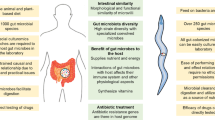
Abstract
The number of bacteria in the human intestine roughly equals the number of cells in the entire human body. This community of bacteria and a much smaller number of unicellular eukaryotes and prokaryotic archaea is referred to as the microbiota. It is becoming increasingly clear that the composition of the microbiota is important for human health and has an impact on obesity, diabetes, various bowel diseases and likely ageing. The microbiota is composed of pathogenic, commensal and beneficial bacteria, the latter often referred to as probiotic. Several studies have reported that the composition of the microbiota changes during ageing. Although recent developments in DNA sequencing technologies have allowed researchers to more accurately determine the composition of the microbiota, little is known about the mechanisms by which the microbiota mechanistically influences the host, not least during ageing. This limits the use of probiotic bacteria to prevent and treat diseases. Researchers are using C. elegans to study both pathogenic and probiotic bacteria, which have opposing effects on lifespan. C. elegans is also successfully being used as screening platform to identify novel strains of probiotic bacteria. Since the natural diet of C. elegans is bacteria and the longevity pathways are well characterized, the nematode is particularly well-suited for this purpose. In this chapter we will review how the microbiota and particularly probiotic bacteria influences ageing in C. elegans.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
18.1 Microbiota, Prebiotics and Probiotic Bacteria
The human gastrointestinal tract was recently estimated to contain ~4 × 1013 bacteria nearly equaling the estimated ~3 × 1013 human cells in a 70 kg “reference” human [1]. Although this 1:1 ratio between bacterial and human cells is lower than the 10:1 estimate previously proposed, and widely referenced, humans do contain a staggering number of microorganisms. In addition to bacteria, the human gastrointestinal tract also hosts unicellular eukaryotes and prokaryotic archaea; collectively these microorganisms are called the microbiota. The combined gene pool of these microorganisms constitutes the microbiome. The largest population of bacteria is found in the gastrointestinal tract, where they vastly outnumber other microorganisms. Therefore, the term microbiota is often used to describe the bacterial community in the intestine .
Dictionary
- Probiotics:
-
Live microorganisms that, when administrated in adequate amounts, confer a health benefit on the host.
- Prebiotics:
-
Supplements that favour growth or activity of probiotic bacteria
- Commensal:
-
Bacteria that are part of the normal microbiota, and which benefit from the symbiosis with the host, but without being beneficial or harmful to the host.
- Microbiota:
-
The communities of bacteria, unicellular eukaryotes and prokaryotic archaea hosted in the human body. These can be commensal, symbiotic/beneficial or pathogenic.
- Microbiome:
-
The collected genomes of the microorganisms in the microbiota. Microbiome and microbiota are sometimes used interchangeable in the literature.
- LAB:
-
Lactic acid bacteria
It is becoming increasingly clear that the composition of our microbiota is an important determinant for our health [2]. For example, the gut microbiota may affect host metabolism and insulin resistance via digestion and nutrient uptake and thus be a causal factor in obesity and diabetes [3–6]. The microbiota has also been implicated in osteosarcopenic obesity [7]. Furthermore, the microbiota helps fight pathogens, reduce inflammation and scavenge toxins and by-products of digestion, and it has a role in numerous bowel diseases [3]. Finally, some studies suggest that the microbiota may even influence neuronal function and development [8].
The term probiotic is defined as “live microorganisms that, when administrated in adequate amounts, confer a health benefit on the host” [9]. Bacteria that have a beneficial effect on their host can thus be called probiotic bacteria. Strains in the Lactobacillus and Bifidobacterium genera are often considered probiotic and a number of studies have shown that certain strains of these species can prevent and treat a range of conditions including intestinal diseases, obesity, metabolic disorders and various infections [10, 11]. Most of these studies are descriptive and mainly identify associations between specific microbes and health or disease rather than causal relationships. Nevertheless, probiotics and prebiotics (supplements favouring growth or activity of probiotic bacteria) are growing industries with many areas of application including drugs, foods, dietary supplements, and animal feed.
18.2 Microbiota and Ageing
The idea that the microbiota could influence ageing was put forward by Ilya Ilyich Metchnikoff more than a century ago [12]. Metchnikoff suggested that health could be improved by altering the microbiota with help of probiotic bacteria found in yogurt . Today many yogurt-based probiotic products are commercially available claiming various beneficial effects, although little is known about their mechanisms of action. However, regarding microbiota influencing human ageing, it seems that Metchnikoff might have been on the right track, since variations in gut microbiota composition between young and elderly have been reported in several studies [13–19]. Most of these studies are of correlative nature and causal mechanisms are largely unknown. The strong track record for uncovering longevity pathways and underlying molecular mechanisms has made C. elegans a popular model system for studying ageing and life history traits. Since bacteria are the natural diet of C. elegans, the nematode is particularly well-suited for understanding the effects of probiotic bacteria on ageing. Although it is a relatively young field of research, several studies have found that feeding C. elegans with probiotic bacteria increases lifespan and resistance towards bacterial infections (Table 18.1). Before we discuss these studies in more detail we need to look at some of the differences and similarities between C. elegans and humans with respect to shaping and hosting a microbiota .
18.3 Diet and Microbiota of C. elegans
In humans the vast majority of bacteria are found in the intestine and likewise in C. elegans the intestine is where most bacteria are found. The intestine is the largest somatic organ in C. elegans (see Chap. 2), and it carries out a variety of functions including nutrient uptake and storage, lipid accumulation, elimination of waste products, and protection against harmful substances and pathogens [20]. Unlike humans, C. elegans is a bacterivore and therefore bacteria are necessary food sources, part of the microbiota and potential pathogens.
