Abstract
Replicating loops represent a class of benchmarks, which is commonly studied in relation with cellular automata. Most of the known loops, for which replication rules exist in two-dimensional cellular space, create the copies of themselves using a certain construction algorithm that is common for all the emerging replicas. In such cases, the replication starts from a single instance of the loop (represented as the initial state of the cellular automaton) and is controlled by the transition function of the automaton according to which the copies of the loop are developed. Despite the fact that universal replicators in cellular automata are possible (for example, von Neumann’s Universal Constructor), the process of replication of the loops is usually specific to the shape of the loop and the replication rules given by the transition function. This work presents a method for the automatic evolutionary design of cellular automata, which allows us to design transition functions for various structures that are able to replicate according to a given specification. It will be shown that new replicating loops can be discovered that exhibit some unconventional features in comparison with the known solutions. In particular, several scenarios will be presented which can, in addition to the replication from the initial loop, autonomously develop the given loop from a seed, with the ability of the loop to subsequently produce its replicas according to the given specification. Moreover, a parallel replicator will be shown that is able to develop the replicas to several directions using different replication algorithms.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Since the introduction of cellular automata (CA) in [20], researchers have dealt, among others, how to effectively design a cellular automaton (and its transition function in particular) to solve various problems. For example, cellular automata have been studied for their ability to perform computations, e.g. using principles from the famous Conway’s Game of Life [1] or by simulating elementary logic functions in non-uniform cellular matrix [15].
One of the topics widely studied in the area of artificial life is the problem of (self-)replicating loops. Since the introduction of probably the most known loop by Langton [8], which is able to replicate in 151 steps in a CA working with 8 states, some other researchers have dealt with this topic trying to simplify the replication process or enhance the abilities of the loop during replication. For example, Byl introduced a smaller loop that is able to replicate in 25 steps using a CA that works with 6 cell states [4]. Later, several unsheathed loops were proposed by Reggia et al. from which the simplest loop consists of 6 cells only and is able to replicate using 8-state CA in 14 steps [12]. On the other hand, Tempesti studied a possibility to introduce construction capabilities into the loops and proposed a 10-state CA that allows to generate patters inside the replicating structures [19]. Perrier et al. created a “self-reproducing universal computer” using 64-state CA by “attaching” executable programs (Turing Machines) on the loops [11]. Although the aforementioned solutions were achieved using analytic methods, the process of determining suitable transition rules for a given problem represents a difficult task and requires an experienced designer (the process of “programming” the CA is not intuitive). As the number of cell states increases, the process of the CA design becomes challenging due to a significant increase of the solution space. Moreover, for some problems no analytic approach has yet been known to the design of the transition rules. In such cases various unconventional techniques have been applied including Genetic Algorithm (GA) [7], possibly in combination with other heuristics.
For example, Mitchell et al. investigated a problem of performing computations in cellular automata using GA [9]. Their work contains a comparison with the original results obtained by Packard in [10] which can be considered as a milestone in applying evolutionary algorithms (EA) to the design and optimisation of cellular automata. In particular, the authors in [9] claim: “Our experiment produced quite different results, and we suggest that the interpretation of the original results is not correct.” It may indicate that the research of cellular automata (and their typical features like emergent behaviour or cooperative cell signalling by means of local rules) using various computing techniques can provide valuable information for advanced studies and applications in this area. Note that Mitchell et al. considered binary (i.e. 2-state) 1D cellular automata only which represent a fundamental concept for advanced models. Sipper proposed a technique called Cellular Programming (a spatially distributed and locally interacting GA) that allows for the automatic design of non-uniform CA that are well suited to various problems [16]. Sapin et al. introduced a GA-based approach to the design of gliders and glider guns in 2D cellular automata [13, 14]. It was shown that a spontaneous emergence of glider guns in CA can occur with a significant number of new gun-based and glider structures discovered by EA. The aim of the glider research was to construct a system for collision-based computationally universal cellular automata that are able to simulate Turing machines [14]. In recent years, several solutions emerged that aim to optimize the CA design by introducing various evolution-based and soft-computing techniques in combination with suitable representations of the transition functions. For example, Elmenreich et al. proposed an original technique for the calculation of the transition function using neural networks (NN) [5]. The goal was to train the NN by means of Evolutionary Programming [6] in order to develop self-organising structures in the CA. A novel technique for encoding the transition functions of CA, called Conditionally Matching Rules, was introduced in [3], and some applications in binary CA with advantages over the conventional (table-based) encoding were presented in [2].
