Abstract
Cochlear implants can restore hearing to completely or partially deaf patients. The intervention planning can be aided by providing a patient-specific model of the inner ear. Such a model has to be built from high resolution images with accurate segmentations. Thus, a precise segmentation is required. We propose a new framework for segmentation of micro-CT cochlear images using random walks combined with a statistical shape model (SSM). The SSM allows us to constrain the less contrasted areas and ensures valid inner ear shape outputs. Additionally, a topology preservation method is proposed to avoid the leakage in the regions with no contrast.
The research leading to these results received funding from the European Union Seventh Frame Programme (FP7/2007-2013) under grant agreement 304857.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Random walks
- Segmentation
- Shape prior
- Iterative segmentation
- Distance map prior
- Statistical shape model
- SSM
- Cochlea segmentation
- Inner ear segmentation
1 Introduction
The HEAR-EUFootnote 1 project aims at reducing the inter-patient variability in the outcomes of surgical electrode implantation by improving CI designs and surgical protocols using computational models [1, 2]. These models are generally built from the segmentations of high resolution images where a large amount of intra-cochlear structures are visible on the image. In this context, we propose a method that enables an accurate segmentation of the inner ear in micro-CT images which contains the hearing organ known as the cochlea. This aids the generation of accurate patient-specific computational models, which can guide implant design, insertion planning and selection of the best treatment strategy for each patient.
There are a few studies on semi- or fully automatic inner ear segmentation from micro-CT data. However, due to the complexity of the anatomical structure, it is generally a manual procedure [3]. One semi-automatic approach to obtain the cochlea is based on 2D snakes [4], but it requires a high degree of user interaction to locate the initial contour and adjustment of the parameters. Another technique is based on statistical shape models (SSMs) [5], where the high resolution segmentations are used to build a statistical model and assist the segmentation of low resolution cochlear images. In order to accurately segment the cochlea in high resolution micro-CT images using the classical SSM approach introduced by Cootes [6], a large number of processed data sets would be required to learn the correct anatomical variability of the data. The scarce availability of micro-CT images means that we have to consider other segmentation strategies.
In order to alleviate these issues, we proposed a new algorithm using random walks with a distance-based shape prior, which is robust independently of the chosen prior and which requires no user interaction [7, 8]. Random walks segmentation is a graph-based segmentation method proposed by Grady [9]. This technique has become very popular because it is robust to noise and weak boundaries and it can be easily extended to 3D and to an arbitrary number of labels. According to the author, random walks can outperform the well-known graph cuts [10] in terms of weak boundaries since the latter tries to minimize the total edge weights in the cut. Thus, graph cuts may return very small segmentations (“small cut” behaviour) in presence of low contrast, a small number of seeds or noise [9]. Additionally, random walks can be straightforwardly generalized to multi-label segmentation unlike graph cuts which usually use complex alpha-beta techniques [11].
Generally, the intensity information is not enough to obtain the object of interest. Thus, a shape prior can be incorporated to be able to separate the target object from the rest of the image. Some techniques to incorporate prior knowledge into random walks have been proposed. Constrained random walks algorithm is developed for pedestrian segmentation [12]. Given binary pedestrian silhouette images as a training data, a pedestrian shape prior model is built by averaging the training data for every pose, as well as averaging all training data to obtain a general prior model. The pedestrian shape models are incorporated into the random walks formulation. The constrained random walks are applied for every shape model separately, and the final segmentation is the one with the highest probability. Baudin et al. proposed a similar work applied to the skeletal muscle [13]. The prior model of the thigh muscles is derived from learning a Gaussian model based on previous segmentations of the thigh muscles in a training set. The main drawback of both methods is the sensitivity to the average model and to the registration inaccuracies. The same may occur in [14] where prior knowledge is obtained from a probabilistic atlas to perform prostate segmentation. In order to allow large scale deformations, Baudin et al. introduced the principal component analysis (PCA) into the random walks formulation [15]. The shape deformation is constrained to remain close to PCA shape space built from training examples. However, the method does not allow representing shapes that differ too much from the standard shapes [16]. According to the authors, PCA can not deal properly with probabilities. Thus, they suggest to find a different shape space more compatible with probabilities such as the barycentric model. A similar work using PCA is presented in [17] which utilizes a PCA-based shape model as a prior but it is also sensitive to the average shape. In order not to be constrained to the average shape, the guided random walks are proposed [18] where the closest subject to the target object in a given database is retrieved to guide the segmentation. If there is not a close shape in the database, the standard random walks are performed. The limitations of this method are that all the samples of the training data must be considered in order to find the closest data set to the target image and that in case there is no good match, it only relies on the standard random walks. Random walks with shape prior have also been used for video tracking and segmentation [19, 20].