18.3.1 Worm Bacterial Diet
When maintained in the laboratory C. elegans nearly always feed on a single bacterial strain, typically the gram-negative bacterium Escherichia coli (E. coli) OP50 . Other E. coli strains are also commonly used for maintenance, e.g. HB101 and HT115 , used for an extra nutritious diet and RNAi, respectively. These different food sources have different effects on lipid deposition, development, metabolism, and lifespan [21–24].
In the wild C. elegans feed on various types of bacteria and thus, they have a diverse bacterial flora in their gut lumen [25–27]. Like all multicellular organisms, nematodes must also choose what to eat when faced with a wide range of bacteria in the wild. C. elegans is able to navigate through these and avoid pathogenic bacteria [28–30] in the search for high quality food, namely bacteria supporting growth, which is partly driven by previous food experience [31]. It has been reported that C. elegans prefers to consume soil bacteria, such as Bacillus mycoides and Bacillus soli [32]. Others have suggested that the feeding preferences of C. elegans are affected by bacterial respiration and growth rates [33] as well as odour attraction [34]. Sensing of food is discussed in more detail in Chap. 17.
E. coli OP50 was originally chosen as food source because it is a uracil auxotroph, growing to a nicely defined lawn on NGM plates making it easier to perform experiments in the laboratory [23]. OP50 is often considered non-pathogenic but studies have suggested that it is in fact mildly pathogenic as the lifespan is increased when C. elegans is fed UV-killed or antibiotic treated OP50 bacteria [35, 36]. The metabolic state of the bacteria is also important for the development and lifespan of C. elegans. Growth in axenic medium is associated with slow and asynchronous development together with reduced fertility, and the worms are believed to enter a state of dietary restriction [37]. Interestingly, addition of live bacteria reverts the development back to normal when worms are cultured axenically. Addition of dead bacteria does not have an effect [38]. Furthermore, respiratory deficient bacteria lacking either Coenzyme Q or ATP synthase prolongs the lifespan [39, 40].
18.3.2 Digestion and Bacterial Colonization
The bacteria consumed by C. elegans are first exposed to the pharyngeal grinder [41] (See Chap. 2). In young animals, the grinder effectively crushes the food, leaving no bacteria to pass through alive. As the worm ages the effectiveness of the pharyngeal grinder is declining and in young adults bacteria starts colonizing the intestine, thereby creating a microbiota [42]. The proliferating bacteria in the intestine will eventually become harmful for its host and old worms can get severe constipation due to bacteria blocking the lumen of the intestine. Hindering bacterial proliferation increases lifespan associated with reduced bacterial packing [35, 36]. It has been suggested that intestinal colonization might be a general mechanism that controls longevity, as it has been demonstrated that long-lived mutants generally have fewer intestinal bacteria than wild-type worms [42]. However, other data suggest that it is unlikely that old animals die solely due to bacterial accumulation in the intestine since this is not observed in all recently dead animals [43].
C. elegans is a popular model for studying innate immunity and host responses to pathogenic bacteria as well as virulence factors [44]. Many different infection models and assays have been established including Staphylococcus aureus [45, 46], Enterococcus faecaelis [47], Pseudomonas aeruginosa [46], and Yersinia pestis [48]. Some pathogenic bacteria cause detrimental infections in the intestine of C. elegans and interestingly several studies have found that treatment with probiotic bacteria can prevent or delay these infections (See Table 18.1).
18.3.3 Food Quality and Dietary Restriction
Dietary restriction has long been known to strongly increase lifespan of many organisms including C. elegans. For a detailed review of the effect of dietary restriction on lifespan see Chap. 16. Different bacterial diets have been found to affect lifespan as well, possibly through dietary restriction or due to different macronutrient composition . Macronutrient analysis of some of the most common feeding strains for C. elegans, OP50 , HT115 , HB101 and DA837, revealed a significant difference in their amount of carbohydrates and fatty acids. Nevertheless, there did not seem to be a significant difference in lifespan of worms grown on these different bacterial diets [22]. Other studies, however, have observed a significant increase in lifespan of worms grown on the E. coli strain HT115 compared to worms grown on E. coli OP50 [21, 24, 49]. Intriguingly, one study has found that feeding with HT115 shortens lifespan compared to an OP50 diet [50]. This could perhaps indicate that the bacterial strains differ between laboratories due to a high forward mutation rate .
18.3.4 The Worm Microbiota
Humans have a very diverse microbiota, and one of the concerns arising from using C. elegans as a model organism is their maintenance in the laboratory on bacterial monocultures, which results in the absence of a complex microbiota in their intestine . However, the use of monocultures can also be seen as an advantage because it is possible to directly link specific bacterial strains to specific host responses (Table 18.1). A few studies have investigated the effect of feeding C. elegans multiple bacterial strains simultaneously [26, 27, 34, 51]. These studies follow the overall strategy that bacterial species residing in the worm intestine can be isolated and analysed. When analysing mixtures of multiple bacterial strains there is currently no way of eliminating a bias towards enrichment of bacteria that grow easily in the laboratory. There is also a risk of completely missing for example anaerobic bacteria that cannot grow in the presence of oxygen.