Whilst the most of the aforementioned studies considered binary CA (i.e. those working with two cell states only), which may be suitable for straightforward hardware implementations (e.g. Sipper’s Firefly machine [17]), multi-state CA can provide a more efficient way for the representation and processing of information in CA thanks to the ability of individual cells to work with more than two states. This feature is important for studying complex systems that are in most cases described by integer (or real-valued) variables. In addition, the introduction of more than two states per cell in the CA may allow to reduce the resources needed to solve a given problem (e.g. the size of the cellular array or dimension of the automaton). For example, Yunès studied computational universality in multi-state one-dimensional cellular automata [21]. A technique for the construction of computing systems in 2D CA was demonstrated by Stefano and Navarra in [18] using rules of a simple game called Scintillae working with 6 cell states. Their approach allows to design components (building blocks) for the construction of bigger systems, e.g. on the basis of gate-level circuits.
The goal of this study is to demonstrate the evolutionary design of 2D cellular automata, using the concept of conditionally matching rules to encode the transition functions, which are able to replicate the given structures with respect to a given arrangement in the cellular array. In particular, uniform, multi-state cellular automata will be treated, the cells of which work with 8 and 10 states. The GA will be applied in order to design suitable transition rules that perform replication of the given structure according to the designer’s specification. It will be shown that novel replication scenarios can be found in CA that can copy the given loop not only from its initial instance but also, from a seed the loop can autonomously grow. Moreover, a parallel replication scheme will be presented, the objective of which is to speed-up the replication process by allowing the structures to replicate to more directions in the 2D CA. The results will demonstrate the ability of the GA to discover different replication scenarios for the replicas developing in parallel in the cellular automaton, which will be encoded in a single evolved transition function.
2 Cellular Automata
The original concept of cellular automaton, introduced in [20], which will be considered in this study, assumes a 2D matrix of cells, each of which at a given moment acquires a state from a finite set of states. The development of the CA is performed synchronously in discrete iterations (time steps) by updating the cell states according to local transition functions of the cells. Uniform cellular automata will be investigated in which the local transition function is identical for all cells and hence it can be considered as a transition function of the CA. The next state of each cell depends on the combination of states in its neighbourhood. In this work, von Neumann neighbourhood will be assumed that includes a given (Central) cell to be updated and its immediate neighbours in the North, South, East and West direction (i.e. it is a case of a 5-cell neighbourhood).
Since the CA behaviour can practically be evaluated in the cellular array of a finite size, boundary conditions need to be specified in order to correctly determine cell states at the edge of the array. Cyclic boundary conditions will be implemented which means that cells at an edge of the CA are “connected” to the appropriate cells on the opposite edge (i.e. these cells are considered as neighbours) in each dimension. In case of the 2D CA the shape of such cellular array can be viewed as a toroid.
The transition function is usually defined as a mapping that for all possible combinations of states in the cellular neighbourhood determines a new state. This mapping can be represented as a set of rules of the form \(N_t\ W_t\ C_t\ E_t\ S_t\rightarrow C_{t+1}\) where \(N_t, W_t, C_t, E_t\) and \(S_t\) denote cell states in the defined neighbourhood at a time t and \(C_{t+1}\) is the new state of the cell to be updated. It means that for every possible combination of states \(N_t\ W_t\ C_t\ E_t\ S_t\) a new state \(C_{t+1}\) needs to be specified. However, if the number of cell states increases, the number of possible transition rules grows significantly which is inconvenient for efficient CA design. Of course, not all transition rules need to be specified explicitly but the problem is how to choose the rules which modify the central cell in the neighbourhood. Therefore, an advanced representation of the transition rules was proposed and denominated as Conditionally Matching Rules [3]. Conditionally matching rules allows us to reduce the size of representation of the transition functions especially with respect to the evolutionary design of cellular automata.
3 Conditionally Matching Rules
The concept of conditionally matching rules (CMR) showed as a very promising technique in comparison with the conventional (table-based) approach considering various experiments with binary cellular automata (e.g. pattern development task [3] or binary multiplication in 2D CA [2]). In this work, evolutionary design of the CMR-based representation will be investigated in order to design cellular automata with up to 10 cell states that support replication of a given structure.