An extension of random walks was presented by Grady in [21] by integrating a non-parametric probability density model which allows localization of disconnected objects and eliminates the requirement of user-specified labels. We use this framework to incorporate prior knowledge into random walks formulation where the region term and the shape prior information given by a SSM constitute the probability density model.
There are some works combining a classical segmentation method with a SSM. Two of the most common methods are based on graph cuts [22,23,24,25,26,27,28] and level sets [29,30,31,32] where they generally use an implicit representation of shapes such as a signed distance map relaxing the need for a costly landmark detection and matching process. In our work, we choose random walks due to the numerous advantages mentioned above.
In this paper, we present an extension of our previous work [7, 8] combining random walks with a SSM to benefit from the strengths of both methods. The region term is combined with a distance-based prior constrained by a SSM. The SSM allows us to constrain the segmentation to a valid inner ear shape to obtain anatomically correct segmentation results. The confidence map adjusts the influence of the prior in certain areas making the method, along with the region term, less sensitive to the average shape. A topology preservation method is also proposed to avoid leakage in the interior and the turns of the cochlea [33]. In the remainder of this paper, we explain the details of the proposed method and show the experimental results on micro-CT images of the inner ear.
2 Random Walks Segmentation
An image can be represented as a graph where the nodes are the pixels of the image, and the weights represent the similarity between nodes. Vertices marked by the user as seeds are denoted by \(V_{m}\) and the rest by \(V_{u}\). Given some seeds, \(v_{j} \in V_{m}\), the random walker assigns to each node, \(v_{i} \in V_{u}\), the probability, \(x_{i}^{s}\), that a random walker starting from that node first reaches a marked node, \(v_j \in V_{m}\) assigned to label \(g^s\). The random walks segmentation is then completed by assigning each free node to the label for which it has the highest probability [9].
An extension to random walks was proposed in [21] by incorporating a probability density model based on the gray-level intensity for each label. Let \(\lambda _{i}^{s}\) be the probability density that the intensity at node \(v_{i}\) belongs to the intensity distribution of label s. The modified random walks segmentation is obtained by solving the following system [21]:
where \(\varLambda = diag(\lambda ^{s}) \), n is the number of labels, \(\gamma \) is a free parameter and L is the Laplacian matrix which can be defined as:

where \(L_{ij}\) is indexed by the vertices \(v_{i}\) and \(v_{j}\) and \(d_{i} = \sum _{j=1}^{n} w_{ij}\). The weight function \( w_{ij}\) can be computed as:
where \(I_{i}\) is the intensity at pixel i and \(\beta \) is a free parameter related to the bandwidth kernel. The weight range is between 0 and 1 and the higher the weight the larger the similarity between pixels [34, 35].
For more details, we refer to [21]. In this work, we use this framework to perform image segmentation but instead of using an intensity-based distribution, we propose a more robust density estimation considering region information as well as shape prior knowledge given by a SSM. We explain them in detail in the remaining part of the section.
2.1 Region Term Formulation
The region term partitions the image in terms of intensities (bright versus dark). A histogram is built from one of the slices of the inner ear. Then, two Gaussian components representing the inner ear including other regions with the same intensity profile and the background are fitted to the histogram with a Gaussian mixture model (GMM). The region-based term can be defined as:

where \(x_{i}\) is the pixel indexed by i, l is the label and \(p(x_{i} |O)\) and \(p(x_{i} | B)\) are the probabilities estimated by the GMM of pixel at i belonging to object and background intensity, respectively.