Studies of C. elegans living on rotten fruit, mimicking their natural environment, have isolated several bacteria species from their intestine indicating that they are capable of hosting a microbiota [26, 27]. If this actually mimics the natural life of C. elegans, this also suggests that the worm would have evolved all the response mechanisms to host a microbiota, containing both beneficial and pathogenic bacteria. This is further supported by the presence of the innate immune system in C. elegans.
In an elegant study it was shown that “you are not what you eat”, at least if you are a C. elegans nematode [27]. Germ free L1 larvae were allowed to develop to adulthood on three types of soil with different bacterial compositions. When the microbiotas of these worms were analysed based on deep sequencing of 16S rDNA it revealed that they resembled each other despite arising from different microbial environments. Thus, it seems that the host plays an active role in shaping its microbiota. From this follows that one should be able to identify mutants with altered microbiotas. Unfortunately, such mutants were not presented in the study. However, with mutants readily available in C. elegans such mutants will likely be identified in the future and help uncover how the host determines its microbiota.
Whereas studies addressing complex microbiotas in C. elegans are still rare, numerous studies have tested the effect of different monocultures including probiotic bacteria .
18.4 Probiotic Bacteria in C. elegans and Their Effect on Longevity
C. elegans has been used to both screen for new potentially probiotic bacteria and to test the effect of known probiotic bacteria on nematode lifespan and resistance to pathogenic infections (Table 18.1). Lactic acid bacteria (LAB) of either the Lactobacillus or the Bifidobacterium genus are the most widely studied species. Although evolving rapidly, the field of studying probiotic bacteria in C. elegans is relatively new. Hence, the mechanistic insights into the effects of feeding probiotic bacteria are still rather limited. However, based on the current knowledge of how probiotic bacteria affect the worm, we have divided the bacteria into five different, but overlapping groups: (I) changes in nutritional value, (II) antimicrobial effect, (III) changes in bacterial metabolism, (IV) direct activation of host signalling pathways and (V) unknown effect (Fig. 18.1 and Table 18.1).
Several strains of probiotic bacteria can be placed in more than one of these groups as they exert multiple effects on the host. For example, many bacterial strains that influence the immune functions of the host are placed both in group II and IV. Other bacterial strains have very specific effects on the host and only belong to one group . As our knowledge improve new groups representing novel mechanism of action are likely to be identified.
18.4.1 Group I: Changes in Nutritional Value
Different LAB strains have been shown to affect worm lifespan by regulating the metabolism of the host. Lactobacillus salivarius was found to increase lifespan probably through dietary restriction [52]. A LAB consortium obtained from cheese containing a mixture of three different species decreased lifespan and regulated expression of genes involved with lipid metabolism [51]. These studies demonstrate the importance of investigating whether an effect on lifespan from feeding probiotic bacteria solely arise from either calorically restricting the worms or from changing the composition of available macronutrients as is the case for OP50 versus HT115 discussed previously (see also Chap. 17). Studies related to this group are very limited, thus it is difficult to conclude on the underlying mechanisms. More work in the future is needed to address this lack of knowledge.
18.4.2 Group II: Antimicrobial Effect
A desired trait of probiotic bacteria is their ability to protect against pathogenic bacteria. This can be accomplished by the probiotic bacteria outcompeting the pathogenic bacteria either by binding to the same host molecules or by altering the pathogens ability to interact with the host. Probiotic bacteria can also inhibit the growth of the pathogen or directly kill it, or they can affect the expression of pathogen toxins. Finally, probiotic bacteria can also activate immune responses in the host, enabling the host to better combat a pathogenic infection. Numerous studies have demonstrated that feeding C. elegans with different probiotic bacteria protects against pathogen infection, through several of the above-mentioned mechanisms. Other studies have demonstrated the ability of probiotics to suppress growth and intestinal colonization of pathogenic bacteria, which increases the survival of the worm following infections [26, 39, 53–55]. Such growth inhibition is strain- specific with regard to both the probiotic and the pathogenic bacteria. For example, L. acidophilus and B. subtilis specifically protects against gram-positive pathogens, but not gram-negative [54, 56].
L. zeae and L. reuteri protect against enterotoxigenic Escherichia coli (ETEC) infection by decreasing expression of certain toxins. However, they do not affect pathogenic colonization in the intestine of the worm [57, 58]. These are examples of probiotics that can directly change virulence factors expressed by pathogenic bacteria. However, so far only one study has been able to identify the bacterial compound that inhibits pathogenic infection. B. subtilis was found to produce an antifungal lipopeptide complex fengycin , which specifically inhibited the growth and intestinal colonization of the pathogenic B. thuringiensis and S. aureus [56]. Bacteria defective in fengycin production could no longer protect against infection, and administration of purified fengycin inhibited the bacterial growth and cured infected nematodes .
Probiotic bacteria have also been demonstrated to activate immune responses in the worm, enabling them to overcome infections. Preconditioning C. elegans with L. acidophilus specifically upregulated expression of genes associated with combating gram-positive pathogen infections through upregulation of the immune pathways containing the mitogen-activated protein kinase PMK-1 orthologous to human p38, the Toll-Interleukin 1 Receptor domain adapter protein TIR-1 and the beta-catenin BAR-1 [54]. P. mendocina also regulates pathogen infection through PMK-1, as its protective effect against P. aeruginosa was abolished in pmk-1 mutants, and downstream targets of PMK-1 were upregulated in response to P. mendocina [26].
