogens. J Agric Food Chem 58:10315–10322
Bakhiet SE, Mahmoud MA (2015) Production of bio-ethanol from molasses by Schizosaccharomyces species. Annu Res Rev Biol 7:45–53
Tai M, Stephanopoulos G (2013) Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng 15:1–9
Goeddel DV, Kleid DG, Bolivar F, Heyneker HL, Yansura DG, Crea R, Hirose T, Kraszewski A, Itakura K, Riggs AD (1979) Expression in Escherichia coli of chemically synthesized genes for human insulin. Proc Natl Acad Sci U S A 76:106–110
Huang C, Lin H, Yang X (2012) Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J Ind Microbiol Biotechnol 39:383–399
Geddes CC, Nieves IU, Ingram LO (2011) Advances in ethanol production. Curr Opin Biotechnol 22:312–319
Jang Y, Park JM, Choi S, Choi YJ, Cho JH, Lee SY (2012) Engineering of microorganisms for the production of biofuels and perspectives based on systems metabolic engineering approaches. Biotechnol Adv 30:989–1000
Anderson J, DiCicco D, Ginder J, Kramer U, Leone T, Raney-Pablo H, Wallington T (2012) High octane number ethanol–gasoline blends: quantifying the potential benefits in the United States. Fuel 97:585–594
United States Environmental Protection Agency (2015) EPA Proposes Renewable Fuel Standards for 2014, 2015, and 2016, and the Biomass-Based Diesel Volume for 2017. In: http://www3.epa.gov/otaq/fuels/renewablefuels/documents/420f15028.pdf. Accessed 21 Sept 2015
Sissine F (2007) Energy independence and security cct of 2007: a summary of major provisions. Library of Congress, Congressional Research Service, Washington DC
de Souza Dias MO, Maciel Filho R, Mantelatto PE, Cavalett O, Rossell CEV, Bonomi A, Leal MRLV (2015) Sugarcane processing for ethanol and sugar in Brazil. Environ Dev 15:35–51
Mumm RH, Goldsmith PD, Rausch KD, Stein HH (2014) Land usage attributed to corn ethanol production in the United States: sensitivity to technological advances in corn grain yield, ethanol conversion, and co-product utilization. Biotechnol Biofuels 7:61-6834-7-61. eCollection 2014
Goldemberg J (2013) Sugarcane ethanol: strategies to a successful program in Brazil. In: Advanced biofuels and bioproducts. Springer, New York, pp 13–20
Wallington T, Anderson J, Mueller S, Kolinski Morris E, Winkler S, Ginder J, Nielsen OJ (2012) Corn ethanol production, food exports, and indirect land use change. Environ Sci Technol 46:6379–6384
IPCC (2014) Climate Change 2014: impacts, adaptation, and vulnerability. Part A: global and sectoral aspects. In: Field CB, Barros VR, Dokken DJ, Mach KJ, Mastrandrea MD, Bilir TE, Chatterjee M, Ebi KL, Estrada YO, Genova RC, Girma B, Kissel ES, Levy AN, MacCracken S, Mastrandrea PR, White LL (eds) Contribution of working group II to the fifth assessment report of the intergovernmental panel on climate change. Cambridge University Press, Canbridge/New York, p 1132
Kircher M (2012) The transition to a bio‐economy: emerging from the oil age. Biofuels Bioprod Biorefin 6:369–375
Dale BE, Anderson JE, Brown RC, Csonka S, Dale VH, Herwick G, Jackson RD, Jordan N, Kaffka S, Kline KL (2014) Take a closer look: biofuels can support environmental, economic and social goals. Environ Sci Technol 48:7200–7203
Wang Q, Li R (2016) Impact of cheaper oil on economic system and climate change: a SWOT analysis. Renew Sustain Energy Rev 54:925–931
Reboredo FH, Lidon F, Pessoa F, Ramalho JC (2016) The fall of oil prices and the effects on biofuels. Trends Biotechnol 34:3–6
Verwaal R, Wu L, Damveld RA, Sagt CMJ (2009) Succinic acid production in a eukaryotic cell. BMC Biotechnol 9:48
Picataggio S, Beardslee T (2013) Biological methods for preparing adipic acid. US Patent No. 8,343,752
Fruchey OS, Manzer LE, Dunuwila D, Keen BT, Albin BA, Clinton NA, Dombek BD (2011) Processes for producing butanediol (BDO), diaminobutane (DAB), succinic dinitrile (SDN) and succinamide (DAM). US Patent Application No. 14/117,141
Gardner TS, Hawkins KM, Meadows AL, Tsong AE, Tsegaye Y (2013) Production of acetyl-coenzyme a derived isoprenoids. US Patent No. 8,603,800
Miller M, Suominen P, Aristidou A, Hause BM, Van Hoek P, Dundon CA (2012) Lactic acid-producing yeast cells having nonfunctional L- or D-lactate:ferricytochrome C oxidoreductase cells. US Patent No. 8,137,953
Pinazo JM, Domine ME, Parvulescu V, Petru F (2015) Sustainability metrics for succinic acid production: a comparison between biomass-based and petrochemical routes. Catal Today 239:17–24
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Turner, T.L., Kim, H., Kong, I.I., Liu, JJ., Zhang, GC., Jin, YS. (2016). Engineering and Evolution of Saccharomyces cerevisiae to Produce Biofuels and Chemicals. In: Zhao, H., Zeng, AP. (eds) Synthetic Biology – Metabolic Engineering. Advances in Biochemical Engineering/Biotechnology, vol 162. Springer, Cham. https://doi.org/10.1007/10_2016_22
Download citation
DOI: https://doi.org/10.1007/10_2016_22
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-55317-7
Online ISBN: 978-3-319-55318-4
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)