A conditionally matching rule represents a generalised rule of a transition function for determining a new cell state. Whilst the common approach specifies a new state for every given combination of states in the cellular neighbourhood, the CMR-based approach allows to encode a wider range of combinations into a single rule. A CMR is composed of two parts: a condition part and a new state. The number of items (size) of the condition part corresponds to the number of cells in the cellular neighbourhood. Let us define a condition item as an ordered pair consisting of a condition function and a state value. The condition function is typically expressed as a function whose result can be interpreted as either true or false. The condition function evaluates the state value in the condition item with respect to the state of the appropriate cell in the cellular neighbourhood. In particular, each item of the condition part is associated with a cell in the neighbourhood with respect to which the condition is evaluated. If the result of such evaluation is true, then the condition item is said to match with the state of the appropriate cell in the neighbourhood. In order to determine a new cell state according to a given CMR, all its condition items must match (in such case the CMR is said to match).
The following condition functions will be considered: \(==0, \ne 0, \le , \ge \). Note that this condition set represents a result of our long-term experimentation and experience with the CMR approach and will be used for all the experiments in this study. The condition \(==0\), respective \(\ne 0\), evaluates whether the corresponding cell state is equal to 0 (i.e. a “dead” state), respective whether it is different from state 0. Note that the state value of the condition item for \(==0\) and \(\ne 0\) is considered implicitly within the condition itself. The conditions \(\le \) and \(\ge \) represent relational operators “less or equal” and “greater or equal” respectively for which the state value of the condition item must be explicitly specified.
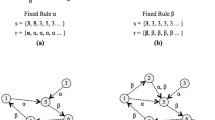
Figure 1 shows an example of conditionally matching rules defined for a 2D CA with the 5-cell neighbourhood together with the illustration of cells the condition items are related to. CMR (A) is a matching CMR since all the conditions of its condition part are evaluated as true with respect to the sample neighbourhood shown in the left part of Fig. 1. On the other hand, CMR (B) does not match because the second condition item \(!=2\) evaluates as false with respect to the west cell that possesses state 2. Similarly, the third condition \(==0\) of CMR (B) is not true as the central cell is in state 2.
Structure of a chromosome for genetic algorithm encoding a CMR-based transition function. cx denote a condition for the cell at position x in the neighbourhood, sx represents the state value to be investigated using the appropriate condition with respect to the state of cell at position x, ns specifies the next state for a given CMR. All the conditions and state values are represented by integer numbers.
A CMR-based transition function can be specified as a finite (ordered) sequence of conditionally matching rules. The following algorithm will be applied to determine a new state of a cell. The CMRs are evaluated sequentially one by one. The first matching CMR in the sequence is used to determine the new state. If no of the CMRs matches, then the cell keeps its current state. These conventions for evaluating and applying the CMRs ensure that the process of calculating the new state is deterministic (it is assumed that the condition functions are deterministic too). Therefore, it is possible to convert the CMR-based transition function to a corresponding table-based representation which preserves the fundamental concept of cellular automata. Moreover, every condition set that includes relation \(==\) allows to formulate transition rules for specific combinations of states if needed (by specifying \(==\) for all condition items of the CMR).
In order to obtain the conventional (table-based) representation of the transition rules from an evolved CMR-based solution, the following algorithm is applied using the same CA that was considered during evolution. Let \(C_t\) and \(C_{t+1}\) denote states of a cell in two successive steps of the CA at time t and \(t+1\) respectively. A transition rule of the form \(N_t\ W_t\ C_t\ E_t\ S_t\rightarrow C_{t+1}\) is generated for the combination of states in the cellular neighbourhood if \(C_t\ne C_{t+1}\). This process is performed after each step and for each cell until the CA reaches a stable or periodic state. The set of rules obtained from this process represents the corresponding conventional prescription of the transition function. Note that only the rules that modify the cell state are generated, all the other rules are implicitly considered to preserve the current state.
4 Evolutionary System Setup
A genetic algorithm is utilized for the evolution of CMR-based transition functions in order to achieve the given behaviour in cellular automata. Each chromosome of the GA represents a candidate transition function encoded as a finite sequence of CMRs. The chromosome is implemented as a vector of integers in which the condition items and next states of the CMRs are encoded. Note that the population consists of chromosomes of a uniform length (given by the number of CMRs) which is specified as a parameter for a specific experiment. The structure of a chromosome is depicted in Fig. 2.
The population of the GA consists of 8 chromosomes that are initialised randomly at the beginning of the evolutionary process. In each generation, four individuals are selected randomly from the current population, the best one of which is considered as a parent. In order to generate an offspring, the parent undergoes a process of mutation as follows. A random integer M in range from 0 to 2 is generated. Then M random positions in the parent chromosome are selected. The offspring is created by replacing the original integers at these positions by new valid randomly generated values. If M equals 0, then no mutation is performed and the offspring is identical to the parent. The process of selection and mutation is repeated until the entire new population is created. Crossover is not applied because no benefit of this operator was observed during the initial experiments. Note that the same GA has successfully been applied since the introduction of CMRs in various case studies [2, 3]. Although no optimal (evolutionary) approach has yet been known for uniform CA, our experiments indicate that small-population EA (i.e. less than 10 individuals) with a simple mutation operator may represent a suitable class of algorithms to obtain working solutions with a reasonable success rate and computational effort. However, the detailed analysis and wider comparison of different techniques is not a subject of this study.
