Abstract
The contemporary forms of image representation vary depending on the application. There are well-known mathematical methods for image representation, which comprise: matrices, vectors, determined orthogonal transforms, multi-resolution pyramids, Principal Component Analysis (PCA) and Independent Component Analysis (ICA), Singular Value Decomposition (SVD), wavelet sub-band decompositions, hierarchical tensor transformations, nonlinear decompositions through hierarchical neural networks, polynomial and multiscale hierarchical decompositions, multidimensional tree-like structures, multi-layer perceptual and cognitive models, statistical models, etc. In this chapter are analyzed the basic methods for hierarchical decomposition of grayscale and color images, and of sequences of correlated images of the kind: medical, multispectral, multi-view, etc. Here is also added one expansion and generalization of the ideas of the authors from their previous publications, regarding the possibilities for the development of new, efficient algorithms for hierarchical image decompositions with various purposes. In this chapter are presented and analyzed the following four new approaches for hierarchical image decomposition: the Branched Inverse Difference Pyramid (BIDP), based on the Inverse Difference Pyramid (IDP); the Hierarchical Singular Value Decomposition (HSVD) with tree-like computational structure; the Hierarchical Adaptive Principle Component Analysis (HAPCA) for groups of correlated images; and the Hierarchical Adaptive Kernel Principal Component Analysis (HAKPCA) for color images. In the chapter are given the algorithms, used for the implementation of these decompositions, and their computational complexity is evaluated. Some experimental results, related to selected applications are also given, and various possibilities for the creation of new hybrid algorithms for hierarchical decomposition of multidimensional images are specified. On the basis of the results obtained from the executed analysis, the basic application areas for efficient image processing are specified, such as: reduction of the information surplus; noise filtration; color segmentation; image retrieval; image fusion; dimensionality reduction for objects classification; search enhancement in large scale image databases, etc.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Hierarchical image decomposition
- Branched inverse difference pyramid
- Hierarchical singular value decomposition
- Hierarchical principal component analysis for groups of images
- Hierarchical adaptive kernel principal component analysis for color images
1.1 Introduction
The methods for image processing, transmission, registration, restoration, analysis and recognition, are defined at high degree by the corresponding mathematical forms and models for their representation. On the other hand, they all depend on the way the image was created, and on their practical use. The primary forms for image representation depend on the used sources, such as: photo and video cameras, scanners, ultrasound sensors, X-ray, computer tomography, etc. The matrix descriptions are related to the primary discrete forms. Each still halftone image is represented by one matrix; the color RGB image—by three matrices; the multispectral, hyper spectral and multi-view images, and also some kinds of medical images (for example, computer tomography, IMR, etc.)—by N matrices (for N > 3), while the moving images are represented through M temporal sequences, of N matrices each. There are already many secondary forms created for image representation, obtained from the primary forms, after reduction of the information surplus, and depending on the application. Various mathematical methods are used to transform the image matrices into reduced (secondary) forms by using: vectors, for each image block, through which are composed vector fields; deterministic and statistical orthogonal transforms; multi-resolution pyramids; wavelet sub-band decompositions; hierarchical tensor transforms; nonlinear decompositions through hierarchical neural networks, polynomial and multiscale hierarchical decompositions, multi-dimensional tree-like structures, multi-layer perceptual and cognitive models, statistical models, fuzzy hybrid methods for image decomposition, etc.
The decomposition methods permit each image matrix to be represented as the sum of the matrix components with different weights, defined by the image contents. Besides, the description of each matrix in the decomposition is much simpler than that of the original (primary) matrix. The number of the matrices in the decomposition could be significantly reduced through analyzing their weights, without significant influence on the approximation accuracy of the primary matrix. To this group could be related the methods for linear orthogonal transforms [1]: the Discrete Fourier Transform (DFT), the Discrete Cosine Transform (DCT), the Walsh-Hadamard Transform (WHT), the Hartley Transform (HrT), the Haar Transform (HT), etc.; the pyramidal decompositions [2]: the Gaussian Pyramid (GP), the Laplacean Pyramid (LP), the Discrete Wavelet Transform (DWT), the Discrete Curvelet Transform (DCuT) [3], the Inverse Difference Pyramid (IDP) [4], etc.; the statistical decompositions [5]: the Principal Component Analysis (PCA), the Independent Component Analysis (ICA) and the Singular Value Decomposition (SVD); the polynomial and multiscale hierarchical decompositions [6, 7]; multi-dimensional tree-like structures [8]; hierarchical tensor transformations [9]; the decompositions based on hierarchical neural networks [10]; etc.
The aim of this chapter is to be analyzed the basic methods and algorithms for hierarchical image decomposition. Here are also generalized the following new approaches for hierarchical decomposition of multi-component matrix images: the Branched Inverse Difference Pyramid (BIDP), based on the Inverse Difference Pyramid (IDP), the Hierarchical Singular Value Decomposition (HSVD)—for the representation of single images; the Hierarchical Adaptive Principal Component Analysis (HAPCA)—for the decorrelation of sequences of images, and the Hierarchical Adaptive Kernel Principal Component Analysis (HAKPCA)—for the analysis of color images.
1.2 Related Work
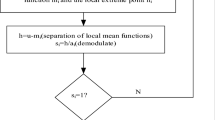
One of the contemporary methods for hierarchical image decomposition is called multiscale decomposition [7]. It is used for noise filtration in the image f, represented by the sum of the clean part u, and the noisy part, v. In accordance to Rudin, Osher and Fatemi (ROF) [11], to define the components u and v it is necessary to calculate the total variation of the functional Q, defined by the relation:
where λ > 0 is a scale parameter; and f ∈ L 2(Ω)—the image function, defined in the space L 2(Ω). The minimization of Q leads to decomposition, in result of which the visual information is divided into a part u that extracts the edges of f, and a part v that captures the texture. Denoising at different scales λ generates a multiscale image representation. In [6], Tadmor, Nezzar and Vese proposed a multiscale image decomposition which offers a hierarchical and adaptive representation for different features in the analyzed images. The image is hierarchically decomposed into the sum of simpler atoms u k , where u k extracts more refined information from the previous scale u k−1. To this end, the atoms u k are obtained as dyadically scaled minimizers of the ROF functionals at increasing λ k scales. Thus, starting with v −1 := f and letting v k denote the residual at a given dyadic scale, λ k = 2k, the recursive step [u k , v k ] = arg{inf[Q T (v k−1, k)]} leads to the desired hierarchical decomposition, f = ΣT(u k ) (here T is a blurring operator).
Another well-known approach for hierarchical decomposition is based on the hierarchical matrices [12]. The concept of hierarchical, or H-matrices, is based on the observation that submatrices of a full rank matrix may be of low rank, and respectively—to have low rank approximations. On Fig. 1.1 is given an example for the representation of a matrix of size 8 × 8 through H-matrices, which contain sub-matrices of three different sizes: 4 × 4, 2 × 2 and 1 × 1.
This observation is used for the matrix-skeleton approximation. The inverses of finite element matrices have, under certain assumptions, submatrices with exponentially decaying singular values. This means that these submatrices have also good low rank approximations. The hierarchical matrices permit decomposition by QR or Cholesky algorithms, which are iterative. Unlike them, the new approaches for hierarchical image decomposition, given in this chapter (BIDP and HSVD—for single images, HAPCA—for groups of correlated images, and HAKPCA—for color images), are not based on iterative algorithms.
1.3 Image Representation Based on Branched Inverse Difference Pyramid
1.3.1 Principles for Building the Inverse Difference Pyramid
In this section is given a short description of the inverse difference pyramid, IDP [4, 13], used as a basis for building its modifications. Unlike the famous Gaussian (GP) and Laplacian (LP) pyramids, the IDP represents the image in the spectral domain. After the decomposition, the image energy is concentrated in its first components, which permits to achieve very efficient compression, by cutting off the low-energy components. As a result, the main part of the energy of the original image is retained, despite the limited number of decomposition components used. For the decomposition implementation various kinds of orthogonal transforms could be used. In order to reduce the number of decomposition levels and the computational complexity, the image is initially divided into blocks and for each is then built the corresponding IDP.
In brief, the IDP is executed as follows: At the lowest (initial) level, on the matrix [B] of size 2n × 2n is applied the pre-selected “Truncated” Orthogonal Transform (TOT) and are calculated the values of a relatively small number of “retained” coefficients, located in the high-energy area of the so calculated transformed (spectrum) matrix [S 0 ]. These are usually the coefficients with spatial frequencies (0, 0), (0, 1), (1, 0) and (1, 1). After Inverse Orthogonal Transform (IOT) of the “truncated” spectrum matrix \( [\hat{S}_{0} ] \), which contains the retained coefficients only, is obtained the matrix \( [\hat{B}_{0} ] \) for the initial IDP level (p = 0), which approximates the matrix [B]. The accuracy of the approximation depends on: the positions of the retained coefficients in the matrix [S 0]; the values, used to substitute the missing coefficients from the approximating matrix \( [\hat{S}_{ 0} ] \) for the zero level, and on the selected orthogonal transform. In the next decomposition level (p = 1), is calculated the difference matrix \( [E_{ 0} ] = [B] - [\hat{B}_{ 0} ] \). The resulting matrix is then split into 4 sub-matrices of size 2n−1 ×2n−1 and on each is applied the corresponding TOT. The total number of retained coefficients for level p = 1 is 4 times larger than that in the zero level. In case, that Walsh-Hadamard Transform (WHT) is used for this level, the values of coefficients (0, 0) in the IDP decomposition levels 1 and higher are always equal to zero, which permits to reduce the number of retained coefficients with ¼. On each of the four spectrum matrices \( [\hat{S}_{1} ] \) for the IDP level p = 1 is applied IOT and as a result, four sub-matrices are obtained, which build the approximating difference matrix \( [\hat{E}_{ 0} ] \). In the next IDP level (p = 2) is calculated the difference matrix \( [E_{ 1} ] = [E_{ 0} ] - [\hat{E}_{ 0} ] \). After that, each difference sub-matrix is divided in similar way as in level 1, into four matrices of size 2n−2 × 2n−2, and for each is performed TOT, etc. In the last (highest) IDP level is obtained the “residual” difference matrix. In case that the image should be losslessly coded, each block of the residual matrix is processed with full orthogonal transform and no coefficients are omitted.
1.3.2 Mathematical Representation of n-Level IDP
The digital image is represented by a matrix of size (2n m) × (2n m). For the processing, the matrix is first divided into blocks of size 2n × 2n and on each is applied the IDP decomposition. The matrix [B(2n)] of each block is represented by the equation:
Here the number of decomposition components, which are matrices of size 2n × 2n, is equal to (r + 2). The maximum possible number of decomposition levels for one block is n + 1 (for r = n − 1). The last component \( [E_{r} ( 2^{n} ) ] \) defines the approximation error for the block \( [B ( 2^{n} ) ] \) for the case, when the decomposition is limited up to level p = r. The first component \( [\hat{B}_{0} ( 2^{n} ) ] \) for the level p = 0 is the coarse approximation of the block [B(2n)]. It is obtained through 2D IOT on the block \( [\hat{S}_{0} (2^{n} ) ] \) in correspondence with the relation:
where \( [T_{0} ( 2^{\text{n}} ) ]^{ - 1} \) is a matrix of size 2n × 2n, used for the inverse orthogonal transform of \( [\hat{S}_{0} ( 2^{\text{n}} ) ] \).
The matrix \( [\hat{S}_{0} ( 2^{\text{n}} ) ]= [m_{0} (u,\,v )s_{0} (u,\,v ) ] \) is the “truncated” orthogonal transform of the block [B(2n)]. Here m 0(u, v) are the elements of the binary matrix-mask [M 0(2n)], used to define the retained coefficients of \( [\hat{S}_{0} ( 2^{\text{n}} ) ] \) in correspondence to the relation:
The values of the elements \( m_{0} (u,\,v ) \) are selected in accordance with the requirement the retained coefficients \( \hat{s}_{0} (u,\,v )= m_{0} (u,\,v )s_{0} (u,\,v ) \) to be these with maximum energy, calculated for all image blocks. The transform \( [S_{0} ( 2^{\text{n}} ) ] \) of the block [B(2n)] is defined through direct 2D OT:
where \( [T_{0} ( 2^{\text{n}} ) ] \) is a matrix of size 2n × 2n for the decomposition level p = 0, used to perform the selected 2D OT, which could be DFT, DCT, WHT, KLT, etc.
The remaining coefficients in the decomposition presented by Eq. 1.1 are the approximating difference matrices \( [\hat{E}_{p - 1} (2^{n - p} ) ] \) for levels p = 1, 2, …, r. They comprise the sub-matrices \( [\hat{E}_{p - 1}^{{k_{p} }} (2^{n - p} ) ] \) of size 2n−p × 2n−p for k p = 1, 2, …, 4p, obtained through quadtree division of the matrix \( [\hat{E}_{p - 1} (2^{n - p} ) ] \). Each sub-matrix \( [\hat{E}_{p - 1}^{{k_{p} }} (2^{n - p} ) ] \) is then defined by the relation:
where 4p is the number of the quadtree branches in the decomposition level p. Here \( [T_{p} (2^{n - p} )]^{ - 1} \) is a matrix of size 2n−p × 2n−p in the level p, used for the inverse 2D OT.