2.2 Shape Prior Knowledge and Statistical Shape Model
Once the region term is obtained, the shape prior is computed to discard areas which do not belong to the inner ear and have similar intensity values. The use of a SSM can provide a realistic prior to initialize the whole segmentation process, and further be a source of plausible shape regularization during each iteration of the random walker. The SSM is used with a procedure, which we refer to as statistical non-rigid registration, described as follows. We perform a non-rigid image registration between a reference data set, \(I_R\), and the target image, \(I_S\), which in the framework of elastix [36] is formulated as an optimization problem. The (parametric) transformation that aligns the two images, \(T_\eta : I_R \rightarrow I_S\) is described by the vector \(\eta \) containing q-parameters which is found by optimization of a cost function, \(\mathcal {C}\).
The chosen transformation is a B-Spline model regularized by a Statistical Deformation Model (SDM) to constrain the non-rigid registration. The SDM was trained by registering a reference data set against 16 different data sets using the registration model described in [37]. The output of each registration is a vector of q-deformation parameters which describes a B-Spline deformation field. Considering the parameters of the B-Spline model to be corresponding variables, a principal component analysis on the 16 fields was made using Statismo [38] to obtain a description of deformation variability in a reduced parameter-space. This type of transformation model is made available through an integration of the Statismo-elastix packages. The cost function, \(\mathcal {C}\), is solely an image similarity measure, in this case using the normalized correlation coefficient. Note, that if the image intensities were normalized to the HU scale, it would be sufficient to use the sum of squared differences. That was, however, not the case for our data. The optimization is solved using Adaptive Stochastic Gradient Descent [39], which is shown to be a good choice for medical image registration with a limited number of parameters [36, 39].
From the statistical non-rigid registration, the deformation between the reference and target images is applied to the segmentation of the reference data set to obtain the shape prior. This prior is constrained to be an anatomically correct cochlea and from its contour we can build a distance map. The idea is that given an estimation of the location and shape of the object to segment, pixels close to the shape contour are more likely to be labelled as foreground and vice versa. The formulation can be defined as follows [40]:
where \(d(i,\varTheta )\) is the distance of a pixel i from a shape \(\varTheta \), being negative inside the shape and positive outside the shape. Here, \(\mu \) is a penalty term determined by the ratio of points outside the shape compared to the points inside the shape and \(d_{r}\) is the “width” of influence of the shape.
Then, the distance-based shape prior term is:

2.3 Random Walks with Region and Prior Knowledge Terms
We combine the region and shape prior terms by a weighted sum. We use a confidence map to adjust the influence of the shape prior according to the strength of the image contour by reducing the weight of this prior where strong contours are present. The formulation is as follows:
where k is the weight of each term and c is the confidence map defined as \(c_{i} = \exp (-k_{v}\sigma _{r}^{2}(i))\) where \(\sigma _{r}^{2}(i)\) is the variance at pixel i computed on a patch with radius r, and \(k_{v}\) is a free parameter that determines the bandwidth of the Gaussian. Equation 8 is used to obtain \(\lambda ^{s}\) in the random walks formulation in Eq. 1 for every label, which results in a segmentation. This segmentation is statistically non-rigidly registered against the reference segmentation to obtain a new prior constrained by the SSM. Note that this second registration is performed in the binary segmentation in contrast to the initial prior whose registration was between the grayscale reference and target images and the resulting deformation was applied to the reference segmentation to obtain the prior. The distance-based prior is then built from Eq. 6 and the random walks segmentation is performed again. This procedure continues until convergence or until the maximum number of iterations is reached. In order to avoid merging the non-contrasted areas of the cochlea, the topology preserving method described in [33] is proposed. The topology preservation method computes the unit outward normal vector of the contour and when two vectors are pointing in opposite directions, the contours in this area are not allowed to merge.
3 Results
In this experiment, 10 micro-CT data sets of the inner ear are used to perform the segmentation in 3D using the proposed method. The original 3D data set was downsampled from a nominal isotropic resolution of 24.5 \(\mu \)m to 49 \(\mu \)m for computational efficiency reasons. Every data set contains around 213 slices with an average size of 413 x 275 pixels. The ground truth is manually annotated. The initial prior is obtained as described in Sect. 2.2. The SSM is built from 17 different data sets (one reference and 16 training samples).
The following parameters were used to produce the results: \(\gamma =0.8\) in Eq. 1, \(d_r=0\) and \(\mu =1.0\) in Eq. 6 and the total number of iterations are 4 with \(k=0.8\) in Eq. 8.