For each experiment, the GA is executed for 3 million generations. If no correct solution is found within this limit, the evolution is terminated. The evaluation of the chromosomes (i.e. the fitness function) and details regarding various experimental settings are described in the next section.
5 Experimental Results
This section summarises statistics of the evolutionary experiments performed and presents some results together with a more detailed analysis. Two sets of experiments are considered, the goal of each is to design CA that is able to replicate the given loop. The first set works with a big loop (the denomination is chosen for the purposes of this work with respect to the loop in the second set of experiments), the objective is to design transition rules that are able to develop a single replica of the loop in a given arrangement against the initial loop. In the second set, a simpler, small loop is treated, the goal is to find replication rules for the development of two independent replicas in parallel on the left and right side of the initial loop. Note that the loops consist of cells in 7 different states (including state 0). In both sets of experiments, the CA working with 8 and 10 cell states are investigated. Moreover, different numbers of CMRs (varying from 20 to 50) encoded in the GA chromosomes are considered. For each setup, 100 independent evolutionary runs are executed. The experiments were executed using the Anselm clusterFootnote 1, the time of a single run (3 million generations) is approximately 12 h.
5.1 Replication Evolution of the Big Loop
A big loop is considered for the replication in the first set of experiments, the structure of which is shown in Fig. 3a. The genetic algorithm is applied to design the transition rules for the CA, which perform the replication of the loop in a maximum of 30 steps. The required CA state, that contains the replica, is depicted in Fig. 3b. The following algorithm is applied to the evaluation of the candidate solutions during evolution and the calculation of the fitness function. A partial fitness function is evaluated after each CA step as the number of cells in correct states with respect to Fig. 3b. The final fitness value of a given candidate solution is defined as the maximum of the partial fitness values. It this case the replication can be considered as a pattern transformation problem from a single (initial) loop onto two loops in a given arrangement. However, the loop is expected to replicate again and again during the subsequent CA development, which will be validated for the results obtained from the evolution. Moreover, an assumption is considered that each newly created loop is shifted by two cells down with respect to its predecessor (as shown in Fig. 3b). Therefore, the solutions obtained are further investigated using a visual software simulator developed by the author of this work in order to check that. The goal of this approach is to determine whether the GA is able to discover various new general replication scenarios. Note that, for the purposes of this study, the term “general” means the ability of a solution to repeatedly produce more replicas of the given loop, not an ability to replicate arbitrary loops.
Table 1 summarises the results of experiments with the big loop and provides an overview of some basic parameters of the CA that can be observed during its development using the evolved transition functions. As evident, the maximum success rate achieved during the experiments is only 12 % which is not very high. Note, however, that the replication of the proposed loop represents a problem for which no working solution was found during our previous experiments using the table-based transition functions.
Develpment of a CA performing replication of the loop from Fig. 3a. The sequence of steps reads from left to right and top to bottom. The upper part of each step of the CA illustrates the replication of the initial loop. The bottom part demonstrates a seed represented by a cell in state 5. Note that after the loop is finished, its replication continues in the same way as from the initial instance (shown by the last CA state).
In addition to the results obtained for the CA working with 8 cell states, some successful solutions have even been obtained for 10 cell states which indicates that the CMRs are an efficient encoding of the transition rules that allows for the design of more complex multi-state CA. The solutions obtained in this work demonstrate a wide range of various replication schemes that can be performed using CA. For example, a solution was found that is able to replicate the loop in 16 steps (the best result achieved for this loop) whilst some CA require 30 steps (the maximal allowed number of steps) in order to finish the replication. Similarly, the number of transition rules generated from the CMRs varies from 84 to more than 1500 rules. These results indicate that cellular automata can in some cases exhibit behaviour that has not yet been discovered which may be beneficial not only for the area of CA but also, for the study of complex systems in general.