These studies of antimicrobial effects of probiotic bacteria are extremely important. There is an alarming spread of multidrug-resistant bacteria, which is claimed by WHO to be a major future threat to global human health. To prevent this dystopian scenario it is necessary to reduce the use of traditional antibiotics and develop new antibiotics. The identification of interactions between specific probiotic and pathogenic bacteria offers the possibility of developing new antibiotics as well as new treatment strategies based upon pro- and prebiotics .
18.4.3 Group III: Changes in Bacterial Metabolism
Changes in metabolism of otherwise commensal bacteria have been found to increase nematode lifespan. Worms fed an E. coli strain mutated in coenzyme Q lived significantly longer than worms fed normal E. coli [40]. The pathways responsible for the lifespan extension in the worm remains elusive but it has been suggested that it could be due to lower intestinal colonization of the Q-less E. coli strain [39]. Bacterial folate synthesis was also found to affect lifespan since an E. coli strain mutated in the aroD gen, required for folate synthesis, extended lifespan in C. elegans [43, 59]. Other studies have identified natural compounds produced by bacteria, which have a positive effect on C. elegans. A study by Gusarov et al. found that worms feeding on B. subtilis lived longer due to bacterial production of NO, compared to a NO deficient B. subtilis strain [60]. This lifespan extension was dependent on both daf-16 and hsf-1, and NO upregulated the expression of the heat shock proteins hsp-16 and hsp-70 and increased thermotolerance. In a recent study it was shown that NO produced by B. subtilis also activates the p38 MAPK and thereby protects against pathogenic bacteria [61]. This is a nice illustration of how commensal bacteria are important for the host.
Although several of these bacteria are not from the traditionally considered probiotic strains, such as LAB and Bifidobacterium, and not directly classified as probiotic, these studies help to shed light on the complicated interplay between the microbiota and the host. It can be speculated, that probiotic bacteria might employ some of the same mechanisms as these commensal bacteria to elicit their beneficial effect on the host .
18.4.4 Group IV: Direct Activation of Host Signalling Pathways
A few studies have identified some of the underlying mechanisms activated in the host by probiotic bacteria that extends C. elegans lifespan. A recurring factor is the bZip transcription factor SKN-1, which seems to be required for the life extending effect of several probiotic bacteria [62–65]. This is not surprising since SKN-1 has been identified as an important protein in regulating several age-related pathways (see Chaps. 9 and 17).
L. gasseri SBT2055 was found to extend lifespan, increase stress resistance and improve several age-related declines [62]. The lifespan extension was dependent on skn-1, and feeding with L. gasseri upregulated the expression of SKN-1 , through the phosphorylation and activation of the p38 MAPK protein PMK-1. Furthermore, age-related and SKN-1 target genes, such as gst-4, sod-1, trx-1, clk-1, hsp-16.2 and hsp-70 were also upregulated in response to feeding with L. gasseri. Reactive oxygen species and the age-related mitochondria decline were also reduced, indicating an overall activation of stress-responses. The probiotic bacteria L. rhamnosus CNCM I-3690 similarly extends nematode lifespan and stress resistance dependent on SKN-1 [64]. Contrary to the study with L. gasseri, which did not require the insulin/IGF-1 receptor homolog DAF-2 and DAF-16 [62], L. rhamnosus requires both DAF-2 , DAF-16 and SKN-1 to extend lifespan [64]. This indicates that the two bacteria activate different signalling pathways in the host as well as some common ones. However, the downstream signalling from SKN-1 was not investigated in the L. rhamnosus study. Instead, they demonstrated that L. rhamnosus had anti-inflammatory properties in cell cultures and mouse models [64].
Bifidobacterium is another LAB genus that has been tested in C. elegans. Feeding with B. infantis extends lifespan but not stress resistance [63]. The lifespan extension was abrogated in skn-1 and pmk-1 mutants, but was still induced in daf-16 mutants, demonstrating a requirement for SKN-1 and PMK-1, but not DAF-16.
A final example of communication between the bacteria and the host, is activation of the C. elegans mitochondrial stress response pathways induced by free oxygen radicals generated by E. coli [66].
A part of the LABs classified as Group II can also belong in Group IV, as some of these probiotic bacteria activate certain signalling pathways in the worm.
18.4.5 Group V: Unknown Effect
This group includes different bacterial species that have a positive or negative effect on nematode lifespan for example B. soli, B. myoides, L.reuteri and L. salivarius [32, 33, 45, 67], but where there is no current knowledge as to which bacterial or host mechanisms cause the effect on lifespan. Further investigations of these bacterial strains will eventually place them in some of the other four groups or perhaps define new groups.
In conclusion, all of these studies demonstrate that the probiotic effects of different bacteria and the host response pathways that are activated appear to be very strain specific. Furthermore, not all LAB strains appear to be probiotic, as a couple of studies have demonstrated that feeding with selected LAB strains can in fact have negative effects on their host, such as decreased lifespan [51, 67]. Therefore, caution is required when handling probiotic bacteria and predicting their effects on the host, as strains of the same genus and species might have widely different effects. However, dealing with species differences is becoming much easier with advanced DNA sequencing enabling better distinction between sub-species .
18.5 Can Worms Teach Us How to Use Probiotics in Human Health and Disease?
It is clear that C. elegans offers a powerful system to study interactions between probiotic bacteria and their host as well as host responses. It is perhaps less clear if these interactions are also going to be relevant for humans and only future experimental testing will tell for sure. However, there are studies strongly indicating that knowledge about probiotics from C. elegans will translate to humans, as is the case for all the other areas of biology covered in the previous chapters of this book.