The elements \( \hat{s}_{p}^{{k_{p} }} (u,\,v )= m_{p} (u,\,v ).\,s_{p}^{{k_{p} }} (u,\,v ) \) of the matrix \( [\hat{S}_{p}^{{k_{p} }} (2^{n - p} ) ] \) are defined by the elements m p (u, v) of the binary matrix-mask [M p (2 n−p)]:
The matrix \( [S_{p}^{{k_{p} }} (2^{n - p} ) ] \) is the transform of \( [E_{p - 1}^{{k_{p} }} (2^{n - p} ) ] \) and is defined through direct 2D OT:
Here \( [T_{p} (2^{n - p} )] \) is a matrix of size 2n−p × 2n−p in the decomposition level p, used for the 2D OT of each block \( [E_{p}^{{k_{p} }} (2^{n - p} ) ] \) (when k p = 1, 2,…, 4p), of the difference matrix for same level, defined by the equation:
In result of the decomposition represented by Eq. 1.1, for each block [B(2n)], are calculated the following spectrum coefficients:
-
all nonzero coefficients of the transform \( [\hat{S}_{0} (2^{n} ) ] \) in the decomposition level p = 0;
-
all nonzero coefficients of the transforms \( [\hat{S}_{p}^{{k_{p} }} (2^{n - p} ) ] \) for k p = 1, 2, …, 4p in the decomposition levels p = 1, 2, …, r.
The spectrum coefficients of same spatial frequency (u, v) from all image blocks are arranged in common data sequences, which correspond to their decomposition level p. The transformation of the 2D data massifs into one-dimensional data sequence is executed, using the recursive Hilbert scan, which preserves very well the correlation between neighboring coefficients.
In order to reduce the decomposition complexity, and in accordance with Eq. 1.1, this could be done recursively, as follows:
For the case, when the number of the retained coefficients for each IDP sub-block k p of size \( 2^{n - p} \, \times \,{\kern 1pt} 2^{n - p} \) is \( \sum\limits_{u = 0}^{{2^{n - p} }} {\sum\limits_{v = 0}^{{2^{n - p} }} {m_{p} (u,\,v )} } = 4, \) then their total number for all levels is:
In this case the total number of “retained” coefficients is 4/3 times higher than that of the pixels in the block, and hence, the IPD is “overcomplete”.
1.3.3 Reduced Inverse Difference Pyramid
For the building of the Reduced IDP (RIDP) [14], the existing relations between the spectrum coefficients from the neighboring IDP levels are used. Let the retained coefficients \( s_{p}^{{k_{p} }} (u,v ) \) with spatial frequencies (0, 0), (1, 0), (0, 1) and (1, 1) for the sub-block k p in the IDP level p, be obtained by using the 2D-WHT. Then, except for level p = 0, the coefficients (0, 0) from each of the four neighboring sub-blocks in same IDP level are equal to zero, i.e.:
From this, it follows that the coefficients \( s_{p + i}^{{k_{p} }} (0,0 ) \) for i = 0, 1, 2, 3 could be cut-off, and as a result they should not be saved or transferred. Hence, the total number of the retained coefficients N R for each sub-block k p in the decomposition levels p = 1, 2,…, n−1 of the RIDP could be reduced by ¼, i.e.
In this case the total number of the “retained” coefficients for all levels is equal to the number of pixels in the block, and hence, the so calculated RIPD is “complete”.
1.3.4 Main Principle for Branched IDP Building
The pyramid BIDP [15, 16] with one or more branches is an extension of the basic IDP. The image representation through the BIDP aims at the enhancement of the image energy concentration in a small number of IDP components. On Fig. 1.2 is shown an example block diagram of the generalized 3-level BIDP. The IDP for each block of size 2n × 2n from the original image, called “Main Pyramid”, is of 3 levels (n = 3, for p = 0, 1, 2). The values of the coefficients, calculated for these 3 levels, compose the inverse pyramid, whose sections are of different color each. The coefficients s(0, 0), s(0, 1), s(1, 0) and s(1, 1) in level p = 0 from all blocks compose corresponding matrices of size m × m, colored in yellow. These 4 matrices build the “Branch for level 0” of the Main Pyramids. Each is then divided into blocks of size 2n−1 ×2n−1, on which in similar way are built the corresponding 3-level IPDs (p = 00, 01, 02). The retained coefficients s(0, 1), s(1, 0) and s(1, 1) in level p = 1 of the Main Pyramids from all blocks build matrices of size 2m × 2m (colored in pink).
Each matrix of size 2m × 2m is divided into blocks of size 2n−1 × 2n−1, on which in similar way are build corresponding 3-level IDPs (p = 10, 11, 12). The retained coefficients, calculated after TOT from the blocks of the Residual Difference in the last level (p = 2) of the Main Pyramids, build matrices of size 4m × 4m; from the first level (p = 00) of the Pyramid Branch 0—matrices of size (m/2n−1 × m/2n−1); and from the first level (p = 10) of the “Pyramid Branch 1”—matrices of size (m/2n−2 × m/2n−2). In order to reduce the correlation between the elements of the so obtained matrices, on each group of 4 spatially neighboring elements is applied the following transform: the first is substituted by their average value, and each of the remaining 3—by its difference to next elements, scanned counter-clockwise. The coefficients, obtained this way from all levels of the Main and Branch Pyramids are arranged in one-dimensional sequences in accordance with Hilbert scan and after that are quantizated and entropy coded using Adaptive RLC and Huffman. The values of the spectrum coefficients are quantizated only in case that the image coding is lossy. In order to retain the visual quality of the restored images, the quantization values are related to the sensibility of the human vision to errors in different spatial frequencies. To reduce these errors, retaining the compression efficiency, in the consecutive BIDP levels could be used various fast orthogonal transforms: for example, in the zero level could be used DCT, and in the next levels—WHT.
1.3.5 Mathematical Representation for One BIDP Branch
In the general case, the branch g of the BIDP is built on the matrix \( [S_{g (u,\,v )} ] \) of size \( 2^{n - g - 1} \, \times {\kern 1pt} \,2^{n - g - 1} , \) which comprises all spectrum coefficients \( s_{p}^{{k_{p} }} (u,\,v ) \) with the same spatial frequency (u, v) from all blocks or sub-blocks k p in the level p = g of the Main IDPs. By analogy with Eq. (1.1), the matrix \( [S_{g (u,v )} ] \) could be decomposed in accordance with the relation, given below:
where
All matrices in Eqs. (1.14)−(1.19) are of size \( 2^{n - g - 1} \times {\kern 1pt} 2^{n - g - 1} , \) and these in Eqs. (1.20) and (1.21)—of size \( 2^{n - g - s - 1} \times {\kern 1pt} 2^{n - g - s - 1} . \) The decomposition from Eq. (1.14) of the matrix \( [S_{g (u,v )} ] \) is named Pyramid Branch (PB g(u,v)). It is a pyramid, whose initial and final levels are g and r correspondingly (g < r). This pyramid represents the branch g of the Main IDPs and contains all coefficients, whose spatial frequency is (u, v).
The maximum number of branches for the levels p = 0, 1, …, n − 1 of the Main IDPs, built on a sub-block of size \( 2^{n - p} \times {\kern 1pt} 2^{n - p} , \) is defined by the general number of retained spectrum coefficients \( M_{p} = {\kern 1pt} 4^{p} \sum\limits_{u = 0}^{{2^{n - p} }} {\sum\limits_{v = 0}^{{2^{n - p} }} {m_{p} (u,v )} } \). For the branch g from the level p = g the corresponding pyramid PB g(uv) is of r levels. The number of the coefficients in this branch of the Main IDPs for p = g, g + 1, …, r, without cutting-off the coefficients, calculated for the spatial frequency (0, 0), is:
In case that the number of the retained spectrum coefficients for each sub-block is set to be \( \sum\limits_{u = 0}^{{2^{n - g} }} {\sum\limits_{v = 0}^{{2^{n - g} }} {m_{g} (u,v )} } = 4, \) then \( M_{g} = {\kern 1pt} 4^{g + 1} \). In this case, from Eq. (1.22) it follows, that the total number of the coefficients in the branch PB g(uv) is \( N_{g,r} = {\kern 1pt} {\kern 1pt} ( 4^{g + 1} / 3) (4^{r + 1} - 4^{g} ). \) Hence, the compression ratio (CR) for PB g(uv) is defined by the relation:
where 4n−g−1 is the number of the elements in one sub-block of size \( 2^{n - g - 1} \times {\kern 1pt} 2^{n - g - 1} \) from PB g(uv).
The compression ratio for the Main IDPs, calculated in accordance with Eq. (1.11), is:
From the comparison of the Eqs. (1.23) and (1.24) it follows, that:
In case that the requirement from Eq. (1.25) for the number of levels r of PB g(u,v) for level g of the Main IDPs is satisfied, the compression ratio for the branch g is higher, than that for each of the basic pyramids. From Eq. (1.25) it follows that the condition r > 1 is satisfied, when n > 4, i.e., when the image is divided into blocks of minimum size of 16 × 16 pixels. For this case, to retain the correlation between their pixels high, is necessary the size of the image (16m) × (16m) to be relatively large. For example, the image should be of size 2k × 2k (for m = 128), or larger. Hence, the BIDP decomposition is efficient mainly for images with high resolution.
The correlation between the elements of the blocks of size \( 2^{n - 1} \times {\kern 1pt} 2^{n - 1} \) from the initial level g = 0 of the Main IDPs is higher than that, between the elements of the sub-blocks of size \( 2^{n - g - 1} \times {\kern 1pt} 2^{n - g - 1} \) from the higher levels g = 1, 2, …, r. Because of this, the branching of the BIDP should always start from the level g = 0.
1.3.6 Transformation of the Retained Coefficients into Sub-blocks of Size 2 × 2
The aim of the transformation is to reduce the correlation between the retained neighboring spectrum coefficients in the sub-blocks of size 2 × 2 in each matrix, built by the coefficients of same spatial frequency (u, v) from all blocks (or respectively—from the sub-blocks k p in the selected level p of the Main IDPs, or their branches). In order to simplify the presentation, the spectrum coefficients in the sub-blocks k p for the level p, are set as follows:
On Fig. 1.3 are shown matrices of size 2 × 2, which contain the retained groups of four spectrum coefficients \( s_{p}^{{k_{p} }} (u,v ) \), which have same frequencies, (0, 0), (1, 0), (0, 1) and (1, 1) correspondingly, placed in four neighboring sub-blocks (k p , k p + 1, k p + 2, k p + 3) of size 2n−p × 2n−p for the level p of the Main IDPs, or their branches.
In correspondence with the symbols, used in Fig. 1.3, the transformation of the groups of four coefficients is represented by the relation below [16]:
Here P i , for i = 1, 2, 3, 4 represent correspondingly:
-
the coefficients A i , for i = 1, 2, 3, 4 with frequencies (0, 0);
-
the coefficients B i , for i = 1, 2, 3, 4 with frequencies (1, 0);
-
the coefficients C i , for i = 1, 2, 3, 4 with frequencies (0, 1);
-
the coefficients D i , for i = 1, 2, 3, 4 with frequencies (1, 1).
In result of the transform, executed in accordance with Eq. (1.27), each coefficient S 1 has higher value, than the remaining three difference coefficients S 2, S 3, and S 4.
The inverse transform executed in respect of Eq. (1.27) gives total restoration of the initial coefficients P i , for i = 1, 2, 3, 4:
Depending on the frequency (0, 0), (1, 0), (0, 1), or (1, 1) of the restored coefficients P 1 ~ P 4, they correspond to A 1 ~ A 4, B 1 ~ B 4, C 1 ~ C 4, or D 1 ~ D 4. The operation, given in Eq. (1.28) is executed through decoding of the transformed coefficients S 1 ~ S 4. The so described features of the coefficients S 1, S 2, S 3, S 4 permit to achieve significant enhancement of their entropy coding efficiency.
The basic quality of the BIDP is that it offers significant decorrelation of the processed image data. As a result, the BIDP permits the following:
-
To achieve highly efficient compression with retained visual quality of the restored image (i.e. visually lossless coding), or efficient lossless coding, depending on the application requirements;
-
Layered coding and transfer of the image data, in result of which is obtained low transfer bit-rate with gradually increased quality of the decoded image;
-
Lower computational complexity than that of the wavelet decompositions [4];
-
Easy adaptation of the coder parameters, so that to ensure the needed concordance of the obtained data stream, to the ability of the communication channel;
-
Resistance to noises in the communication channel, or due to compression/decompression. The reason for this is the use of TOT in the decoding of each image block;
-
Retaining the quality of the decoded image after multiple coding/decoding;
The BIDP could be further developed and modified in accordance to the requirements of various possible applications. One of these applications for processing of groups of similar images, for example, is a sequence of Computer Tomography (CT) images, Multi-Spectral (MS) images, etc.