Figure 4 shows a CA development performed by one of the successful transition functions obtained for the replication of the given loop. It is one of the best solutions discovered in this work with respect to the number of steps needed to create a copy of the loop. The transition function was found with 30 CMRs in the GA chromosomes and the corresponding conventional representation contains 238 transition rules. If the development of the initial loop is considered (see the upper parts of each step in Fig. 4), the CA needs 21 steps to create a complete replica. As shown by the last step, more replicas can be created in the same way according to the original specification if the CA development continues. However, a more detailed investigation of this result showed that the complete initial loop is not strictly needed in order to successfully perform the replication. For example, the loop is able to emerge even from a single seed – the lower parts of each step presented in Fig. 4 shows a development of the loop from a single initial cell (a seed) in state 5. As marked by the up-most black arrow a complete loop is developed from the seed after 18 steps which is by 3 steps faster compared to the development from the initial loop. This behaviour is caused by a need of the initial loop to generate a cell in state 5 (i.e. the same state as the seed) from which the replica can be developed (it takes 3 steps – see the top-right CA state in Fig. 4). The process of finishing the replica is identical with the development from the seed. Note that the ability of the transition function to develop and replicate the loop from a seed was not explicitly required in the fitness evaluation. Hence it can be considered as an additional, unconventional feature of this solution.
Another result is presented in the form of an evolved transition function (Fig. 5) and the appropriate CA development (Figs. 6 and 7). This cellular automaton demonstrates a development process from a seed that at first creates rather a chaotic structure even larger than the required loop itself. A “mature” loop is developed from this structure during the subsequent CA development that is able to replicate itself. Whilst the replication of the initial loop takes 25 steps (marked by the black arrow in Fig. 6), the development of the chaotic structure needs 36 steps. Starting by step 37 (Fig. 7) the loop is developed from that structure in the same way as from the initial loop. It was verified that the loops are able to replicate repeatedly if the CA development continues.
Part 1 of the replication according to the transition function from Fig. 5. The sequence of steps reads from left to right and top to bottom. The development shows a replication of the initial loop (the upper part of each step) and a growth of a non-specific structure from a seed allowing to create the loop autonomously (the lower part of each step). The seed is represented by a cell in state 7.
Part 2 of the replication according to the transition function from Fig. 5. The sequence of steps reads from left to right and top to bottom. The development shows an autonomous growth of the loop from a non-specific structure that emerged in the last step of Fig. 6 (the bottom part of each step). It was verified that the loop is able to replicate in the same way as the initial loop during the subsequent CA development.
For both the presented solutions the transition function was identified as redundant (i.e. not all the conventional transition rules generated from the CMR representation are needed for the replication of the initial loop required by the fitness function). A more detailed analysis showed that this redundancy is caused by the finite CA size with cyclic boundary conditions and by generating the transition rules from the CMRs until the CA reaches a stable or periodic state. Although this approach leads to more complex table-based transition functions, in this case it showed as very beneficial for achieving some additional features that were not required during evolution (especially the ability to develop the loops from a seed). Advanced experiments with the resulting CA showed that if the transition functions are optimized (i.e. only the rules for the development of a single replica from the initial loop are considered), the CA in most cases loose the ability of the development from the seed. It was also determined that the seed-based development does not work in case of the known replicating loops (e.g. Langton’s or Byl’s loop). In the future, this ability may be beneficial for the advanced study of complex systems in which a given (complex) configuration needs to be achieved—distributed—from a single cell or a simple initial configuration. In addition to the results presented herein, various other solutions were found that are able to replicate a given structure. It indicates that the replication in CA is not limited to known schemes only but can be performed in many different ways.
5.2 Parallel Replication of the Small Loop
The second set of experiments presents the evolution of parallel replication techniques of a small loop with its structure shown in Fig. 8a. As with the evolution of the big loop, the CA behaviour is evaluated for 30 steps using the partial fitness calculated after each step with respect to the target arrangement of the replicas shown in Fig. 8b, and the final fitness value is given by the maximum of the partial fitness values. In this case, however, two replicas are required with the arrangement on the left and right side of the original loop. The hypothesis evaluated herein is that if suitable transition functions exist for the development of the replicas, then at least a subset of the results will produce the replicas repeatedly in the given directions during the subsequent CA development (i.e. for the purposes of this study, such the solutions will be considered as general). Since the loop is not fully symmetric with respect to the cell states on the sides of the loop, it is expected that different replication algorithms (i.e. sequences of the CA steps) need to be designed to produce the replicas.
Table 2 summarises the results of experiments with the small loop and provides an overview of some basic parameters of the CA that can be observed during its development using the evolved transition functions. Although the shape of the small loop is simpler than the big loop, the requirement of two independent replicas increases the overall complexity of this task, the maximum success rate achieved does not exceed 12 %. Despite this fact, the evolution provided some solutions that perfectly fulfil the target specification and, in addition, also exhibit the capability of the seed-based development which was not explicitly required.