Recently, a new probiotic LAB strain was identified using C. elegans as a screening platform [64]. A L. rhamnosus strain enhanced survival and stress resistance in C. elegans, and further experiments established that this LAB strain significantly reduced inflammation in a coculture system with human epithelial cells. Furthermore, this bacterial strain also enhanced the performance of a murine colitis model [64]. From this example it is clear that certain probiotics have effects on the host that are conserved across species and that genetic analysis in C. elegans can inform on the underlying biology.
Another study demonstrated that several LAB strains found to protect C. elegans from Salmonella Typhimurium DT104-induced death also protected pigs from diarrhoea and improved their growth performance, whereas LAB strains found not to protect C. elegans from pathogen-induced death did not protect the pigs either [68]. Again, this is an example that clearly illustrates that probiotic bacteria operate via a conserved mechanism in different hosts. Therefore, it is also likely that additional probiotic strains can be isolated using a similar approach.
There is further evidence that C. elegans can be used to identify probiotics which are functional in other organisms. A study comparing C. elegans and a porcine intestinal epithelial cell line as screening platforms for selecting probiotic bacteria was to a large extent able to identify the same probiotic bacteria in the two systems [58], although a few strains were only selected by one system. Furthermore, one selected probiotic strain induced similar host defence responses in both models [58].
Taken together, these studies demonstrate the relevance of C. elegans as a screening model organism when identifying novel probiotics for applications in livestock and humans. In this context, the ability to inexpensively generate germ free individuals as well as maintaining larges cultures are strong benefits of the C. elegans model. However, there are also some limitations that should be kept in mind.
A concern when using C. elegans in host-microbe interactions is that bacteria have never been observed to infect the intestinal cells of the worm (see Chap. 2). Rather it seems that bacteria only colonize the intestinal lumen . This is in contrast to the human intestine , where pathogenic bacteria can transverse the intestinal barrier and colonize the intestinal cells. Especially for studying the antimicrobial effect of a probiotic strain, the worm response might be different from that seen in humans, due to the difference in intestinal colonization. Furthermore, human studies have found probiotic bacteria to have an effect on several different tissues in the human body that are not found in the worm. For obvious reasons these tissue cannot be studied directly in C. elegans .
18.6 Concluding Remarks and Predictions for the Future
The main reasons for using C. elegans to study probiotics are the easily accessible genetic and biochemical methods combined with the fact that effects on organismal lifespan can be determined. Furthermore, as the worms eat bacteria as natural food sources, and since bacterial mutagenesis can be done fairly simply, C. elegans presents a system where both host and food can be mutagenized to identify which genes are required for the probiotic effect in both species, and within a relatively short time frame. For example, the effect of bacterially synthesized folate on C. elegans lifespan was identified by using a mutagenized E. coli strain [59] and the effect of vitamin B12 on worm development and fertility was demonstrated by a combination of a mutagenized bacterial screen, a drug screen and different C. elegans mutants [69]. Both studies demonstrate the feasibility of mutagenizing both the host and its microbiota to use genetics to provide the answers.
Probiotic bacteria and their effect on the host is a relatively young field , consequently many of the published studies have been descriptive and only reported correlations rather than mechanistic insight. Although these studies demonstrate that C. elegans is a useful screening platform and model organism for studying probiotic bacteria, it is clear that causal underlying mechanisms need to be identified. C. elegans presents a unique opportunity for uncovering the specific interaction between host and microbes, and not just correlations. Given the number of mutants readily available from the Caenorhabditis Genetics Center (CGC), and that they can be generated by means of CRISPR, there are really no excuses for not undertaking proper genetic dissections of the microbiota–host interactions.
Some of the discrepancies in terms of effect on lifespan due to feeding with specific bacterial strains are likely to stem from differences in experimental design . We suggest that the probiotic field should learn from the drug screening field where large efforts have been invested in standardizing experimental setups across laboratories. Having well established standardized protocols will increase consistency and reproducibility, make data interpretation more straight forward and help advance the field. For example, in some of the reported studies it is unclear if live, UV- arrested or dead bacteria are being used. From a probiotic point of view the use of live versus dead bacteria is interesting as there is a formal requirement for a microorganism to be alive in order to be classified as probiotic [9]. However, studies have shown that dead bacteria can also have beneficial effects on the host in various species [62, 70]. Although, strictly speaking, these are not probiotics they can still teach us about the molecular mechanisms of host–microbe interactions and should not be excluded from further analysis. The use of dead bacteria to modify host responses might also offer simpler treatment strategies in human and livestock compared to using live bacteria. Studying the effect of mixed cultures is an exciting area of research that needs to be developed further. The human microbiota is composed of many different bacteria species with complex interactions that influence the host in different ways. We need to develop C. elegans protocols to study feeding with mixed cultures as well as robust downstream analysis of the host responses , in order to make discoveries more applicable to human testing. The ease with which germ free L1 larvae can be generated following hypochlorite treatment is a huge advantage for these studies. Currently, experiments with mixed cultures are biassed due to different growth rates of the involved bacteria. Methods allowing in vivo detection of single or few bacteria in live animals would be a tremendous step towards more unbiased evaluations of the microbiota.
In summary, we predict that C. elegans will help understand the interactions between microbes and their host, and elucidate the host responses. This will lead the way to new treatment strategies for numerous different human diseases affected by the microbiota .