1.3.7 Experimental Results
The experimental results, given below, were obtained from the investigation of image database, which contained medical images stored in DICOM (dcm) format, of various size and kind, grouped in 24 classes. The database was created at the Medical University of Sofia, and comprises the following image kinds: CTI—computer tomography images; MGI—mammography images; NMI—nuclear magnetic resonance images; CRI—computer radiography images, and USI—ultrasound images. For the investigation, the DICOM images were first transformed into non-compressed (bmp format), and then they were processed by using various lossless compression algorithms. A part of the obtained results is given in Table 1.1.
Here are shown the results for the lossless compression of the bmp files of still images, and of image sequences, after their transformation into files of the kind jp2 and tk. The image file format jp2 is based on the standard JPEG2000LS, and the tk format—on the algorithms BIDP for single images, combined with the adaptive run-length lossless coding (ARLE), based on the histogram statistics [17]. For the execution of the 2D-TOT/IOT in the initial levels of all basic pyramids and their branches was used the 2D-DCT, and in their higher levels—the 2D-WHT transform. The number of the pyramid levels for the blocks of the smallest treated images (of size 512 × 512), is two, and for the larger ones, it is three. The basic IDP pyramids have one branch only, comprising coefficients with spatial frequency (0, 0) for their initial levels.
From the analysis of the obtained results, the following conclusions could be done:
-
1.
The new format tk surpasses the jp2, especially for images, which contain objects, placed on a homogenous background. From the analyzed 24 classes of images, 17 are of this kind. Some examples are shown in Table 1.1;
-
2.
Together with the enlargement of the analyzed images, the compression ratio for the lossless tk compression grows up, compared to that of the jp2;
-
3.
The data given in Table 1.1 show that the mean compression ratio for all DICOM images after their transformation into the format tk is 41:1, while for the jp2 this coefficient is 26:1. Hence, the use of the tk format for all 24 classes ensures compression ratio which is ≈40 % higher than that of the jp2 format.
The experimental results, obtained for the comparison of the coding efficiency for several kinds of medical images through BIDP and JPEG2000 confirmed the basic advantages of the new approach for hierarchical pyramid decomposition, presented here.
1.4 Hierarchical Singular Value Image Decomposition
The SVD is a statistical decomposition for processing, coding and analysis of images, widely used in the computer vision systems. This decomposition was an object of vast research, presented in many monographs [18–22] and papers [23–26]. This is optimal image decomposition, because it concentrates significant part of the image energy in minimum number of components, and the restored image (after reduction of the low-energy components), has minimum mean square error. One of the basic problems, which limit, to some degree, the use of the “classic” SVD, is related to its high computational complexity, which grows up together with the image size.
To overcome this problem, several new approaches are already offered. The first is based on the SVD calculation through iterative methods, which do not require defining the characteristic polynomials of a pair of matrices. In this case, the SVD is executed in two stages: in the first, each matrix is first transformed into triangular form with the QR decomposition, and then—into bidiagonal, through the Householder transforms [27]. In the second stage on the bidiagonal matrix is applied an iterative method, whose iterations stop when the needed accuracy is achieved. For this could be used the iterative method of Jacobi [21], in accordance with which for the calculation of the SVD with bidiagonal matrix is needed the execution of a sequence of orthogonal transforms with rotation matrix of size 2 × 2. The second approach is based on the relation of the SVD with the Principal Component Analysis (PCA). It could be executed through neural networks [28] of the kind generalized Hebbian or multilayer perceptron networks, which use iterative learning algorithms. The third approach is based on the algorithm, known as Sequential KL/SVD [29]. The basic idea here is as follows: the image matrix is divided into blocks of small size, and on each is applied the SVD, based on the QR decomposition [21]. At first, the SVD is calculated for the first block from the original image (the upper left, for example), and then is used iterative SVD calculation for each of the remaining blocks by using the transform matrices, calculated for the first block (by updating the process). In the flow of the iteration process are deleted the SVD components, which correspond to very small eigen values.
For the acceleration of the SVD calculation several methods are already developed [30–32]. The first, is based on the algorithm, called Randomized SVD [30], a number of matrix rows (or columns) is randomly chosen. After scaling, they are used to build a small matrix, for which is calculated the SVD, and it is later used as an approximation of the original matrix. In [31] is offered the algorithm QUIC-SVD, suitable for matrices of very large size. Through this algorithm is achieved fast sample-based SVD approximation with automatic relative error control. Another approach is based on the sampling mechanism, called the cosine tree, through which is achieved best-rank approximation. The experimental investigation of the QUIC-SVD in [32] presents better results than those, from the MATLAB SVD and the Tygert SVD. The so obtained 6–7 times acceleration compared to the SVD depends on the pre-selected value of the parameter δ which defines the upper limit of the approximation error, with probability (1 − δ).
Several SVD-based methods developed, are dedicated to enhancement of the image compression efficiency [33–37]. One of them, called Multi-resolution SVD [33], comprises three steps: image transform, through 9/7 biorthogonal wavelets of two levels, decomposition of the SVD-transformed image, by using blocks of size 2 × 2 up to level six, and at last—the use of the algorithms SPIHT and gzip. In [34] is offered the hybrid KLT-SVD algorithm for efficient image compression. The method K-SVD [35] for facial image compression, is a generalization of the K-means clusterization method, and is used for iterative learning of overcomplete dictionaries for sparse coding. In correspondence with the combined compression algorithm, in [36] is proposed a SVD based sub-band decomposition and multi-resolution representation of digital colour images. In the paper [37] is used the decomposition, called Higher-Order SVD (HOSVD), through which the SVD matrix is transformed into a tensor with application in the image compression.
In this chapter, the general presentation of one new approach for hierarchical decomposition of matrix images is given, based on the multiple application of the SVD on blocks of size 2 × 2 [38]. This decomposition, called Hierarchical SVD (HSVD), has tree-like structure of the kind “binary tree” (full or truncated). The SVD calculation for blocks of size 2 × 2 is based on the adaptive KLT [5, 39]. The HSVD algorithm aims to achieve a decomposition with high computational efficiency, suitable for parallel and recursive processing of the blocks through simple algebraic operations, and offers the possibility for enhancement of the calculations through cutting-off the tree branches, whose eigen values are small or equal to zero.
1.4.1 SVD Algorithm for Matrix Decomposition
In the general case, the decomposition of each image matrix [X(N)] of size N × N could be executed by using the direct SVD [5], defined by the equation below:
The inverse SVD is respectively:
In the relations above, the terms \( [U (N ) ]= [\vec{U}_{1} ,\,\vec{U}_{2}, \ldots,\,\vec{U}_{N} ] \) and \( [V (N ) ]= [\vec{V}_{1} ,\,\vec{V}_{2}, \ldots ,\,\vec{V}_{N} ] \) are matrices, composed respectively by the vectors \( \vec{U}_{s} \) and \( \vec{V}_{s} \) for s = 1, 2, …, N; \( \vec{U}_{s} \) are the eigenvectors of the matrix \( \left[ {Y\left( N \right)} \right] = \left[ {X\left( N \right)} \right]\left[ {X\left( N \right)} \right]^{t} \) (left-singular vectors of the [X(N)]), and \( \vec{V}_{s} \)—the eigenvectors of the matrix \( \left[ {Z\left( N \right)} \right] = \left[ {X\left( N \right)} \right]^{t} \left[ {X\left( N \right)} \right] \) (right-singular vectors of the [X(N)]), for which:
\( [\varLambda (N ) ]{\kern 1pt} {\kern 1pt} = diag{\kern 1pt} {\kern 1pt} [\lambda_{1} ,\lambda_{2} ,..,\lambda_{\text{N}} ] \) is a diagonal matrix, composed by the eigenvalues \( \lambda_{s} \) which are identical for the matrices \( [Y (N ) ]{\kern 1pt} \) and \( [Z (N ) ]{\kern 1pt} \).
From Eq. (1.29) it follows that for the description of the decomposition for a matrix of size N × N, N × (2N + 1) parameters are needed in total, i.e. in the general case the SVD is a decomposition of the kind “overcomplete”.
1.4.2 Particular Case of the SVD for Image Block of Size 2 × 2
In this case, the direct SVD for the block [X] of size 2 × 2 (for N = 2) is represented by the relation:
or
where \( [C_{1} ]= \sqrt {\lambda_{1} } \vec{U}_{1} \vec{V}_{1}^{t} ; \) \( [C_{2} ]= \sqrt {\lambda_{2} } \vec{U}_{2} \vec{V}_{2}^{t} ; \) a, b, c, d are the elements of the block [X]; \( \lambda_{1} , \) \( \lambda_{2} \) are the eigenvalues of the symmetrical matrices [Y] and [Z], defined by the relations below:
-
\( \vec{U}_{1} \) and \( \vec{U}_{2} \) are the eigenvectors of the matrix [Y], for which: \( [Y ]\vec{U}_{s} = \;\lambda_{s} \vec{U}_{s} \) (s = 1, 2);
-
\( \vec{V}_{1} \) and \( \vec{V}_{2} \) are the eigenvectors of the matrix [Z], for which: \( [Z ]{\kern 1pt} {\kern 1pt} \vec{V}_{s} {\kern 1pt} = \,\lambda_{s} \vec{V}_{s} \) (s = 1, 2).
-
\( \left[ {\,U} \right] = [\vec{U}_{1} ,\vec{U}_{2} ] \) and \( \left[ {\,V} \right]{\kern 1pt}^{t} = \left[ {\begin{array}{*{20}c} {\vec{V}_{1}^{t} } \\ {\vec{V}_{2}^{t} } \\ \end{array} } \right] \) are matrices, composed by the eigen vectors \( \vec{U}_{s} \) and \( \vec{V}_{s} \).
In accordance with the solution given in [38] for the case when N = 2, the couple direct/inverse SVD for the matrix [X(2)] could be represented as follows:
where
Figure 1.4 shows the algorithm for direct SVD for the block [X] of size 2 × 2, composed in accordance with the relations (1.36), (1.38) and (1.39). This algorithm is the basic building element—the kernel, used to create the HSVD algorithm.
In accordance with Eq. (1.32) the matrix [X] is transformed into the vector \( \vec{X} = [a,b,c,d{\kern 1pt} \, ]^{{{\kern 1pt} t}} , \) whose components are arranged by using the “Z”-scan. The components of the vector \( \vec{X} \) are the input data for the SVD algorithm. After its execution, are obtained the vectors \( \vec{C}_{1} \) and \( \vec{C}_{2} , \) from whose components are defined the elements of the matrices [C 1] and [C 2] of size 2 × 2, by using the “Z”-scan again. In this case however, this scan is used for the inverse transform of all vectors \( \vec{C}_{1} \), \( \vec{C}_{2} \) in the corresponding matrix [C 1], [C 2].
1.4.3 Hierarchical SVD for a Matrix of Size 2n × 2n
The hierarchical n-level SVD (HSVD) for the image matrix [X(N)] of size 2n × 2n pixels ( N = 2n) is executed through multiple applying the SVD on image sub-blocks (sub-matrices) of size 2 × 2, followed by rearrangement of the so calculated components.
In particular, for the case, when the image matrix [X(4)] is of size 22 × 22 ( N = 22 = 4), then the number of the hierarchical levels of the HSVD is n = 2. The flow graph, which represents the calculation of the HSVD, is shown on Fig. 1.5. In the first level (r = 1) of the HSVD, the matrix [X(4)] is divided into four sub-matrices of size 2 × 2, as shown in the left part of Fig. 1.5. Here the elements of the sub-matrices on which is applied the SVD2×2 in the first hierarchical level, are colored in same color (yellow, green, blue, and red). The elements of the sub-matrices are:
On each sub-matrix [X k (2)] of size 2 × 2 (k = 1, 2, 3, 4), is applied SVD2×2, in accordance with Eqs. (1.36)−(1.39). As a result, it is decomposed into two components:
where \( \sigma_{1,k} = \sqrt {\frac{{\omega_{1,k} + A_{1,k} }}{2}} , \) \( \sigma_{2,k} = \sqrt {\frac{{\omega_{2,k} - A_{2,k} }}{2}} , \) \( [T_{1,k} (2 ) ]= \vec{U}_{1,k} \vec{V}_{1,k}^{t} , \) \( [T_{2,k} (2 ) ]= \vec{U}_{2,k} \vec{V}_{2,k}^{t} . \)
Using the matrices \( [C_{m,k} (2 ) ] \) of size 2 × 2 for k = 1, 2, 3, 4 and m = 1, 2, are composed the matrices \( [C_{m} (4 ) ] \) of size 4 × 4:
Hence, the SVD decomposition of the matrix [X] in the first level is represented by two components:
In the second level (r = 2) of the HSVD, on each matrix \( [C_{m} (4 ) ] \) of size 4 × 4 is applied four times the SVD2×2. Unlike the transform in the previous level, in the second level, the SVD2×2 is applied on the sub-matrices [C m,k (2)] of size 2 × 2, whose elements are mutually interlaced and are defined in accordance with the scheme, given in the upper part of Fig. 1.5. The elements of the sub-matrices, on which is applied the SVD2×2 in the second hierarchical level are colored in same color (yellow, green, blue, and red). As it is seen on the figure, the elements of the sub-matrices of size 2 × 2 in the second level are not neighbors, but placed one element away in horizontal and vertical directions. As a result, each matrix \( [C_{m} (4 ) ] \) is decomposed into two components:
Then, the full decomposition of the matrix [X] is represented by the relation:
Hence, the decomposition of an image of size 4 × 4 comprises four components in total.