A sequence of CA steps demonstrating the parallel replication of the small loop according to the evolved transition function from Fig. 10. The states are ordered from left to right and top to bottom. The bottom part of each state shows the replication from the initial loop, the top part of each state demostrates the development and replication of the loop from a seed.
A transition function designed by evolution for the parallel replication of the small loop from Fig. 8a.
A sample of the CA development from the seed according to the transition function from Fig. 10: (a) the initial seed, (b) the small loop is developed from the seed after step 24, leaving an undesirable structure on its left side, (c) the loop creates its first replica after step 47, the undesirable structure prevents from the replication on the left side, (d) the replication to the right in progress after step 51, the structure on the left no longer changes.
Figure 9 shows a CA that performs a successful parallel replication of the small loop. The CA works with 8 cell states and, in addition to the replication of the initial loop, is also able to perform the development and replication of the loop from a seed. This is one of the most efficient and compact solution obtained in this study regarding the number of CA steps and the number of transition rules. The corresponding table-based transition function consists of 154 rules as shown in Fig. 10. The CA needs to perform 23 steps in order to finish the replicas of the initial loop. However, if a cell is initialised as a seed by one of the states 1, 3, 5, 6, or 7, the small loop autonomously grows into its full shape and subsequently is able to replicate according to the original specification. The analysis of the seed-based development showed that the loop needs 19 steps to fully develop from state 1, 18 steps from states 3, 6 and 7, and 24 steps from state 5. An interesting behaviour of the CA can be observed after finishing the seed development when the loop ought to be replicated. In particular, the loop replicates according to the given specification from states 1, 3, 6, and 7. However, the state-5 seed creates an undesirable structure that prevents the loop replication to the left side, i.e. the loop developed from state 5 can replicate to the right side only (see Fig. 11). This indicates that a wide range of states used as the seed allows emerging the loop using various processes (i.e. sequences of CA states), which are totally different from the processes of replication from the complete loop. Although the state-5 seed does not enable to replicate the loop to both sides, the solution can be considered as robust because the undesirable structure does not cause the destruction of the loop that can subsequently replicate to the right side.
As an example of our research regarding the optimisation of replication techniques in cellular automata, a symmetric loop is considered as shown in Fig. 8c. Although the evaluation method applied to design the CA for this loop is out of the scope of this study, a result of a successful parallel replication will be presented, which demonstrates the potential of the GA in combination with the CMR encoding to discover novel techniques in cellular automata. As in the previous example, the goal of the experiment was to design transition rules for the parallel replication of the loop to the given directions. Since the loop is symmetric with respect to the arrangement of the cell states, it would be possible to adapt a single replication algorithm to perform the replication process simultaneously to various directions. Such adaptation is based on “rotating” the transition rules according to the ordering of cells in the cellular neighbourhoods with respect to the given directions as known from Byl’s loop [4]. However, if the evaluation of the candidate solutions during evolution is performed with respect to the number and arrangement of the replicas only, then the GA can discover various independent replication algorithms as shown in Fig. 12. The corresponding transition function contains 137 table-based rules and is shown in Fig. 13. In this solution, not only the algorithms for the replication to the left and right side differ significantly, the number of steps needed to create the replica on the left side is nearly the double of the number of steps required for the replication to the right side. As evident from Fig. 12, the first replica of the initial loop is created on the right side after the 15th step, the first replica on the left side needs 26 steps to be completed. After the 27th step, the second replica on the right side is completed whilst the second copy on the left side has just started to develop. Such a process has never been observed before as regards the known replicating loops and hence it can be considered as an unconventional feature of the solution obtained in this experiment.
The transition function designed by evolution for the parallel replication of the symmetric loop from Fig. 8c.
5.3 Summary and Discussion
Both the proposed loops proved the ability to replicate according to the given specification. It is worth to note that although the development of the loop from a seed was not explicitly required, the evaluation of the results obtained for both the loops showed that this ability is not rare. This means that the seed-based development may be evolved directly (without any initial loop available) in order the given loop can emerge autonomously. Some experiments were performed in order to validate this hypothesis, with the following observations. The GA is able to discover transition rules for the development of the given loop from the seed. However, no solution has yet been achieved that would be able to subsequently replicate the loop. One of the reasons for this issue may be the fact that the exact place in the cellular space, where the loop is developed from the seed, is hard to predict (it depends on the state of the seed, shape of the loop and the transition rules). Therefore, it is not evident how the replicas ought to be specified within the target CA state for the continuous replication. More research is needed in order to determine the necessary information provided to the GA, which would enable to solve this problem.