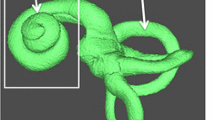
Some inner ear segmentation results using our approach are illustrated in Fig. 1. In this example, we can observe from the 3D volume that the topology of the inner ear shape is preserved and that the contour of the segmentation is adjusted to the edges of the image whereas the interior of the cochlea and less contrasted areas are conserved due to the shape prior and topology preservation method.
To quantify the segmentation quality for the proposed method, we compute the well-known Dice with respect to a manual ground truth. The formulation is defined as \(Dice= \frac{2TP}{2TP+FN+FP}\) where TP and FP stand for true positive and false positive and TN and FN for true negative and false negative. We compare our approach with the initial shape prior (corresponding to using the SSM alone) described in Sect. 2.2. The proposed method achieves a mean Dice index of 0.947 and the initial shape prior reaches a mean Dice index of 0.856. The reason for a lower value is that 17 samples are obviously not enough to cover the true variability in inner ear shapes in high resolution images. The results are presented in Fig. 2 where we can observe a high improvement from using the SSM alone. In contrast with the SSM method alone, the Dice similarity coefficients computed from the segmentation results of the proposed technique have a smaller standard deviation having a small range of Dice values between [0.94,0.95] except for one single case that it has a 0.92 of Dice. The reason of these satisfactory results is that the exterior of the cochlea can be efficiently separated as there is enough contrast between the cochlea and background and the small and invisible regions can be extracted with the guidance of the prior. The topology preservation method prevents leakage in the non-contrasted areas. In high gradient areas of the image (edges) around the prior, the confidence map reduces the influence of the prior coping with the possible artefacts and inaccuracies in the prior shape. It is clear that for internal regions, this method relies on the prior but the SSM constrains the shape of these areas and for the exterior of the inner ear, the region term with the prior can provide promising results.
4 Conclusion
We presented a new framework for the inner ear segmentation in micro-CT using the random walks algorithm which is able to deal with weak boundaries efficiently. The combination of the distance map prior with a region term into random walks provides accurate segmentations of the inner ear. The SSM allows us to constrain the interior part of the cochlea to a valid shape while the exterior of the contour evolves along the shape prior. In this work, the SSM is implemented as a non-rigid registration with learnt statistical shape regularization. The experiments suggest that the proposed approach is robust and accurate for the inner ear segmentation in micro-CT images. As future work, we would like to do an exhaustive analysis and thorough study of this method as well as a comparison with other methods.
Notes
References
Ceresa, M., et al.: Patient-specific simulation of implant placement and function for cochlear implantation surgery planning. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8674, pp. 49–56. Springer, Heidelberg (2014). doi:10.1007/978-3-319-10470-6_7
Ceresa, M., Mangado, N., Andrews, R.J., Ballester, M.A.G.: Computational models for predicting outcomes of neuroprosthesis implantation: the case of cochlear implants. Mol. Neurobiol. 52(2), 934–941 (2015)
Braun, K., Böhnke, F., Stark, T.: Three-dimensional representation of the human cochlea using micro-computed tomography data: presenting an anatomical model for further numerical calculations. Acta Oto-Laryngologica 132(6), 603–613 (2012)
Poznyakovskiy, A.A., Zahnert, T., Kalaidzidis, Y., Lazurashvili, N., Schmidt, R., Hardtke, H.-J., Fischer, B., Yarin, Y.M.: A segmentation method to obtain a complete geometry model of the hearing organ. Hear. Res. 282(1), 25–34 (2011)