The matrix [X(8)] is of size 23 × 23 ( N = 23 = 8 for n = 3), and in this case, the HSVD is executed through multiple calculation of the SVD2×2 on blocks of size 2 × 2, in all levels (the general number of the decomposition components is eight). In the first and second levels, the SVD2×2 is executed in accordance with the scheme, shown on Fig. 1.5. In the third level, the SVD2×2 is mainly applied on sub-matrices of size 2 × 2. Their elements are defined in similar way, as shown on Fig. 1.5, but the elements of same color (i.e., which belong to same sub-matrix) are moved three elements away in the horizontal and vertical direction.
The described HSVD algorithm could be generalized for the cases when the image [X(2n )] is of size 2n × 2n pixels. Then the relation (1.45) becomes as shown below:
The maximum number of the HSVD decomposition levels is n, the maximum number of the decomposition components (1.46) is 2n, and the distance in horizontal and vertical direction between the elements of the blocks of size 2 × 2 in the level r is correspondingly (2r−1 − 1) elements, for r = 1, 2,…, n.
1.4.4 Computational Complexity of the Hierarchical SVD of Size 2n × 2n
1.4.4.1 Computational Complexity of the SVD of Size 2 × 2
The computational complexity could be defined by using the Eq. (1.36), taking into account the number of multiplication and addition operations, needed for the preliminary calculation of the components \( \omega , \) \( \mu , \) \( \delta , \) \( \nu , \) \( \eta , \) \( A, \) B, θ 1, θ 2, σ 1, σ 1, defined by the Eqs. (1.38) and (1.39). Then:
-
The number of the multiplications, needed for the calculation of Eq. (1.36) is Σ m = 39;
-
The number of the additions, needed for the calculation of Eq. (1.36) is Σ s = 15.
Then the total number of the algebraic operations executed with floating point for SVD of size 2 × 2 is:
1.4.4.2 Computational Complexity of the Hierarchical SVD of Size 2n × 2n
The computational complexity is defined on the basis of SVD2×2. In this case, the number M of the sub-matrices of size 2 × 2, which comprise the image of size 2n × 2n, is 2n−1 × 2n−1 = 4n−1, and the number of the decomposition levels is n.
-
The number of SVD2×2 in the first level is M 1 = M = 4n−1;
-
The number of SVD2×2 in the second level is M 2 = 2 × M = 2 × 4n−1;
-
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
-
The number of SVD2×2 in the level n is M n = 2n−1 × M = 2n−1 × 4n−1;
The total number of SVD2×2 is correspondingly M Σ = M(1 + 2 + … + 2n−1) = 4n−1(2n − 1) = 22n−2(2n − 1). Then the total number of the algebraic operations for the HSVD of size 2n × 2n is:
1.4.4.3 Computational Complexity of the SVD of Size 2n × 2n
For the calculation of the matrices [Y(N)] and [Z(N)] of size N × N for N = 2n are needed in total \( \varSigma_{m} = 2^{2n + 2} \) multiplications and \( \varSigma_{s} = 2^{n + 1} (2^{n} - 1 ) \) additions. The total number of the operations is:
In accordance with [40], the number of the operations O(N) for the iterative calculation of all N eigenvalues and the eigen N-component vectors of the matrix of size N × N for N = 2n with L iterations, is correspondingly:
From Eq. (1.31) it follows, that two kinds of eigen vectors (\( \vec{U}_{s} \) and \( \vec{V}_{s} \)) should be calculated, so the number of the needed operations in accordance with Eq. (1.51) should be doubled. From the analysis of the Eq. (1.29) it follows that:
-
The number of the needed multiplications for all components is: \( \varSigma_{m} = {\kern 1pt} 2^{n} (2^{2n} + {\kern 1pt} 2^{2n} ){\kern 1pt} = {\kern 1pt} 2^{3n + 1} ; \)
-
The number of the needed additions for all components is: \( \varSigma_{s} = 2^{n} - 1. \)
Then the total number of the needed operations for the calculation of Eq. (1.29) is:
Hence, the total number of the algebraic operations, needed for the execution of the SVD of size 2n × 2n is:
1.4.4.4 Relative Computational Complexity of the HSVD
The relative computational complexity of the HSVD could be calculated on the basis of Eqs. (1.53) and (1.48), using the relation below:
For n = 2, 3, 4, 5 (i.e., for image blocks of size 4 × 4, 8 × 8, 16 × 16 and 32 × 32 pixels), the values of ψ 1(n, L) for L = 10 are given in Table 1.2.
For big values of n the relation ψ 1(n, L) does not depend on n and trends towards:
Hence, for big values of n, when the number of the iterations L ≥ 4, the relation \( \psi_{1} (n,L )> 1 \), and the computational complexity of the HSVD is lower than that of the SVD. Practically, the value of L is significantly higher than 4. For big values of n the coefficient ψ 1(n, 10) = 3.1 and the computational complexity of the HSVD is three times lower than that of the SVD.
1.4.5 Representation of the HSVD Algorithm Through Tree-like Structure
The tree-like structure of the HSVD algorithm of n = 2 levels, shown on Fig. 1.6, is built on the basis of the Eq. (1.45), for image block of size 4 × 4. As it could be seen, this is a binary tree. For a block of size 8 × 8, this binary tree should be of n = 3 levels.
Each tree branch has a corresponding eigen value \( \lambda_{s,k} \), or resp. \( \sigma_{s,k} {\kern 1pt} \, = \,{\kern 1pt} \sqrt {\lambda_{s,k} } \) for the level 1, and \( \lambda_{s,k} (m ) \) or resp. \( \sigma_{s,k} (m )\,{\kern 1pt} = \,{\kern 1pt} \sqrt {\lambda_{s,k} (m )} \)—for the level 2 (m = 1, 2). The total number of the tree branches shown on Fig. 1.6, is equal to six. It is possible to cut off some branches, if for them the following conditions are satisfied: \( \sigma_{s,k} \vee \sigma_{s,k} (m )= 0 \) or \( \sigma_{s,k} \le \varDelta_{s,k} \vee \sigma_{s,k} (m )\, \le \varDelta_{s,k} (m ), \) i.e., when they are equal to 0, or are smaller than a small threshold \( \varDelta_{s,k} , \) resp. \( \varDelta_{s,k} (m ). \) To cut down one HSVD component [C i ] in one level, it is necessary all values of σ i , which participate in this component, to be equal to zero, or very close to it. In result, the decomposition in the corresponding branch could be stopped before the last level n. As a consequence, it follows that the HSVD algorithm is adaptive with respect of the contents of each image block. In this sense, the algorithm HSVD is adaptive and could be easily adjusted to the requirements of each particular application.
From the analysis of the presented HSVD algorithm it follows that its basic advantages compared to the “classic” SVD are:
-
1.
The computational complexity of the full-tree HSVD algorithm (without truncation) for a matrix of size 2n × 2n, compared to SVD for a matrix of same size, is at least three times lower;
-
2.
The HSVD is executed following the tree-like scheme of n levels, which permits parallel and recursive processing of image blocks of size 2 × 2 in each level. The corresponding SVD is calculated by using simple algebraic relations;
-
3.
The HSVD algorithm retains the quality of the SVD in respect of the high concentration of the main part of the image energy in the first components of the decomposition. After removal of the low-energy components, the restored matrix has minimum mean square error and is the optimal approximation of the original;
-
4.
The tree-like structure of the HSVD algorithm (a binary tree of n levels) makes more feasible the ability to stop the decomposition earlier in some of the tree branches, for which the corresponding eigen value is zero, or approximately zero. As a result, the computational complexity of the HSVD is additionally reduced, compared to the classic SVD;
-
5.
The HSVD algorithm could be easily generalized for matrices of size different from 2n × 2n. In these cases each matrix should be divided into blocks of size 8 × 8, to which is applied the HSVD (i.e., a decomposition of eight components). In case that after the division the blocks at the image borders are incomplete, they should be extended through extrapolation. Such approach is suitable in case, that the number of the decomposition components, which is limited up to 8, is sufficient for the application. If more components are needed, their number could be increased, by dividing the image into blocks of size 16 × 16, or larger;
-
6.
The HSVD algorithm opens new possibilities for fast image processing in various application areas, as: compression, filtration, segmentation, merging, watermarking, extraction of minimum number of features for pattern recognition, etc.
1.5 Hierarchical Adaptive Principal Component Analysis for Image Sequences
Image sequences are characterized with the huge volumes of visual information and very high spatial and spectral correlation. The decorrelation of this visual information is the first and basic stage of the processing, related to various publication areas, such as: compression and transfer/storage, analysis, objects recognition, etc. For the decorrelation of correlated image sequences, are developed significant number of methods for interframe prediction with movement compensation for temporal decorrelation of moving images and for transform-coding techniques for intra-frame and inter-frame decorrelation. One of the most efficient methods for decorrelation of groups of images is based on the Principal Component Analysis (PCA), known also as Hotelling transform, and Karhunen-Loeve Transform (KLT). This transform is the object of large number of investigations, presented in many scientific monographs [11, 40–47] and papers [12, 48–53]. The KLT is related to the class of linear statistical orthogonal transforms for groups of vectors, obtained, for example, from the pixels of one image, or from a group of matrix images. The PCA has significant role in image analysis and processing, and also in the systems for computer science and pattern recognition. It has a wide variety of application areas: for the creation of optimal models in the image color space [46], for compression of signals and groups of correlated images [41–44, 47], for the creation of objects descriptors in the reduced features’ space [50, 51], for image fusion [52] and segmentation [53], image steganography [54], etc.
The PCA has some significant properties: (1) it is an optimal orthogonal transform for a group of vectors, because as a result of the transform, the maximum part of their energy is concentrated in a minimum number of their components; (2) after reduction of the low energy components of the transformed vectors, the corresponding restored vectors have minimum mean square error (MSE); (3) the components of the transformed vectors are not correlated. In particular, in case that the probability distribution of the vectors is Gaussian, their components become decorrelated and independent after PCA. The Independent Components Analysis (ICA) [55] is very close to the PCA in respect of their calculation and properties.
For PCA implementation the pixels of same spatial position in a group of N images compose an N-dimensional vector. The basic difficulty of the PCA implementation is related to the large size of the covariance matrix. For the calculation of its eigenvectors is necessary to calculate the roots of a polynomial of nth degree (characteristic equation) and to solve a linear system of N equations [21, 56]. For large values of N, the computational complexity of the algorithm for calculation of the transform matrix is significantly increased.
One of the basic problems, which limit the use of the PCA, is due to its high computational complexity, which grows up together with the number of the vectors’ components. Various approaches are offered to overcome this problem. One of them is based on the PCA calculation through iterative methods, which do not require the definition of the characteristic polynomial of the vectors’ covariance matrix. In this case the PCA is executed in 2 stages: in the first, the original image matrix is transformed into a three-diagonal form through QR decomposition [21], and after that—into a bi-diagonal, by using the Householder’s transforms [27]. In the second stage, on the bi-diagonal matrix are applied iterative methods, for which the iterations are stopped, after the needed accuracy is achieved. The iterative PCA calculation through the methods of Jacobi and Givens [21, 56], is based on the execution of a sequence of orthogonal transforms with rotational matrices of size 2 × 2.
One well known approach is based on the PCA calculation by using neural networks [28] of the kind Generalized Hebbian, or Multilayer Perceptron Networks. They both use iterative learning algorithms, for which the number of needed operations can reach several hundreds.
The third approach is based on the Sequential KLT/SVD [29], already commented in the preceding section. In [28, 29] is presented one more approach, based on the recursive calculation of the covariance matrix of the vectors, its eigen values and eigen vectors. In the papers [57, 58] is introduced hierarchical recursive block processing of matrices.
The next approach is based on the so-called Distributed KLT [59, 60], where each vector is divided into sub-vectors and on each is applied Partial KLT. Then is executed global iterative approximation of the KLT, through Conditional KLT, based on side information. This approach was further developed in [61], where is offered one algorithm for adaptive two-stage KLT, combined with JPEG2000, and aimed at the compression of hyper-spectral (HS) images. Similar algorithm for enhanced search is the “Integer Sub-optimal KLT” (Int SKLT) [62], which uses the lifting factorization of matrices. This algorithm is basic for the KLT, executed through a multilevel strategy, also called Divide-and-Conquer (D&C) [63]. In correspondence with this approach, the KLT for a long sequence of images is executed after dividing it into smaller groups, for which the corresponding KLT have lower computational complexity. By applying the KLT on each group, is obtained local decorrelation only. For this reason, the eigen images for the first half of each group in the first decomposition level are used as an input for the next (second) level of the multi-level transform, etc. In the case, when the KLT group contains 2 components only, the corresponding multilevel transform is called Pair-wise Orthogonal Transform (POT) [64]. The experimental results obtained for this transform, when used for HS images, show that it is more efficient than the Wavelet Transform (WT) in respect of Rate-Distortion performance, computational cost, component scalability, and memory requirements.
Another approach is based on the Iterative Thresholding Sparse PCA (ITSPCA) [65] algorithm, aiming at the reduction of the features’ space dimension, with minimum dispersion loss.