References
Sender R, Fuchs S, Milo R (2016) Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164(3):337–340. doi:10.1016/j.cell.2016.01.013
Quigley EM (2013) Gut bacteria in health and disease. Gastroenterol Hepatol (NY) 9(9):560–569
Tremaroli V, Backhed F (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489(7415):242–249. doi:10.1038/nature11552
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature 457(7228):480–484. doi:10.1038/nature07540
Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101(44):15718–15723. doi:10.1073/pnas.0407076101
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444(7122):1027–1031. doi:10.1038/nature05414
Inglis JE, Ilich JZ (2015) The microbiome and osteosarcopenic obesity in older individuals in long-term care facilities. Curr Osteoporos Rep 13(5):358–362. doi:10.1007/s11914-015-0287-7
Foster JA, Lyte M, Meyer E, Cryan JF (2015) Gut microbiota and brain function: an evolving field in neuroscience. Int J Neuropsychopharmacol. doi:10.1093/ijnp/pyv114
Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME (2014) Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11(8):506–514. doi:10.1038/nrgastro.2014.66
Savino F, Cordisco L, Tarasco V, Palumeri E, Calabrese R, Oggero R, Roos S, Matteuzzi D (2010) Lactobacillus reuteri DSM 17938 in infantile colic: a randomized, double-blind, placebo-controlled trial. Pediatrics 126(3):e526–e533. doi:10.1542/peds.2010-0433
Szajewska H, Gyrczuk E, Horvath A (2013) Lactobacillus reuteri DSM 17938 for the management of infantile colic in breastfed infants: a randomized, double-blind, placebo-controlled trial. J Pediatr 162(2):257–262. doi:10.1016/j.jpeds.2012.08.004
Metchnikoff E, Mitchell PC (1910) The prolongation of life. G. P. Putnam’s Sons, New York
Jeffery IB, Lynch DB, O’Toole PW (2016) Composition and temporal stability of the gut microbiota in older persons. ISME J 10(1):170–182. doi:10.1038/ismej.2015.88
Collino S, Montoliu I, Martin FP, Scherer M, Mari D, Salvioli S, Bucci L, Ostan R, Monti D, Biagi E, Brigidi P, Franceschi C, Rezzi S (2013) Metabolic signatures of extreme longevity in northern Italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLoS ONE 8(3), e56564. doi:10.1371/journal.pone.0056564
Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488(7410):178–184. doi:10.1038/nature11319
Kinross J, Nicholson JK (2012) Gut microbiota: dietary and social modulation of gut microbiota in the elderly. Nat Rev Gastroenterol Hepatol 9(10):563–564. doi:10.1038/nrgastro.2012.169
Sepp E, Kolk H, Loivukene K, Mikelsaar M (2014) Higher blood glucose level associated with body mass index and gut microbiota in elderly people. Microb Ecol Health Dis 25. doi:10.3402/mehd.v25.22857
Hopkins MJ, Sharp R, Macfarlane GT (2002) Variation in human intestinal microbiota with age. Dig Liver Dis 34(Suppl 2):S12–S18
O’Toole PW, Jeffery IB (2015) Gut microbiota and aging. Science 350(6265):1214–1215. doi:10.1126/science.aac8469
Cabreiro F, Gems D (2013) Worms need microbes too: microbiota, health and aging in C. elegans. EMBO Mol Med 5(9):1300–1310. doi:10.1002/emmm.201100972
Brejning J, Norgaard S, Scholer L, Morthorst TH, Jakobsen H, Lithgow GJ, Jensen LT, Olsen A (2014) Loss of NDG-4 extends lifespan and stress resistance in C. elegans. Aging Cell 13(1):156–164. doi:10.1111/acel.12165
Brooks KK, Liang B, Watts JL (2009) The influence of bacterial diet on fat storage in C. elegans. PLoS ONE 4(10), e7545. doi:10.1371/journal.pone.0007545
Clark LC, Hodgkin J (2014) Commensals, probiotics and pathogens in the C. elegans model. Cell Microbiol 16(1):27–38. doi:10.1111/cmi.12234
Pang S, Curran SP (2014) Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab 19(2):221–231. doi:10.1016/j.cmet.2013.12.005
Felix MA, Duveau F (2012) Population dynamics and habitat sharing of natural populations of C. elegans and C. briggsae. BMC Biol 10:59. doi:10.1186/1741-7007-10-59
Montalvo-Katz S, Huang H, Appel MD, Berg M, Shapira M (2013) Association with soil bacteria enhances p38-dependent infection resistance in C. elegans. Infect Immun 81(2):514–520. doi:10.1128/IAI.00653-12
Berg M, Stenuit B, Ho J, Wang A, Parke C, Knight M, Alvarez-Cohen L, Shapira M (2016) Assembly of the C. elegans gut microbiota from diverse soil microbial environments. ISME J. doi:10.1038/ismej.2015.253
Beale E, Li G, Tan MW, Rumbaugh KP (2006) C. elegans senses bacterial autoinducers. Appl Environ Microbiol 72(7):5135–5137. doi:72/7/5135 [pii] 10.1128/AEM.00611-06
Sicard M, Hering S, Schulte R, Gaudriault S, Schulenburg H (2007) The effect of Photorhabdus luminescens (Enterobacteriaceae) on the survival, development, reproduction and behaviour of C. elegans (Nematoda: Rhabditidae). Environ Microbiol 9(1):12–25. doi:EMI1099 [pii] 10.1111/j.1462-2920.2006.01099.x