Noble, J.H., Labadie, R.F., Majdani, O., Dawant, B.M.: Automatic segmentation of intracochlear anatomy in conventional CT. IEEE Trans. Biomed. Eng. 58(9), 2625–2632 (2011)
Cootes, T.F., Taylor, C.J., Cooper, D.H., Graham, J.: Active shape models-their training and application. Comput. Vis. Image Underst. 61(1), 38–59 (1995)
Pujadas, E.R., Kjer, H.M., Piella, G., Ceresa, M., Ballester, M.A.G.: Random walks with shape prior for cochlea segmentation in ex vivo \(\mu \)CT. Int. J. Comput. Assist. Radiol. Surg. 11(9), 1647–1659 (2016)
Pujadas, E.R., Kjer, H.M., Piella, G., Ballester, M.A.G.: Iterated random walks with shape prior. Image Vis. Comput. 54, 12–21 (2016)
Grady, L.: Random walks for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 28(11), 1768–1783 (2006)
Boykov, Y., Veksler, O.: Graph cuts in vision and graphics: theories and applications. In: Paragios, N., Chen, Y., Faugeras, O. (eds.) Handbook of Mathematical Models in Computer Vision, pp. 79–96. Springer, Heidelberg (2006)
Boykov, Y., Veksler, O., Zabih, R.: Fast approximate energy minimization via graph cuts. IEEE Trans. Pattern Anal. Mach. Intell. 23(11), 1222–1239 (2001)
Li, K.-C., Su, H.-R., Lai, S.-H.: Pedestrian image segmentation via shape-prior constrained random walks. In: Ho, Y.-S. (ed.) PSIVT 2011. LNCS, vol. 7088, pp. 215–226. Springer, Heidelberg (2011). doi:10.1007/978-3-642-25346-1_20
Baudin, P.-Y., Azzabou, N., Carlier, P.G., Paragios, N.: Prior knowledge, random walks and human skeletal muscle segmentation. In: Ayache, N., Delingette, H., Golland, P., Mori, K. (eds.) MICCAI 2012. LNCS, vol. 7510, pp. 569–576. Springer, Heidelberg (2012). doi:10.1007/978-3-642-33415-3_70
Li, A., Li, C., Wang, X., Eberl, S., Feng, D.D., Fulham, M.: Automated segmentation of prostate MR images using prior knowledge enhanced random walker. In: 2013 International Conference on Digital Image Computing: Techniques and Applications (DICTA), pp. 1–7. IEEE (2013)
Baudin, P.-Y., Azzabou, N., Carlier, P.G., Paragios, N.: Manifold-enhanced segmentation through random walks on linear subspace priors. In: Proceedings of the British Machine Vision Conference (2012)
Baudin, P.-Y.: De la segmentation au moyen de graphes d’images de muscles striés squelettiques acquises par RMN. Ph.D. thesis, Ecole Centrale Paris (2013)
Ting, Y., Xiaoming Liu, S., Lim, N.K., Tu, P.H.: Automatic surveillance video matting using a shape prior. In: 2011 IEEE International Conference on Computer Vision Workshops (ICCV Workshops), pp. 1761–1768. IEEE (2011)
Eslami, A., Karamalis, A., Katouzian, A., Navab, N.: Segmentation by retrieval with guided random walks: application to left ventricle segmentation in MRI. Med. Image Anal. 17(2), 236–253 (2013)
Lee, Y.-T., Te-Feng, S., Hong-Ren, S., Lai, S.-H., Lee, T.-C., Shih, M.-Y.: Human segmentation from video by combining random walks with human shape prior adaption. In: 2013 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA), pp. 1–4. IEEE (2013)
Papoutsakis, K.E., Argyros, A.A.: Object tracking and segmentation in a closed loop. In: Bebis, G., et al. (eds.) ISVC 2010. LNCS, vol. 6453, pp. 405–416. Springer, Heidelberg (2010). doi:10.1007/978-3-642-17289-2_39
Grady, L.: Multilabel random walker image segmentation using prior models. In: IEEE Computer Society Conference on Computer Vision and Pattern Recognition, CVPR 2005, vol. 1, pp. 763–770. IEEE (2005)
Cremers, D., Grady, L.: Statistical priors for efficient combinatorial optimization via graph cuts. In: Leonardis, A., Bischof, H., Pinz, A. (eds.) ECCV 2006. LNCS, vol. 3953, pp. 263–274. Springer, Heidelberg (2006). doi:10.1007/11744078_21
Malcolm, J., Rathi, Y., Tannenbaum, A.: Graph cut segmentation with nonlinear shape priors. In: 2007 IEEE International Conference on Image Processing, vol. 4, pp. 365–368. IEEE (2007)