In order to perform a general evaluation of the results obtained within the context of computational features of cellular automata and with respect to the existing replicating loops, the following issues need to be clarified:
-
1.
The objective was not to design self-replication. The loops with the ability to self-replicate contain the information of how to create a copy encoded in their “body” as a suitable arrangement of cell states. The transition rules interpret this information and calculate the appropriate state transitions of the CA in order to perform the replication process. In this work, however, the initial loop is considered as an object of a given shape that ought to be transformed onto a CA state that contains the copy of the loop. The goal was to find both the transition rules and the sequence of the CA states that lead to the emergence of the replica.
-
2.
The resulting CA do not represent universal computing models (it was not a goal of the experiments). It means that a specific transition function, that was obtained as a result of a successful evolution, is dedicated to replicate the given loop only that was a subject of evaluation in the fitness function. Nevertheless, as the results showed, some transition functions are able to create the loops from a seed which was not explicitly required within the fitness evaluation.
Although the shape of the proposed loops was inspired by the existing (self-replicating) loops and the GA provided some successful results to replicate the loops with respect to the given specifications, no working solution has yet been achieved by the GA to replicate the existing loops (e.g. Byl’s loop) with the exact shape and arrangement of the replicas. This issue can be caused by the fact that some of the self-replicating loops are dynamical structures even after the replica is finished (e.g. Byl’s loop exhibits such feature). However, only static replicas were considered in our experiments. Another aspect may be the size of the loop. Large loops require a considerable number of steps to finish the replica (e.g. Langton’s loop needs 151 steps), which makes the evaluation of such solutions very time-consuming. Finally, the information encoded in the loop body, that specifies the self-replication features, actually determines the replication algorithm (i.e. the CA development) which is specific for the given loop. If no more valid replication algorithms exist in the solution space for a given loop, then the GA may not be able to find the solution in a reasonable time.
6 Conclusions
In summary, the results presented in this work shows several facts related to the problem of replication in cellular automata. First, there are plenty of transition functions that are able to replicate a given loop. The experiments showed that it is possible to discover such functions routinely by means of the genetic algorithm even for complex multi-state cellular automata (herein demonstrated for CA working with 8 and 10 cell states). This was enabled by the utilisation of conditionally matching rules as a technique for the representation of the transition functions. Second, some unconventional features of the solutions were identified that cannot be observed in the known replicating loops and have never been published before. Specifically, in case of some solutions obtained, the CA can be initialised by a single-cell seed in a non-zero state, which allows developing the given loop that is subsequently able to replicate. Note that this ability was identified as an extra feature of the resulting cellular automata, which was not explicitly required by the specification for the evolutionary algorithm. This shows that some cellular automata are able, using a minimum information encoded in the initial state, to autonomously develop a complex emergent behaviour that is fully determined by the transition function and the state of a single cell only. Another feature, that was achieved by the evolution, is a parallel replication of the given loop into more directions, using different algorithms to create the replicas. The results showed that this behaviour is needed if the arrangement of the cell states in the loop is not fully symmetrical. However, an unconventional parallel replication can be observed even in case of a symmetric loop, where the difference is both in the way of the replication and the number of steps needed to create the replicas. Again, the evolution itself discovered such the behaviour just on the basis of the given target pattern containing the replicas of the initial loop.
The results obtained bring some open questions, the answers of which could be beneficial for the research of cellular automata in general. For example, can the seed-based development create a configuration in the CA that supports self-replication (or other useful features)? Are there other (simple) structures that support development of more complex (self-)replicating objects? Can evolutionary techniques be applied to the design of computationally universal CA-based models? Not only these questions represent ideas for our future work.