Zhang Y, Lu H, Bargmann CI (2005) Pathogenic bacteria induce aversive olfactory learning in C. elegans. Nature 438(7065):179–184
Shtonda BB, Avery L (2006) Dietary choice behavior in C. elegans. J Exp Biol 209(Pt 1):89–102. doi:209/1/89 [pii] 10.1242/jeb.01955
Abada EA, Sung H, Dwivedi M, Park BJ, Lee SK, Ahnn J (2009) C. elegans behavior of preference choice on bacterial food. Mol Cells 28(3):209–213. doi:10.1007/s10059-009-0124-x
Yu L, Yan X, Ye C, Zhao H, Chen X, Hu F, Li H (2015) Bacterial respiration and growth rates affect the feeding preferences, brood size and lifespan of C. elegans. PLoS ONE 10(7), e0134401. doi:10.1371/journal.pone.0134401
Choi JI, Yoon KH, Subbammal Kalichamy S, Yoon SS, Il Lee J (2016) A natural odor attraction between lactic acid bacteria and the nematode C. elegans. ISME J 10(3):558–567. doi:10.1038/ismej.2015.134
Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C (2002) Genetic analysis of tissue aging in C. elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161(3):1101–1112
Gems D, Riddle DL (2000) Genetic, behavioral and environmental determinants of male longevity in C. elegans. Genetics 154(4):1597–1610
Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR (2002) Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in C. elegans. Exp Gerontol 37(12):1371–1378. doi:S0531556502001730 [pii]
Lenaerts I, Walker GA, Van Hoorebeke L, Gems D, Vanfleteren JR (2008) Dietary restriction of C. elegans by axenic culture reflects nutritional requirement for constituents provided by metabolically active microbes. J Gerontol A Biol Sci Med Sci 63(3):242–252. doi:63/3/242 [pii]
Gomez F, Monsalve GC, Tse V, Saiki R, Weng E, Lee L, Srinivasan C, Frand AR, Clarke CF (2012) Delayed accumulation of intestinal coliform bacteria enhances life span and stress resistance in C. elegans fed respiratory deficient E. coli. BMC Microbiol 12:300. doi:10.1186/1471-2180-12-300
Larsen PL, Clarke CF (2002) Extension of life-span in C. elegans by a diet lacking coenzyme Q. Science 295(5552):120–123. doi:10.1126/science.1064653, 295/5552/120 [pii]
Avery L, Thomas JH (1997) Feeding and defecation. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR (eds) C. elegans II, 2nd edn. Cold Spring Harbor, Plainview
Portal-Celhay C, Bradley ER, Blaser MJ (2012) Control of intestinal bacterial proliferation in regulation of lifespan in C. elegans. BMC Microbiol 12:49. doi:10.1186/1471-2180-12-49
Virk B, Jia J, Maynard CA, Raimundo A, Lefebvre J, Richards SA, Chetina N, Liang Y, Helliwell N, Cipinska M, Weinkove D (2016) Folate acts in E. coli to accelerate C. elegans aging independently of bacterial biosynthesis. Cell Rep 14(7):1611–1620. doi:10.1016/j.celrep.2016.01.051
Powell JR, Ausubel FM (2008) Models of C. elegans infection by bacterial and fungal pathogens. Methods Mol Biol 415:403–427. doi:10.1007/978-1-59745-570-1_24
Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300(5627):1921. doi:10.1126/science.1080147, 300/5627/1921 [pii]
Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2(11), e183. doi:06-PLGE-RA-0292R2 [pii] 10.1371/journal.pgen.0020183
Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, Murray BE, Ausubel FM, Calderwood SB (2002) Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in C. elegans and mice. Infect Immun 70(10):5647–5650
Darby C, Chakraborti A, Politz SM, Daniels CC, Tan L, Drace K (2007) C. elegans mutants resistant to attachment of Yersinia biofilms. Genetics 176(1):221–230. doi:genetics.106.067496 [pii] 10.1534/genetics.106.067496
Reinke SN, Hu X, Sykes BD, Lemire BD (2010) C. elegans diet significantly affects metabolic profile, mitochondrial DNA levels, lifespan and brood size. Mol Genet Metab 100(3):274–282. doi:10.1016/j.ymgme.2010.03.013
MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJ (2013) Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153(1):240–252. doi:10.1016/j.cell.2013.02.049
Zanni E, Laudenzi C, Schifano E, Palleschi C, Perozzi G, Uccelletti D, Devirgiliis C (2015) Impact of a complex food microbiota on energy metabolism in the model organism C. elegans. Biomed Res Int 2015:621709. doi:10.1155/2015/621709
Zhao Y, Zhao L, Zheng X, Fu T, Guo H, Ren F (2013) Lactobacillus salivarius strain FDB89 induced longevity in C. elegans by dietary restriction. J Microbiol 51(2):183–188. doi:10.1007/s12275-013-2076-2
Lee J, Yun HS, Cho KW, Oh S, Kim SH, Chun T, Kim B, Whang KY (2011) Evaluation of probiotic characteristics of newly isolated Lactobacillus spp. immune modulation and longevity. Int J Food Microbiol 148(2):80–86. doi:10.1016/j.ijfoodmicro.2011.05.003
Kim Y, Mylonakis E (2012) C. elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances gram-positive immune responses. Infect Immun 80(7):2500–2508. doi:10.1128/IAI.06350-11