Zhu-Jacquot, J., Zabih, R.: Graph cuts segmentation with statistical shape priors for medical images. In: Third International IEEE Conference on Signal-Image Technologies and Internet-Based System, SITIS 2007, pp. 631–635. IEEE (2007)
El-Zehiry, N., Elmaghraby, A.: Graph cut based deformable model with statistical shape priors. In: 19th International Conference on Pattern Recognition, ICPR 2008, pp. 1–4. IEEE (2008)
Vu, N., Manjunath, B.S.: Shape prior segmentation of multiple objects with graph cuts. In: 2008 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2008, pp. 1–8. IEEE (2008)
Chen, X., Udupa, J.K., Alavi, A., Torigian, D.A.: GC-ASM synergistic integration of graph-cut and active shape model strategies for medical image segmentation. Comput. Vis. Image Underst. 117(5), 513–524 (2013)
Chang, J.C., Chou, T.: Iterative graph cuts for image segmentation with a nonlinear statistical shape prior. J. Math. Imaging Vis. 49(1), 87–97 (2014)
Leventon, M.E., Eric, W., Grimson, L., Faugeras, O.: Statistical shape influence in geodesic active contours. In: 2000 Proceedings of IEEE Conference on Computer Vision and Pattern Recognition, vol. 1, pp. 316–323. IEEE (2000)
Tsai, A., Yezzi, A., Wells, W., Tempany, C., Tucker, D., Fan, A., Grimson, W.E., Willsky, A.: A shape-based approach to the segmentation of medical imagery using level sets. IEEE Trans. Med. Imaging 22(2), 137–154 (2003)
Cremers, D.: Dynamical statistical shape priors for level set-based tracking. IEEE Trans. Pattern Anal. Mach. Intell. 28(8), 1262–1273 (2006)
Cremers, D., Rousson, M., Deriche, R.: A review of statistical approaches to level set segmentation: integrating color, texture, motion and shape. Int. J. Comput. Vis. 72(2), 195–215 (2007)
Pujadas, E.R., Kjer, H.M., Vera, S., Ceresa, M., Ballester, M.A.G.: Cochlea segmentation using iterated random walks with shape prior. In: SPIE Medical Imaging, p. 97842U. International Society for Optics and Photonics (2016)
Pujadas, E.R., Reisert, M.: Shape-based normalized cuts using spectral relaxation for biomedical segmentation. IEEE Trans. Image Process. 23(1), 163–170 (2014)
Ruiz, E., Reisert, M.: Image segmentation using normalized cuts with multiple priors. In: SPIE Medical Imaging, p. 866937. International Society for Optics and Photonics (2013)
Klein, S., Staring, M., Murphy, K., Viergever, M.A., Pluim, J.P.W.: Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29(1), 196–205 (2010)
Kjer, H.M., Fagertun, J., Vera, S., Gil, D., Ballester, M.Á.G., Paulsen, R.R.: Free-form image registration of human cochlear \(\mu \)CT data using skeleton similarity as anatomical prior. Pattern Recogn. Lett. 76, 76–82 (2015)
Lüthi, M., Blanc, R., Albrecht, T., Gass, T., Goksel, O., Büchler, P., Kistler, M., Bousleiman, H., Reyes, M., Cattin, P., Vetter, T.: Statismo - a framework for PCA based statistical models. Insight J. 1, 1–18 (2012)
Klein, S., Pluim, J.P.W., Staring, M., Viergever, M.A.: Adaptive stochastic gradient descent optimisation for image registration. Int. J. Comput. Vis. 81(3), 227–239 (2009)
Kohli, P., Rihan, J., Bray, M., Torr, P.H.S.: Simultaneous segmentation and pose estimation of humans using dynamic graph cuts. Int. J. Comput. Vis. 79(3), 285–298 (2008)
Acknowledgments
The research leading to these results received funding from the European Union Seventh Frame Programme (FP7/2007–2013) under grant agreement 304857, HEAR-EU Project.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Ruiz Pujadas, E., Kjer, H.M., Piella, G., Ballester, M.A.G. (2016). Statistical Shape Model with Random Walks for Inner Ear Segmentation. In: Reuter, M., Wachinger, C., Lombaert, H. (eds) Spectral and Shape Analysis in Medical Imaging. SeSAMI 2016. Lecture Notes in Computer Science(), vol 10126. Springer, Cham. https://doi.org/10.1007/978-3-319-51237-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-51237-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-51236-5
Online ISBN: 978-3-319-51237-2
eBook Packages: Computer ScienceComputer Science (R0)