A fast calculation algorithm (Fast KLT) is known for the particular case, when the images are represented through first order Markov model [66].
In correspondence with the algorithm for PCA randomization [67], on the basis of an accidental choice are selected a certain number of rows (or columns) of the covariance matrix, and on the basis of this approximation, the computational complexity of the KLT is reduced.
In the works [68, 69], are presented hybrid methods for compression of multi-component images through KLT, combined with Wavelets, Adaptive Mixture of Principal Components Model, and JPEG2000.
The analysis of the famous KLT methods shows that: (1) In case of iterative calculations, the number of iterations depends on the covariance matrix of the vectors. In many cases this number is very high, which makes the real-time KLT calculation extremely difficult; (2) In case that the method for multilevel D&C is used, the eigen images from the second half of each group are not transformed in the next levels and as a result, they are not completely decorrelated. Moreover—the selection of the length of each group of images is not optimized.
One of the possible approaches for reducing the computational complexity of PCA for N-dimensional group of images is based on the so-called Hierarchical Adaptive PCA (HAPCA) [70]. Unlike the famous Hierarchical PCA (HPCA) [58], this transform is not related to the image sub-blocks, but to the whole image from one group. For this, the HPCA is implemented through dividing the images into groups of length, defined by their correlation range. Each group is divided into sub-groups of 2 or 3 images each, on which is applied Adaptive PCA (APCA) [71–73], of size 2 × 2 or 3 × 3. This transform is performed using equations, which are not based on iterative calculations, and as a result, they have lower computational complexity. To obtain decorrelation for the whole group of images, it is necessary to use APCA of size 2 × 2 or 3 × 3, which will be applied in several consecutive stages (hierarchical levels), with rearranging of the obtained intermediate eigen images after each stage. In result, is obtained a decorrelated group of eigen images, on which could be applied other combined approaches to obtain efficient compression through lossless or lossy coding.
1.5.1 Principle for Decorrelation of Image Sequences by Hierarchical Adaptive PCA
The new principle was developed for the transformation of image sequences using the adaptive PCA (APCA) with transform matrix of size 2 × 2 or 3 × 3. The sequence is divided into groups, whose length is harmonized with their correlation range. The corresponding algorithm comprises the following steps: (1) correlation analysis of the image sequence, in result of which is defined the length N of each group; (2) dividing the processed group into sub-groups of two or three images each, depending on the length of the group, (3) adding (when necessary) new interpolated images, which supplements the last sub-group up to two or three images; (4) defining the number of the hierarchical transform levels on the basis of the mutual decorrelation, which should be achieved, (5) executing of the HAPCA algorithm for each group from the image sequence. For this, on each sub-group of two or three images from the first hierarchical level of HAPCA, is applied Adaptive PCA (APCA) with matrix of size 2 × 2 or 3 × 3. In result, are obtained 2 or 3 eigen images. After that, the eigen images are rearranged so that the first sub-group of 2 eigen images to comprise the first images from each group, the second group of 2 or 3 eigen images—the second images from each group, etc. To each group of intermediate eigen images in the first hierarchical level is applied in similar way the next APCA with a 2 × 2 or 3 × 3 matrix, on each sub-group of 2 or 3 eigen images. In result are obtained the corresponding new intermediate eigen images in the second hierarchical level. Then the eigen images are rearranged again so, that the first group of 2 or 3 eigen images contains the first images from each group before the rearrangement; the second group of 2 or 3 eigen images—the second image before the rearrangement, etc.
1.5.2 Description of the Hierarchical Adaptive PCA Algorithm
1.5.2.1 Calculation of Eigen Images Through APCA with a 2 × 2 Matrix
For any 2 digital images of size S = M × N pixels each, shown on Fig. 1.7, are calculated the vectors \( \vec{C}_{s} = \left[ {{\kern 1pt} C_{1s} ,C_{2s} } \right]\,^{t} \) for s = 1, 2, …, S.
Each vector is transformed into the corresponding vectors \( \vec{L}_{s} = [L_{1s} ,L_{2s} ]^{t} \) through direct APCA using the matrix [Φ] of size 2 × 2 in correspondence with the relation:
where
On the other hand, the components of the vectors \( \vec{\varPhi }_{1} ,\;\vec{\varPhi }_{2} \) could be defined using the rotation angle θ of the coordinate system (L 1, L 2) towards the original coordinate system \( (C_{1} ,C_{2} ) \), resulting from the APCA execution. Then:
where
As a result of the transform from Eq. (1.56) on all S vectors, are obtained the corresponding two eigen images [L 1], [L 2], shown in the right part of Fig. 1.7. The transformation from Eq. (1.56) is reversible, and the inverse APCA is represented by the relation:
On Fig. 1.8 is shown the algorithm for direct/inverse APCA for a group of two images.
1.5.2.2 Hierarchical APCA Algorithm for a Group of 8 Images
On Fig. 1.9 is shown the 3-level HAPCA algorithm for the case, when the number of the correlated images in one group (GOI) is N = 8; in one sub-group it is N sg = 2; and the number of the sub-groups is N g = 4, i.e. N = N sg × N g .
As it is shown on Fig. 1.9, on each sub-group of two images from the first hierarchical level of HAPCA is applied APCA with a 2 × 2 matrix. In result are obtained two “eigen” images, colored in yellow and blue correspondingly. After that, the “eigen” images are rearranged so that the first sub-group of two “eigen” images comprises the first images from each group, the second group of two “eigen” images—the second images from each group, etc. For each GOI of 8 intermediate eigen images in the first hierarchical level, is applied in similar way the next APCA, with a 2 × 2 matrix, on each sub-group of two eigen values. In result are obtained two new “eigen” images (i.e. the “eigen” images of the group of two intermediate eigen images), colored in yellow, and blue correspondingly in the second hierarchical level. Then the eigen images are rearranged again, so that the first group of two “eigen” images to contain the first images from each group before the rearrangement; the second group of two “eigen” images—the second image before the rearrangement, etc. In result, is achieved significant decorrelation for the processed group of images, which is a reliable basis for their efficient compression/restoration. For this is necessary to have information about the transform matrix, used for each couple of images in all hierarchical levels—12 matrices for one GOI altogether (when N = 8).
1.5.2.3 Calculation of Eigen Images Through APCA with a 3 × 3 Matrix
From the three digital images of S pixels each, are obtained the vectors \( \vec{C}_{s} = \, [C_{1s} ,C_{2s} ,C_{3s} ]^{t} \) for s = 1, 2, …, S. The vectors \( \vec{C}_{s} \) are transformed into the vectors \( \vec{L}_{s} = \,{\kern 1pt} {\mathbf{[}}L_{1s} ,L_{2s} ,L_{3s} {\mathbf{]}}^{t} \) through direct APCA, given in Eq. (1.56), and using the matrix [Φ] of size 3 × 3.
The elements Φ ij of the matrix [Φ] and the vector \( \vec{\mu }\, = \, [\bar{C}_{1} ,\bar{C}_{2} ,\bar{C}_{ 3} ]^{t} \) for \( \bar{C}_{ 1} = E (C_{1s} ), \) \( \bar{C}_{ 2} = E (C_{2s} ), \) \( \bar{C}_{ 3} = E (C_{3s} ) \) are defined below:
where
The inverse APCA, using the matrix [Φ] of size 3 × 3, is defined by Eq. (1.58).
1.5.2.4 Hierarchical APCA Algorithm for a Group of 9 Images
In this case, the HAPCA algorithm for a group of nine images N = 9 is executed in similar way, as that, shown on Fig. 1.9 for a group of eight images (N = 8). Each GOI is divided into N g = 3 sub-groups, each containing N sg = 3 images, and the number of the HAPCA decomposition levels is n = 2. In the first HAPCA level, in accordance with Eq. (1.56) on the vectors \( \vec{C}_{s} = \, [C_{1s} ,C_{2s} ,C_{3s} ]^{t} \) for each of the three sub-groups the APCA is executed. In this case, the elements of the matrix [Φ] of size 3 × 3 are defined by Eq. (1.59). In result, for each sub-group the vectors \( \vec{L}_{s} = \,{\kern 1pt} {\mathbf{[}}L_{1s} ,L_{2s} ,L_{3s} {\mathbf{]}}^{t} \) are calculated. After the rearrangement of the vectors components from all sub-groups and their second division into sub-groups of same size (N g = 3), are obtained the corresponding input vectors \( \vec{L}_{s}^{1} (r )\, = \, [L_{1s}^{1} (r ),L_{2s}^{1} (r ),L_{3s}^{1} (r ) ]^{t} \) for the next HAPCA level, etc.
1.5.3 Setting the Number of the Levels and the Structure of the HAPCA Algorithm
1.5.3.1 Number of the HAPCA Levels
The minimum number of levels n min needed for the execution of the HAPCA algorithm for a group of N images could be defined through the analysis of the mutual correlation of the group of transformed N-dimensional vectors, obtained after each hierarchical level. For this, after the execution of the first HAPCA level for the transformed vectors \( \vec{L}_{s} \) for each sub-group (with two or three components), are obtained the N-dimensional vectors \( \vec{L}_{s}^{1} \, = \, [L_{1s}^{1} ,L_{2s}^{1} , \ldots ,L_{Ns}^{1} ]^{t} \). After the rearrangement of the components of each vector \( \vec{L}_{s}^{1} \), it is transformed into the vector \( \vec{L}_{s}^{1} (r )\, = \, [L_{1s}^{1} (r ),L_{2s}^{1} (r ), \ldots ,L_{Ns}^{1} (r ) ]^{t} \). The decision to continue with the next (second) HAPCA is based on the analysis of the covariance matrix \( {\kern 1pt} [ {\text{K}}_{L}^{1} (r ) ] \) of the rearranged vectors \( \vec{L}_{s}^{1} (r ) \) for s = 1, 2, …, S, from which could be calculated the achieved decorrelation in the first level. In case that full decorrelation is achieved, the matrix \( {\kern 1pt} [ {\text{K}}_{L}^{1} (r ) ] \) is diagonal. The HAPCA algorithm could be stopped before the second level even if the decorrelation is not full, provided that the relation below is satisfied:
Here \( k_{i,j} (r ) \) is the element (i, j) of the matrix \( {\kern 1pt} [K_{L}^{1} (r ) ] \), and δ is a threshold with preliminary set small value. In case that the condition from Eq. (1.60) is not satisfied, the processing continues with the second HAPCA level. After all calculations are finished, the condition in Eq. (1.60) is checked again, but here \( k_{i,j} (r ) \) are the elements of the matrix \( {\kern 1pt} [ {\text{K}}_{L}^{2} (r ) ] \) of the rearranged vectors \( \vec{L}_{s}^{2} (r ) \) in the second level, etc.
1.5.3.2 Structure of the HAPCA Algorithm
The structure of the HAPCA algorithm for one group (GOI) depends on the number of images (N) in it. This number is defined through correlation analysis of the whole image sequence, and most frequently it is in the range from 4, up to 16. In some cases, the number of images in the group is not divisible by the number of the images in a sub-group (N g ), which should be two or three, and then the number of the images N has to be extended by adding m int interpolated images to the GOI. In result, the new value N e = N + m int becomes divisible by two or three. In Table 1.3, are given the basic parameters of HAPCA for one GOI: N—number of images in the group, n—the number of transform levels, N sg —the number of the sub-groups, N g —the number of the images in one sub-group, N e —the number of images in the extended GOI, and m int —the number of the interpolated images in the extended GOI.
The number of the levels n in Table 1.3 is defined through correlation analysis of the whole GOI, and the values of N sg and N g —on the basis of the requirement for minimum value of the number of interpolated images, m int .