Wang J, Nakad R, Schulenburg H (2012) Activation of the C. elegans FOXO family transcription factor DAF-16 by pathogenic Bacillus thuringiensis. Dev Comp Immunol 37(1):193–201. doi:10.1016/j.dci.2011.08.016
Iatsenko I, Yim JJ, Schroeder FC, Sommer RJ (2014) B. subtilis GS67 protects C. elegans from Gram-positive pathogens via fengycin-mediated microbial antagonism. Curr Biol 24(22):2720–2727. doi:10.1016/j.cub.2014.09.055
Zhou M, Yu H, Yin X, Sabour PM, Chen W, Gong J (2014) Lactobacillus zeae protects C. elegans from enterotoxigenic Escherichia coli-caused death by inhibiting enterotoxin gene expression of the pathogen. PLoS ONE 9(2), e89004. doi:10.1371/journal.pone.0089004
Zhou M, Zhu J, Yu H, Yin X, Sabour PM, Zhao L, Chen W, Gong J (2014) Investigation into in vitro and in vivo models using intestinal epithelial IPEC-J2 cells and C. elegans for selecting probiotic candidates to control porcine enterotoxigenic Escherichia coli. J Appl Microbiol 117(1):217–226. doi:10.1111/jam.12505
Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, Briggs Z, Hodge E, Edwards R, Ward J, Gems D, Weinkove D (2012) Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biol 10:67. doi:10.1186/1741-7007-10-67
Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E (2013) Bacterial nitric oxide extends the lifespan of C. elegans. Cell 152(4):818–830. doi:10.1016/j.cell.2012.12.043
Xiao Y, Liu F, Zhang Z, Tang J, Zou CG, Zhang KQ (2016) Gut-colonizing bacteria promote C. elegans innate immunity by producing nitric oxide. Cell Rep 14(6):1301–1307. doi:10.1016/j.celrep.2016.01.032
Nakagawa H, Shiozaki T, Kobatake E, Hosoya T, Moriya T, Sakai F, Taru H, Miyazaki T (2016) Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in C. elegans. Aging Cell 15(2):227–236. doi:10.1111/acel.12431
Komura T, Ikeda T, Yasui C, Saeki S, Nishikawa Y (2013) Mechanism underlying prolongevity induced by bifidobacteria in C. elegans. Biogerontology 14(1):73–87. doi:10.1007/s10522-012-9411-6
Grompone G, Martorell P, Llopis S, Gonzalez N, Genoves S, Mulet AP, Fernandez-Calero T, Tiscornia I, Bollati-Fogolin M, Chambaud I, Foligne B, Montserrat A, Ramon D (2012) Anti-inflammatory Lactobacillus rhamnosus CNCM I-3690 strain protects against oxidative stress and increases lifespan in C. elegans. PLoS ONE 7(12), e52493. doi:10.1371/journal.pone.0052493
Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D (2013) Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153(1):228–239. doi:10.1016/j.cell.2013.02.035
Govindan JA, Jayamani E, Zhang X, Mylonakis E, Ruvkun G (2015) Dialogue between E. coli free radical pathways and the mitochondria of C. elegans. Proc Natl Acad Sci U S A 112(40):12456–12461. doi:10.1073/pnas.1517448112
Fasseas MK, Fasseas C, Mountzouris KC, Syntichaki P (2013) Effects of Lactobacillus salivarius, Lactobacillus reuteri, and Pediococcus acidilactici on the nematode C. elegans include possible antitumor activity. Appl Microbiol Biotechnol 97(5):2109–2118. doi:10.1007/s00253-012-4357-9
Wang C, Wang J, Gong J, Yu H, Pacan JC, Niu Z, Si W, Sabour PM (2011) Use of C. elegans for preselecting Lactobacillus isolates to control Salmonella Typhimurium. J Food Prot 74(1):86–93. doi:10.4315/0362-028X.JFP-10-155
Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJ (2014) Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156(4):759–770. doi:10.1016/j.cell.2014.01.047
Adams CA (2010) The probiotic paradox: live and dead cells are biological response modifiers. Nutr Res Rev 23(1):37–46. doi:10.1017/S0954422410000090
Park MR, Yun HS, Son SJ, Oh S, Kim Y (2014) Short communication: development of a direct in vivo screening model to identify potential probiotic bacteria using C. elegans. J Dairy Sci 97(11):6828–6834. doi:10.3168/jds.2014-8561
Ikeda T, Yasui C, Hoshino K, Arikawa K, Nishikawa Y (2007) Influence of lactic acid bacteria on longevity of C. elegans and host defense against salmonella enterica serovar enteritidis. Appl Environ Microbiol 73(20):6404–6409. doi:AEM.00704-07 [pii] 10.1128/AEM.00704-07
Park MR, Oh S, Son SJ, Park DJ, Kim SH, Jeong DY, Oh NS, Lee Y, Song M, Kim Y (2015) Bacillus licheniformis isolated from traditional Korean food resources enhances the longevity of C. elegans through serotonin signaling. J Agric Food Chem 63(47):10227–10233. doi:10.1021/acs.jafc.5b03730
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Christensen, K.V., Morch, M.G., Morthorst, T.H., Lykkemark, S., Olsen, A. (2017). Microbiota, Probiotic Bacteria and Ageing. In: Olsen, A., Gill, M. (eds) Ageing: Lessons from C. elegans. Healthy Ageing and Longevity. Springer, Cham. https://doi.org/10.1007/978-3-319-44703-2_18
Download citation
DOI: https://doi.org/10.1007/978-3-319-44703-2_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-44701-8
Online ISBN: 978-3-319-44703-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)