References
Berlekamp, E.R., Conway, J.H., Guy, R.K.: Winning Ways for Your Mathematical Plays, vol. 4, 2nd edn. A K Peters/CRC Press, Boca Raton (2004)
Bidlo, M.: Evolving multiplication as emergent behavior in cellular automata using conditionally matching rules. In: 2014 IEEE Congress on Evolutionary Computation, pp. 2001–2008. IEEE Computational Intelligence Society (2014)
Bidlo, M., Vasicek, Z.: Evolution of cellular automata with conditionally matching rules. In: 2013 IEEE Congress on Evolutionary Computation (CEC 2013), pp. 1178–1185. IEEE Computer Society (2013)
Byl, J.: Self-reproduction in small cellular automata. Phys. D Nonlinear Phenom. 34(1–2), 295–299 (1989)
Elmenreich, W., Fehérvári, I.: Evolving self-organizing cellular automata based on neural network genotypes. In: Bettstetter, C., Gershenson, C. (eds.) IWSOS 2011. LNCS, vol. 6557, pp. 16–25. Springer, Heidelberg (2011). doi:10.1007/978-3-642-19167-1_2
Fogel, L.J., Owens, A.J., Walsh, M.J.: Artificial Intelligence Through Simulated Evolution. Wiley, New York (1966)
Holland, J.H.: Adaptation in Natural and Artificial Systems. University of Michigan Press, Ann Arbor (1975)
Langton, C.G.: Self-reproduction in cellular automata. Phys. D Nonlinear Phenom. 10(1–2), 135–144 (1984)
Mitchell, M., Hraber, P.T., Crutchfield, J.P.: Revisiting the edge of chaos: evolving cellular automata to perform computations. Complex Syst. 7(2), 89–130 (1993)
Packard, N.H.: Adaptation toward the edge of chaos. In: Kelso, J.A.S., Mandell, A.J., Shlesinger, M.F. (eds.) Dynamic Patterns in Complex Systems, pp. 293–301. World Scientific, Singapore (1988)
Perrier, J.-Y., Sipper, M., Zahnd, J.: Toward a viable, self-reproducing universal computer. Phys. D 97, 335–352 (1996)
Reggia, J.A., Armentrout, S.L., Chou, H.-H., Peng, Y.: Simple systems that exhibit self-directed replication. Science 259(5099), 1282–1287 (1993)
Sapin, E., Adamatzky, A., Collet, P., Bull, L.: Stochastic automated search methods in cellular automata: the discovery of tens of thousands of glider guns. Natural Comput. 9(3), 513–543 (2010)
Sapin, E., Bull, L.: Searching for glider guns in cellular automata: exploring evolutionary and other techniques. In: Monmarché, N., Talbi, E.-G., Collet, P., Schoenauer, M., Lutton, E. (eds.) EA 2007. LNCS, vol. 4926, pp. 255–265. Springer, Heidelberg (2008). doi:10.1007/978-3-540-79305-2_22
Sipper, M.: Quasi-uniform computation-universal cellular automata. In: Morán, F., Moreno, A., Merelo, J.J., Chacón, P. (eds.) ECAL 1995. LNCS, vol. 929, pp. 544–554. Springer, Heidelberg (1995). doi:10.1007/3-540-59496-5_324
Sipper, M. (ed.): Evolution of Parallel Cellular Machines. LNCS, vol. 1194. Springer, Heidelberg (1997)
Sipper, M., Goeke, M., Mange, D., Stauffer, A., Sanchez, E., Tomassini, M.: Online evolware. In: IEEE International Conference on Evolutionary Computation, pp. 181–186 (1997)
Stefano, G., Navarra, A.: Scintillae: how to approach computing systems by means of cellular automata. In: Sirakoulis, G.C., Bandini, S. (eds.) ACRI 2012. LNCS, vol. 7495, pp. 534–543. Springer, Heidelberg (2012). doi:10.1007/978-3-642-33350-7_55
Tempesti, G.: A new self-reproducing cellular automaton capable of construction and computation. In: Morán, F., Moreno, A., Merelo, J.J., Chacón, P. (eds.) ECAL 1995. LNCS, vol. 929, pp. 555–563. Springer, Heidelberg (1995). doi:10.1007/3-540-59496-5_325
von Neumann, J.: Theory of self-reproducing automata. In: Burks, A.W. (ed.) Essays on Cellular Automata. University of Illinois Press, Urbana and London (1966)
Yunès, J.-B.: Achieving universal computations on one-dimensional cellular automata. In: Bandini, S., Manzoni, S., Umeo, H., Vizzari, G. (eds.) ACRI 2010. LNCS, vol. 6350, pp. 660–669. Springer, Heidelberg (2010). doi:10.1007/978-3-642-15979-4_74
Acknowledgements
This work was supported by The Ministry of Education, Youth and Sports of the Czech Republic from the National Programme of Sustainability (NPU II); project IT4Innovations excellence in science - LQ1602.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Bidlo, M. (2017). Evolution of Cellular Automata-Based Replicating Structures Exhibiting Unconventional Features. In: Merelo, J.J., et al. Computational Intelligence. IJCCI 2015. Studies in Computational Intelligence, vol 669. Springer, Cham. https://doi.org/10.1007/978-3-319-48506-5_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-48506-5_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-48504-1
Online ISBN: 978-3-319-48506-5
eBook Packages: EngineeringEngineering (R0)