1.5.3.3 Computational Complexity of HAPCA
The computational complexity of the n-levels HAPCA algorithm can be calculated and compared with the classic PCA for a covariance matrix of size N × N for group of N images with N sg sub-groups for the APCA of size 2 × 2 or 3 × 3. In case of classic PCA, this number is n = N sg = 1, because there are no hierarchical levels or sub-groups. For this, both algorithms are compared regarding the number of operations O (additions and multiplications) [74] needed for the calculation of the following components:
-
Covariance matrices [K C ] of size N × N for the classic PCA algorithm and for the APCA with size of the transform matrix 2 × 2 or 3 × 3 [73]:
$$ O_{cov} (N )= ( 1 / 2 )N (N + 1 ) [N (N - 1 )+ 2 (N + 2 ) ]\; {\text{for}}\;{\text{the}}\;{\text{classic}}\;{\text{PCA}}; $$(1.61)$$ O_{cov} (2 )= 3 0\; {\text{for}}\;{\text{APCA}}\;{\text{of}}\;{\text{size}}\; 2\times 2\;(N = 2); $$(1.62)$$ O_{cov} (3 )= 9 6\; {\text{for}}\;{\text{APCA}}\;{\text{of}}\;{\text{size}}\; 3\times 3\;(N = 3). $$(1.63) -
Calculation of the eigen values of the corresponding [K C ] covariance matrix when the QR decomposition and the Householder transform of (N − 1) steps are used for the classic PCA [73]:
$$ O_{val} (N )= (N{\kern 1pt} { - 1)(}\frac{ 4}{ 3}N^{2} + \frac{17}{6}N + 7 )\; {\text{for}}\;{\text{classic}}\;{\text{PCA}}; $$(1.64)$$ O_{val} (2 )\approx 1 2\; {\text{for}}\;{\text{APCA}}\;{\text{of}}\,{\text{size}}\; 2\times 2\;(N = 2); $$(1.65)$$ O_{val} (3 )= 55\; {\text{for}}\;{\text{APCA}}\;{\text{of}}\;{\text{size}}\; 3\times 3\;(N = 3). $$(1.66) -
Calculation of the eigen vectors of the corresponding [K C ] covariance matrix in case that iterative algorithm with 4 iterations is used for the classic PCA [73]:
$$ O_{vec} (N )= N [ 2N ( 4N + 5 )- 1 ]\; {\text{for}}\;{\text{classic}}\,{\text{PCA}}; $$(1.67)$$ O_{vec} (2 )= 102\; {\text{for}}\;{\text{APCA}}\;{\text{of}}\;{\text{size}}\; 2\times 2\;(N = 2); $$(1.68)$$ O_{vec} (3 )= 303\; {\text{for}}\;{\text{APCA}}\;{\text{of}}\;{\text{size}}\; 3\times 3\;(N = 3). $$(1.69) -
The number of operations needed for the calculation of a group of N eigen images (each of S pixels), obtained in result of direct PCA transform for zero mean vectors, is:
$$ O (N,S )= SN ( 2N - 1 )\; {\text{for}}\;{\text{classic}}\;{\text{PCA}}; $$(1.70)$$ O (2,S )= 6S\; {\text{for}}\;{\text{APCA}}\;{\text{of}}\;{\text{size}}\; 2\times 2\;(N = 2); $$(1.71)$$ O (3,S )= 15S\;{\text{for}}\;{\text{APCA}}\;{\text{of}}\;{\text{size}}\; 3\times 3\;(N = 3). $$(1.72) -
Using Eqs. (1.61)−(1.72) the total number of operations (TO) needed for both algorithms (the classic PCA and the HAPCA-based algorithms with APCA of size 2 × 2 or 3 × 3) is:
$$ TO_{PCA} (N,{\kern 1pt} S ){\kern 1pt} = {\kern 1pt} \frac{ 1}{ 2}N (N + {\kern 1pt} 1 ) [N - {\kern 1pt} 1 )+ {\kern 1pt} 2 (N + 2 )]{\kern 1pt} + {\kern 1pt} (N{\kern 1pt} { - }{\kern 1pt} 1 ) (\frac{ 4}{ 3}N^{2} + {\kern 1pt} \frac{17}{6}N + 7 ){\kern 1pt} + {\kern 1pt} N [ 2N ( 4N + {\kern 1pt} 5 ){\kern 1pt} - {\kern 1pt} 1{\kern 1pt} ]{\kern 1pt} + SN ( 2N - {\kern 1pt} 1 ) ; $$(1.73)$$ TO_{HAPCA - 2} (N,S ){\kern 1pt} = {\kern 1pt} nN_{sg} ( 3 0+ 1 2+ 1 0 2+ 6S )= {\kern 1pt} nN_{g} ( 1 4 4\, + 6S ) ; $$(1.74)$$ TO_{HAPCA - 3} (N,S ){\kern 1pt} = {\kern 1pt} nN_{sg} ( 9 6+ 55 + 303 + 15S )= {\kern 1pt} nN_{g} ( 4 5 4+ 15S ). $$(1.75)
Having obtained the total number of operations required by the algorithms (1.73)−(1.75), we can compare the computational complexity of both the classic PCA and the proposed algorithms. The reduction of the number of operations needed for these algorithms can be described by the coefficient:
is the ratio of the number of operations for the classic PCA and the proposed HAPCA-2 algorithm (with APCA of size 2 × 2), and:
is the ratio of the number of operations for the classic PCA and the proposed HAPCA-3 algorithm (with APCA of size 3 × 3).
For example, for N = 8, n = 3 and N g = 4, from Eq. (1.76) is obtained:
For N = 9, n = 2 and N g = 3, from Eq. (1.77) it follows:
If S = 218, then \( \eta_{2} ( 8 ,{\kern 1pt} {\kern 1pt} 2^{ 1 8} ){\kern 1pt} {\kern 1pt} = 1.66 \) and \( \eta_{3} ( 9 ,{\kern 1pt} {\kern 1pt} 2^{ 1 8} ){\kern 1pt} {\kern 1pt} = 1.7, \) i.e., the coefficient η(S) is at least 1.66 times larger than 1 for images of size 512 × 512, or higher (in average, about 2 times). For higher values of N (for example, between 9 and 16), and for big values of S, the coefficient η(S) > 2.
1.5.4 Experimental Results
The presented experimental results are for sequences of multispectral (MS) images. As an example, was used the test MS sequence “balloons” shown on Fig. 1.10; on Fig. 1.11 is shown the corresponding color image with RGB values, obtained after lighting with neutral daylight. This sequence is from the free-access image database of the Columbia University, USA (http://www1.cs.columbia.edu/CAVE/databases/multispectral/). It contains N = 15 MS images of size 512 × 512 pixels, 16 bpp. On it was applied the 3-level HAPCA algorithm. The sequence was divided into N sg = 5 sub-groups, each of N g = 3 MS images, and the number of the vectors in each sub-group is S = 218.
On Fig. 1.12 are shown the corresponding eigen MS images, obtained after applying the 3-level HAPCA algorithm on the group of images. As it could be seen from the results shown on Fig. 1.13, the main part of the energy of these 15 images is concentrated on the first eigen MS image, and the energy of the next eigen images decreases rapidly.
The graphics on Fig. 1.13 represent the power distribution of all 15 eigen images in levels 1, 2, 3 before and after the rearrangement. In the first three eigen MS images are concentrated 99, 88 % of the total power of all 15 images in the GOI.
The basic qualities of the HAPCA algorithm for processing of groups of MS images are:
-
1.
Lower computational complexity than PCA for the whole GOI, due to the lower complexity of APCA with matrices of size 2 × 2 and 3 × 3 compared to the case, when for the calculation of the PCA matrix are used iterative methods;
-
2.
HAPCA could be used not only for efficient compression of sets of MS images, but also for sequences of medical CT images, video sequences, obtained from stationary TV camera, compression of multi-view images, image fusion, face recognition, etc.;
-
3.
There is also a possibility for further development of the HAPCA algorithms, through: the use of Integer PCA for lossless coding of MS images; HAPCA with a matrix of size N × N (N—a digit, divisible by 2 or 3), but without using numerical methods, etc.
1.6 Hierarchical Adaptive Kernel Principal Component Analysis for Color Image Segmentation
The color image segmentation is of high significance in computer vision as the first stage of the processing, concerning the detection and extraction of objects with predefined color, the shape of the visible part of the surface, and the texture. The existing color image segmentation techniques can be classified into seven main approaches based on: edge detection, region growing, neural network based, fuzzy logic, histogram analysis, Support Vector Machine and principal color [75–79]. One of the contemporary methods for color image segmentation is based on the adaptive models in the perceptual color space, using neural networks as multilayer perceptrons with multi-sigmoid activation function [80]. Recently special attention attracted the methods for human skin segmentation in color images [81–85]. These methods are mainly based on different color spaces, adaptive color space switching, skin color models and detection techniques.
The color space representation based on the PCA [86–88] offers significant advantages in the efficient image processing, as image compression and filtration, color segmentation, etc. In this section, a new approach for adaptive object color segmentation is presented through combining the linear and nonlinear PCA. The basic problem of PCA, which makes its application for efficient representation of the image color space relatively difficult, is related to the hypothesis for Gaussian distribution of the primary RGB vectors. One of the possible approaches for solving the problem is the use of PCA variations, such as: the nonlinear Kernel PCA (КPCA) [88], Fast Iterative KPCA [89], etc. In this section, for the color space representation an adaptive method for transform selection is used: linear PCA or nonlinear KPCA. The first transform (the linear PCA) could be considered as a particular case of the КPCA. The linear PCA is carried out on the basis of the already described Color Adaptive PCA (CAPCA) [85, 87]. The choice of CAPCA or КPCA is made through evaluation of the kind of distribution of the vectors, which describe the object color: Gaussian or not.
1.6.1 Mathematical Representation of the Color Adaptive Kernel PCA
In the general case, through KPCA is executed nonlinear transform of the original centered vectors \( \vec{X}_{s} \) over S pixels (\( \vec{X}_{s} = \sum\limits_{s = 1}^{S} {\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} } \)) into the high-dimensional space, and then, for the obtained transformed vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \), the PCA is applied. The aim is, in the new multidimensional space the vertices of the vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) to be concentrated in an area, which is accurately enough enveloped by a hyperellipsoid, whose axes are the eigenvectors of the covariance matrix of the vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \). Figure 1.14 illustrates the idea of the new 3D color space of eigenvectors \( \vec{\nu }_{1} ,\vec{\nu }_{2} ,\vec{\nu }_{3} \) [85].
In particular, it is possible that the vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) in the transformed space are represented by their projections on the first eigenvector \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {\nu }_{1} \) of their covariance matrix, as shown in Fig. 1.15. For the example, shown in this figure, on the eigenvector \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {\nu }_{1} \) is projected the basic part of the multitude of all transformed vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \). The original 3D color vectors \( \vec{C}_{s} \) are first centered:
where \( \vec{m}_{C} \) is the mean value of the color vector and then follows some kind of nonlinear transform, which uses the selected nonlinear function Φ (.). In result, the corresponding N-dimensional vectors, \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) (N ≥ 3) are obtained. The value of N depends on the selected function Φ (.), used for the nonlinear transform [88].
The covariance matrix \( [ {\tilde{\text{K}}}_{\text{x}} ] \) of the transformed color vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) is of size N × N and can be calculated in accordance with the relation:
where \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) = [\varPhi (x_{s1} ),\varPhi (x_{s2} ),..,\varPhi (x_{sN} )]^{t} \) for s = 1, 2,…, S.
For each eigenvalue \( \tilde{\lambda }_{i} \) and eigenvector \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {\nu }_{i} = [\nu_{i1} ,\nu_{i2} , \ldots ,\nu_{iN} ]^{t} \) of the matrix \( [ {\tilde{\text{K}}}_{\text{x}} ] \) the following relation is performed:
After substitution in Eq. (1.82) using Eq. (1.81), is got:
In result of the transformation of Eq. (1.83), known as the “kernel trick” [90], for the ith eigenvector is obtained:
where for \( \tilde{\lambda }_{i} \, \ne \,0 \) the coefficient \( \alpha_{si} = \frac{{\varPhi (\vec{X}_{s} )^{t} .\vec{\nu }_{i} }}{{S\tilde{\lambda }_{i} }}. \)
From this, it follows, that:
Substituting Eq. (1.84) in Eq. (1.83), is obtained:
or
from which follows:
After multiplying the left side of the above equation with the vector \( \varPhi (\vec{X}_{s} )^{t} , \) is obtained:
The dot product of the vectors \( \varPhi (\vec{X}_{s} ) \) and \( \varPhi (\vec{X}_{l} ) \) could be represented through the kernel function \( k(\vec{X}_{s} ,\vec{X}_{l} ), \) defined by the relation:
Here, the term \( k(\vec{X}_{s} ,\,\vec{X}_{l} ) \) represents the elements (s, l) of the Gram matrix [K] of size S × S, called “kernel matrix”. After substituting Eq. (1.88) in Eq. (1.87), is obtained:
Under the condition, that the matrix [K] is positively defined (i.e. when it eigenvalues are positive) is got a shorter representation than in Eq. (1.89). Then:
From this relation it follows, that \( S\tilde{\lambda }_{i} \) are the eigenvalues of the matrix [K], and \( \vec{\alpha }_{i} = [\alpha_{i1} ,\alpha_{i2} , \ldots ,\alpha_{iS} ]^{t} \) are the corresponding eigenvectors of same matrix. Taking into account the requirement \( \vec{\nu }_{i}^{t} \vec{\nu }_{i} = 1 \), from Eq. (1.84) is obtained the relation:
After substituting Eq. (1.90) in Eq. (1.91) is obtained \( S\tilde{\lambda }_{i} \vec{\alpha }_{i}^{t} \vec{\alpha }_{i} = 1 \), from which is defined the square of the module of the vector \( \vec{\alpha }_{i} = [\alpha_{i1} ,\alpha_{i2} , \ldots ,\alpha_{iS} ]^{t} \):
In the general case, the vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) in Eq. (1.88) are not centered. In order to apply the PCA on them, they should be centered in advance, and in result are obtained the vectors:
where \(\vec{m}_{{{\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\smile}$}}{\varPhi } }}} = E\left\{ {\varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} )} \right\} = \frac{1}{S}\sum\limits_{s = 1}^{S} {\varPhi (\vec{X}_{s} )} . \)
The covariance matrix \( [\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\smile}$}}{\text{K}}] \) of the centered vectors \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) is of size S × S and is defined by the relation:
The matrix kernel is:
The relation between the covariance matrices \( [ \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\smile}$}}{{\text{K} ]}} \) and [K] is:
where [I1/s] is a matrix of size S × S, whose elements are equal to 1/S.
The projection of the vector \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) on the eigenvector \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {\nu }_{i} \) in the S-dimensional space is:
Using the projections \( Pr_{si} \) of the vector \( \varPhi (\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {X}_{s} ) \) on each of the first k ≤ N eigenvectors \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {\nu }_{i} \) (for i = 1, 2,…, k), could be taken the decision for the classification of the sth pixel to the dominant color of the selected object, using some of the well-known classifiers, as: SVM, LDA, k-nearest neighbors, neural networks, etc. [89]
To carry out the KPCA, one could use different kinds of kernel functions, such as the polynomial, the Gaussian, the sigmoid, etc. By substituting \( \varPhi (\vec{X}_{s} )= \vec{x} \) and \( \varPhi (\vec{X}_{l} )= \vec{y} \) the polynomial kernel function of degree d is defined by the relation:
For d = 2 and if assumed that for the transformation of the 3-component vectors \( \vec{X}_{s} = [x_{s1} , x_{s2} ,x_{s3} ]^{t} \) and \( \vec{X}_{l} = [x_{l1} , x_{l2} ,x_{l3} ]^{t} \) into N-component is used the nonlinear function Ф(.), then:
where the vectors components are defined by the relations:
for r 1 , r 2 = 0, 1, p 1 , p 2 = 1, 2, 3, i = 1, 2, … N and s, l = 1, 2, …, S.
In this case the maximum value of N is N = 9. In order to reduce the needed calculations, it is suitable to use smaller number of the possible 9 components of the quadratic function Ф(.).
For example, if assumed N = 3 and if only mixed products of the vectors components \( \vec{X}_{s} \) and \( \vec{X}_{l} \) are chosen, then from Eq. (1.100) it follows:
Then the corresponding kernel function of vectors \( \varPhi (\vec{X}_{s} ) \) and \( \varPhi (\vec{X}_{l} ) \) is represented by the polynomial below:
In particular, for d =1, \( \varPhi (\vec{X}_{s} )= \vec{X}_{s} \) and \( \varPhi (\vec{X}_{l} )= \vec{X}_{l} \) the corresponding kernel function is linear:
From the above, it follows that KPCA is transformed into linear PCA (i.e. PCA is a particular case of KPCA).
1.6.2 Algorithm for Color Image Segmentation by Using HAKPCA
The general algorithm for objects segmentation in the extended color space, based on the Hierarchical Adaptive Kernel PCA (HAKPCA) and the classifier of the reduced vectors, is shown on Fig. 1.16.
In the preprocessing block, each color vector \( \vec{C}_{s} = [{\kern 1pt} R_{s} ,G_{s} ,B_{s} ]^{t} \) is transformed into the corresponding expanded vector \( \vec{P}_{s} \). If the chosen kernel-function is polynomial, and the 3D color space is transformed into a 9-dimensional, then the components p is of vectors \( \vec{P}_{s} \) could be defined as follows:
In order to put all components p is in the range [0, 255], for i = 4, 5, …, 9: \( R_{s}^{2} ,G_{s}^{2} ,B_{s}^{2} , \) \( R_{s} G_{s} , \) \( B_{s} G_{s} , \) \( R_{s} B_{s} \), are normalized in the range 0−255. The vectors \( \vec{P}_{s} \) are then transformed by the 2-level HAKPCA, whose algorithm is shown in Fig. 1.17. As a result of the transform are obtained the 2-component vectors \( \vec{E}_{s} = [E_{1s} ,E_{2s} ]^{t} \), which are used to substitute the input 9-components vectors \( \vec{P}_{s} = [P_{1s} ,P_{2s} ,P_{3s} ,P_{4s} ,P_{5s} ,P_{6s} ,P_{7s} ,P_{8s} ,P_{9s} ]^{t} \). In this way the performance of the classifier is also simplified, because it has to process the vectors \( \vec{E}_{s} \) in the two-component, instead of the nine-dimensional space.
At its output are separated all pixels in the image, whose corresponding vectors \( \vec{E}_{s} \) are in the area of the cluster, belonging to the object. With this, the color segmentation is finished. In accordance with the algorithm shown in Fig. 1.17, for the 2-level HAKPCA [91], the nine components of each input vector \( \vec{P}_{s} \) are divided into three groups, which contain the three-components vectors
At the first level of HAKPCA, on each group of the three-component vectors \( \vec{P}_{ks} = {\kern 1pt} {\kern 1pt} [P_{k1s} ,P_{k2s} ,P_{k3s} ]^{t} \) for k = 1, 2, 3, is performed color APCA with a transform matrix of size 3 × 3. The so obtained vectors from each group comprise three “eigen” images, shown in Fig. 1.18. These images are rearranged in accordance to the rule:
where \( \lambda_{i} \ge 0 \) for l = 1, 2, …, 9 are eigen values of the covariance matrices of the three-component vectors \( \vec{P}_{ks} \) for each group (k = 1, 2, 3) in the first level of HAКPCA, arranged as monotonously decreasing sequence of eigen values. After that these components are divided again, this time into 3 groups, of 3 images each.
The vectors, obtained from the pixels with same coordinates in the images from each group, are of 3 components. For the second level of HAKPCA for each group of 3-component vectors is executed direct APCA with a transform matrix of size 3 × 3.
The algorithm for direct/inverse transform of the components [R], [G], [B] of the color image, calculated using APCA in three “eigen” images [L 1], [L 2], [L 3], is shown on Fig. 1.19. The vectors from each group build the three eigen images, which are rearranged again in accordance with Eq. (1.25). As a result, the nine eigen images E 1 ~ E 9 are obtained, from which are retained the first two (E 1 and E 2) only, which carry the main information, needed for the color objects segmentation.
As a result, the computational complexity of HAKPCA is lower than that of the KPCA, for the case, when it is used to transform directly the 9-component vectors \( \vec{P}_{s} \). In this way, the general computational complexity of HAKPCA with a classifier, needed for the processing of the vectors \( \vec{P}_{s} \) is lower than that, needed for the processing of same vectors with KPCA with a classifier. From the pixels with same coordinates in the images E 1 and E 2 are obtained the vectors \( \vec{E}_{s} = {\mathbf{[}}E_{1s} ,\,E_{2s} {\mathbf{]}}^{t} , \) which are then used by the classifier.
1.6.3 Experimental Results
To verify the feasibility of the proposed algorithm, skin pigmentation images were tested and evaluated. Figures 1.20 and 1.21 show the original tested images and their color vectors distribution in the RGB space, respectively. It can be seen that their color distributions are considered as non-linear Gaussian ones.
a, b Color vectors distribution in RGB space for original images in Fig. 1.20 a, b
These images are passed through the HAKPCA algorithm (shown on Fig. 1.17). The obtained transformed vectors \( \vec{E}_{s} \) in the new color space \( E_{1s} ,E_{2s} ,E_{3s} \) are plotted in the 3D domain shown in Fig. 1.22. It is easy to notice that the proposed techniques concentrate the energy of the different skin color into very small and close components of transformed vectors.
The HAKPCA transformed coefficients are then used to train a classifier. For briefing, fuzzy K-means clustering is used. The segmentation results are shown in Fig. 1.23a, b respectively.
The proposed approach depends mainly on the evaluation of the color vectors distribution. For a non-Gaussian distribution of the vectors, is used HAKPCA. The selected nonlinear transform results in negligible expansion of the original color space, which increases slightly the number of needed calculations. The main advantage of the new approach is that in result of its adaptation in respect to the color vectors distribution, it could be used as universal tool for efficient image processing. One more advantage of HAKPCA towards the KPCA is the lower computational complexity.
On the basis of the presented approach, new algorithm for objects color segmentation was developed, which was distinguished by its high accuracy. This algorithm could be used in the CBIR systems for extraction of objects with preset color, in the computer vision systems for detection and tracking of objects in correspondence to their color under changing surveillance conditions, for automatic control of various manufacturing processes, etc.
1.7 Conclusions
The new approaches for image decomposition, presented in this chapter, are distinguished from the well-known methods by their low computational complexity and high efficiency, which makes them very attractive for future applications in computer vision, video communications, image content protection through digital Watermarking [92], image search in large databases, etc. Besides, the methods for hierarchical decomposition could be also used for the creation of new hybrid algorithms [93, 94] for processing of some specific kinds of images (static, or sequences), of the kind: medical, multi- and hyper spectral, multiview, panoramic satellite photos, etc. Depending on the requirements of the applications, in respect of their efficiency and the computation time needed for the processing, for each image kind could be selected the most suitable from the four decompositions (BIDP, HSVD, HAPCA and HAKPCA) i.e., they can complement each other and have their own place and significance.
The future development of the presented new algorithms will be focused at the investigation of the possibilities for their integration in the contemporary systems for parallel processing, analysis and image recognition.
References
Rao, R., Kim, D., Hwang, J.: Fast Fourier Transform: Algorithms and Applications. Springer, Heidelberg (2010)
Gonzalez, R., Woods, R.: Digital Image Processing, 3rd edn. Prentice Hall, NJ (2008)
Do, M., Vetterli, M.: Contourlets: a directional multiresolution image representation. In: Proceedings of International Conference on Image Processing, 2002, vol. 1, pp. 357−360 (2002)
Kountchev, R., Vl, Todorov, Kountcheva, R.: Linear and non-linear inverse pyramidal image representation: algorithms and applications, Chapter. In: Kountchev, R., Nakamatsu, K. (eds.) Advances in Reasoning-based Image Processing, Analysis and Intelligent Systems, pp. 35–89. Springer, Berlin (2012)
Orfanidis, S.: SVD, PCA, KLT, CCA, and all that. In: Optimum Signal Processing, pp. 332−525. Rutgers University (2007)
Ahuja, N.: On approaches to polygonal decomposition for hierarchical image representation. Comput. Vis. Graph. Image Proc. 24, 200−214 (1983)
Tadmor, E., Nezzar, S., Vese, L.: Multiscale hierarchical decomposition of images with applications to deblurring, denoising and segmentation. Commun. Math. Sci. 6, 281–307 (2008)
Hidane, M., Lezoray, O., Ta, V., Elmoataz, A.: Nonlocal multiscale hierarchical decomposition on graphs. In: Daniilidis K, Maragos P, Paragios N (eds.) Computer Vision—ECCV 2010, Part IV, pp. 638–650, LNCS 6314. Springer, Berlin (2010)
Wu, Q., Xia, T., Yu, Y.: Hierarchical tensor approximation of multidimensional images. In: IEEE International Conference on Image Processing (ICIP’07), San Antonio, TX, 6, IV-49, IV-52 (2007)
Cherkashyn, V., He, D., Kountchev, R.: Image decomposition on the basis of an inverse pyramid with 3-layer neural networks. J. Commun. Comput. 6(11), 1–9 (2009)
Mach, T.: Eigenvalue algorithms for symmetric hierarchical matrices. Dissertation, Chemnitz University of Technology, March 13, 175p (2012)
Rudin, L., Osher, S., Fatemi, E.: Nonlinear total variation based noise removal algorithms. Physica D 60, 259–268 (1992)
Kountchev, R.: New Approaches in Intelligent Image Processing, 322p. WSEAS Press, Athene (2013)
Kountchev, R., Kountcheva, R.: Image representation with reduced spectrum pyramid. In: Tsihrintzis, G., Virvou, M., Howlett, R., Jain, L. (eds.) New Directions in Intelligent Interactive Multimedia, pp. 275−284. Springer, Berlin, Heidelberg (2008)
Kountchev, R., Kountcheva, R.: Compression of CT images with modified inverse pyramidal decomposition. In: 12th WSEAS International Conference on Signal Processing, Computational Geometry and Artificial Vision (ISCGAV’12), pp. 74–79, Istanbul, Turkey (2012)
Draganov, I., Kountchev, R., Georgieva, V.: Compression of CT images with branched inverse pyramidal decomposition. In: Iantovics, B., Kountchev, R. (eds.) Advanced Intelligent Computational Technologies and Decision Support Systems, vol. 486, VII, pp. 71–81. Springer (2014)
Kountchev, R., Todorov, V., Kountcheva, R.: New method for adaptive lossless compression of still images based on the histogram statistics. In: Tsihrintzis, G., Virvou, M., Jain, L., Howlett, R. (eds.) Intelligent Interactive Multimedia Systems and Services, Proceedings of the 4th International Conference on Intelligent Interactive Multimedia Systems and Services (KES-IIMSS 2011), pp. 61−70. Springer (2011)
Deprettere, E.: SVD and Signal Processing. Elsevier, New York (1988)
Vaccaro, R.: SVD and Signal Processing II. Elsevier, New York (1991)
Moonen, M., Moor, B.: SVD and Signal Processing III. Elsevier, New York (1995)
Carlen, E.: Calculus++. In: The Symmetric Eigenvalue Problem. Georgia Tech (2003)
Andrews, H., Patterson, C.: Singular value decompositions and digital image processing. IEEE Trans. Acoust. Speech Signal Process. ASSP-24, 26–53 (1976)
Gerbrands, J.: On the relationships between SVD KLT, and PCA, Pattern Recogn. 14(6), 375–381 (1981)
Kalman, D.: A singularly valuable decomposition: the SVD of a matrix. Coll. Math. J 27(1), 2–23 (1996)
Rehna, V., Jeyakumar, M.: Singular value decomposition based image coding for achieving additional compression to JPEG images. Int. J. Image Process. Vis. Sci. 1(2), 56–61 (2012)
Sadek, R.: SVD based image processing applications: state of the art, contributions and research challenges. Int. J. Adv. Comput. Sci. Appl. 3(7), 26–34 (2012)
Householder, A.: The Theory of Matrices in Numerical Analysis. Dover, New York (1975)
Diamantaras, K., Kung, S.: Principal Component Neural Networks. Wiley, New York (1996)
Levy, A., Lindenbaum, M.: Sequential Karhunen-Loeve basis extraction and its application to images. IEEE Trans. Image Process. 9(8), 1371–1374 (2000)
Drinea, E., Drineas, P., Huggins, P.: A randomized singular value decomposition algorithm for image processing applications. In: Manolopoulos, Y., Evripidou, S. (eds.) Proceedings of the 8th Panhellenic Conference on Informatics, pp. 278–288, Nicosia, Cyprus (2001)
Holmes, M., Gray, A., Isbell, C.: QUIC-SVD: fast SVD using cosine trees. In: Proceedings of NIPS, pp. 673–680 (2008)
Foster, B., Mahadevan, S., Wang, R.: A GPU-based approximate SVD algorithm. In: 9th International Conference on Parallel Processing and Applied Mathematics, 2011, LNCS 7203, pp. 569–578 (2012)
Yoshikawa, M., Gong, Y., Ashino, R., Vaillancourt, R.: Case study on SVD multiresolution analysis, CRM-3179, pp. 1–18 (2005)
Waldemar, P., Ramstad, T.: Hybrid KLT-SVD image compression. In: IEEE International Conference on Acoustics, Speech, and Signal Processing, pp. 2713–2716. IEEE Computer Society Press, Los Alamitos (1997)
Aharon, M., Elad, M., Bruckstein, A.: The K-SVD: an algorithm for designing of overcomplete dictionaries for sparse representation. IEEE Trans. Signal Process. 54, 4311–4322 (2006)
Singh, S., Kumar, S.: Singular value decomposition based sub-band decomposition and multi-resolution (SVD-SBD-MRR) representation of digital colour images. Pertanika J Sci. Technol. (JST) 19(2), 229–235 (2011)
De Lathauwer, L., De Moor, B., Vandewalle, J.: A multilinear singular value decomposition. SIAM J. Matrix Anal. Appl. 21, 1253–1278 (2000)
Kountchev, R., Kountcheva, R.: Hierarchical SVD-based image decomposition with tree structure. Int. J. Reasoning-Based Intell. Syst. (IJRIS) 7(1/2), 114–129 (2015)
Kountchev, R., Nakamatsu, K.: One approach for grayscale image decorrelation with adaptive multi-level 2D KLT. In: Graña, M., Toro, C., Posada, J., Howlett, R., Jain, L. (eds.) Advances in Knowledge-Based and Intelligent Information and Engineering Systems, pp. 1303–1312. IOS Press (2012)
Fieguth, P.: Statistical image processing and multidimensional modeling. Springer, Science+Business Media (2011)
Reed, T.: Digital Image Sequence Processing, Compression, and Analysis. CRC Press (2004)
Shi, Y., Sun, H.: Image and Video Compression for Multimedia Engineering: Fundamentals, Algorithms, and Standards, 2nd edn. CRC Press, Taylor & Francis Group, LLC (2008)
Mukhopadhyay, J.: Image and Video Processing in the Compressed Domain. CRC Press (2011)
Hyvarinen, A., Hurri, J., Hoyer, P.: Natural Image Statistics, A Probabilistic Approach to Early Computational Vision. Springer (2009)
Jolliffe, I.: Principal Component Analysis, 2nd edn. Springer-Verlag, NY (2002)
Dony, R.: Karhunen-Loeve Transform. In: Rao, K., Yip, P. (eds.) The Transform and Data Compression Handbook. CRC Press, Boca Raton (2001)
Fleury, M., Dowton, A., Clark, A.: Karhunen-Loeve Transform—Image Processing. University of Essex, Wivenhoe (1997)
Miranda, A., Borgne, Y., Bontempi, G.: New routes from minimal approximation error to principal components, pp. 1−14. Kluwer Academic Publishers (2007)
Landqvist, R., Mohammed, A.: Comparative performance analysis of three algorithms for principal component analysis. Radioengineering 15(4), 84–90 (2006)
Tipping, M., Bishop, C.: Probabilistic principal component analysis. J. Roy. Stat. Soc. B, 61(3), 611−622 (1999)
Liwicki, S., Tzimiropoulos, G., Zafeiriou, S., Pantic, M.: Euler principal component analysis. Int. J. Comput. Vis. 101(3), 498–518 (2013)
Ujwala, P., Uma, M.: Image fusion using hierarchical PCA. In: International Conference on Image Information Processing (ICIIP’11). pp. 1–6 (2011)
Abadpour, A., Kasaei, S.: Color PCA eigen images and their application to compression and watermarking. Image Video Comput. 26, 878–890 (2008)
Chadha, A., Satam, N., Sood, R., Bade, D.: Image steganography using Karhunen-Loève transform and least bit substitution. Int. J. Comput. Appl. 79(9), 31–37 (2013)
Naik, G., Kumar, D.: An overview of independent component analysis and its applications. Informatica 35, 63–81 (2011)
Press, W., Teukolsky, S., Vetterling, W.: Numerical Recipes in C, The Art of Scientific Computing, 2nd edn. Cambridge University Press (2001)
Erdogmus, D., Rao, Y., Peddaneni, H., Hegde, A., Principe, J.: Recursive PCA using eigenvector matrix perturbation. EURASIP J. Adv. Signal Process. 13, 2034–2041 (2004)
Hanafi, M., Kohler, A., Qannari, E.: Shedding new light on hierarchical principal component analysis. J. Chemom. 24(11–12), 703–709 (2010)
Amar, A., Leshem, A., Gastpar, M.: Recursive implementation of the distributed Karhunen-Loève transform. IEEE Trans. Signal Process. 58(10), 5320–5330 (2010)
Thirumalai, V.: Distributed compressed representation of correlated image sets. Thesis no 5264, Lausanne, EPFL (2012)
Saghri, J., Schroeder, S., Tescher, A.: An adaptive two-stage KLT scheme for spectral decorrelation in hyperspectral bandwidth compression. SPIE Proc. 7443, 72–84 (2009)
Wongsawat, Y., Oraintara, S., Rao, K.: Integer sub-optimal Karhunen Loeve transform for multi-channel lossless EEG Compression. In: 14th European Signal Processing Conference (EUSIPCO’06), Florence, Italy (2006)
Blanes, I., Sagristà, J., Marcellin, M., Rapesta, J.: Divide-and-conquer strategies for hyperspectral image processing. IEEE Signal Process. Mag. 29(3), 71–81 (2012)
Blanes, I., Sagristà, J.: Pairwise orthogonal transform for spectral image coding. IEEE Trans. Geosci. Remote Sens. 49(3), 961–972 (2011)
Ma, Z.: Sparse principal component analysis and iterative thresholding. Ann. Stat. 41(2), 772–801 (2013)
Jain, A.: A fast Karhunen-Loeve transform for a class of random processes. IEEE Trans. Commun. COM-24, 1023–1029 (1976)
Rokhlin, V., Szlam, A., Tygert, M.: A randomized algorithm for principal component analysis. SIAM J. Matrix Anal. Appl. 31, 1100–1124 (2009)
Kambhatla, N., Haykin, S., Dony, R.: Image compression using KLT, wavelets and an adaptive mixture of principal components model. J. VLSI Signal Process. 18, 287–296 (1998)
Bita, I., Barret, M., Pham, D.: On optimal transforms in lossy compression of multi-component images with JPEG2000. Sig. Process. 90(3), 759–773 (2010)
Kountchev, R., Kountcheva, R.: PCA-based adaptive hierarchical transform for correlated image groups. In Proceedings of the International Conference on Telecommunications in Modern Satellite, Cable and Broadcasting Services (TELSIKS’13), pp. 323−332. IEEE, Serbia (2013)
Kountchev, R.: Applications of the hierarchical adaptive PCA for processing of medical CT images. Egypt. Comput. Sci. J. 37(3), 1–25 (2013)
Kountchev, R., Kountcheva, R.: Decorrelation of multispectral images, based on hierarchical adaptive PCA. Int. J. WSEAS Trans. Signal Process. 3(9), 120–137 (2013)
Kountchev, R., Kountcheva, R.: Hierarchical adaptive KL-based transform: algorithms and applications. In: Favorskaya, M., Jain, L. (eds.) Computer Vision in Advanced Control Systems: Mathematical Theory, pp. 91−136. Springer, Heidelberg (2015)
Arora, S., Barak, B.: Computational complexity: A modern approach. Cambridge University Press (2009)
Jie, X., Fei, S.: Natural color image segmentation. In: Proceedings of IEEE IC on Image Processing (ICIP’03), pp. 973–976, Barcelona, Spain (2003)
Navon, E., Miller, O., Averabuch, A.: Color image segmentation based on adaptive local thresholds. Image Vis. Comput. 23, 69–85 (2005)
Deshmukh, K., Shinde, G.: An adaptive color image segmentation. Electron. Lett. Comput. Vis. Image Anal. 5(4), 12–23 (2005)
Yu, Z., Wong, H., Wen, G.: A modified support vector machine and its application to image segmentation. Image Vis. Comput. 29, 29–40 (2011)
Wang, X., Wang, T., Bu, J.: Color image segmentation using pixel wise support vector machine classification. Pattern Recogn. 44, 777–787 (2011)
Bhoyar, K., Kakde, O.: Skin color detection model using neural networks and its performance evaluation. J. Comput. Sci. 6(9), 963–968 (2010)
Stern, H., Efros, B.: Adaptive color space switching for face tracking in multi-colored lighting environments. In: Proceedings of the International Conference on Automatic Face and Gesture Recognition, pp. 249–255 (2002)
Kakumanu, P., Makrogiannis, S., Bourbakis, N.: A survey of skin-color modeling and detection methods. Pattern Recogn. 40, 1106–1122 (2007)
Ionita, M., Corcoran, P.: Benefits of using decorrelated color information for face segmentation/tracking. Adv. Optical Technol. 2008(583687) (2008)
Hassanpour, R., Shahbahrami, A., Wong, S.: Adaptive Gaussian mixture model for skin color segmentation. World Acad. Sci. Eng. Technol. 41, 1–6 (2008)
Hikal, N., Kountchev, R.: Skin color segmentation using adaptive PCA and modified elliptic boundary model. In: International Proceedings of the IEEE International Conference on Advanced Computer Science and Information Systems (IEEE ICACSIS’11), pp. 407–412, Jakarta, Universitas Indonesia (2011)
Abadpour, A., Kasaei, S.: Principal color and its application to color image segmentation. Scientia Iranica 15(2), 238–245 (2008)
Kountchev, R., Kountcheva, R.: Image color space transform with enhanced KLT. In: Nakamatsu, K., Wren, G., Jain, L., Howlett, R. (eds.) New Advances in Intelligent Decision Technologies, pp. 171−182. Springer-Verlag (2009)
Gunter, S., Schraudolph, N., Vishwanathan, S.: Fast iterative kernel principal component analysis. J. Mach. Learn. Res. 8, 1893–1918 (2007)
Scholkopf, B., Smola, A., Muller, K.: Kernel principal component analysis. In: Scholkopf, B., Burges, C., Smola, A. (eds.) Advances in Kernel Methods—Support Vector Learning, pp. 327–352. MIT Press, Cambridge, MA (1999)
Lee, J., Verleysen, M.: Nonlinear Dimensionality Reduction. Springer (2007)
Kountchev, R., Hikal, N., Kountcheva, R.: A new approach for color image segmentation with hierarchical adaptive kernel PCA. In: 18th International Conference on Circuits, Systems, Communications and Computers (CSCC’14), Proceedings of Advances in Information Science and Applications, pp. 38–43, Santorini Island, I (2014)
Kountchev, R., Milanova, M., Todorov, V., Kountcheva, R.: Image watermarking based on pyramid decomposition with CH transform, In: Chan, Y., Talburt, J., Talley, T. (eds.) Data Engineering: Mining, Information, and Intelligence, pp. 353–387. Springer (2010)
Draganov, I.: Inverse pyramid decomposition of wavelet spectrum for image compression. Int. J. Reasoning-based Intell. Syst. (IJRIS) 6(1/2), 19–23 (2014)
Martino, F., Loia, V., Sessa, S.: A fuzzy hybrid method for image decomposition. Int. J. Reasoning-based Intell. Syst. (IJRIS) 1(1/2), 77–84 (2009)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Kountchev, R., Kountcheva, R. (2016). New Approaches for Hierarchical Image Decomposition, Based on IDP, SVD, PCA and KPCA. In: Kountchev, R., Nakamatsu, K. (eds) New Approaches in Intelligent Image Analysis. Intelligent Systems Reference Library, vol 108. Springer, Cham. https://doi.org/10.1007/978-3-319-32192-9_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-32192-9_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-32190-5
Online ISBN: 978-3-319-32192-9
eBook Packages: EngineeringEngineering (R0)