Abstract
Research involving the recovery of viruses from various water sources can be traced back to the 1950s when scientists were eager to better understand the occurrence and transmission of disease-causing viruses in water; most significant viruses at the time were poliovirus and hepatitis virus. LoGrippo and Berger (1952) utilized ion-exchange resins to concentrate poliovirus, and shortly thereafter Kelly (1953) demonstrated the use of these same resins for the concentration of coxsackie viruses from sewage samples. This seminal research led to development of the first methods that relied on manipulation of virus surface charge for recovering viruses from large volumes of water. Virus adsorption to filter media followed by elution by a pH-adjusted solution (Wallis and Melnick 1967) became collectively known as VIRADEL (virus adsorption elution) method. Shortly after VIRADEL methods were introduced, alternative methods to concentrate viruses based on size exclusion (i.e. ultrafiltration based on nominal molecular weight cut-off) were investigated. Belfort et al. (1975) concentrated viruses from 5 L of water using hollow fiber membranes and from this initial report through the present day, numerous studies have shown hollow-fiber ultrafiltration is effective for the recovery of viruses from water (Fong and Lipp 2005; Gensberger and Kostić 2013; Hill et al. 2007).
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Foodborne viruses
- Waterborne viruses
- Water
- Recovery methods
- VIRADEL
- Charged membrane filtration
- Ultrafiltration
- Virus concentration methods
1 Introduction
Research involving the recovery of viruses from various water sources can be traced back to the 1950s when scientists were eager to better understand the occurrence and transmission of disease-causing viruses in water; most significant viruses at the time were poliovirus and hepatitis virus. LoGrippo and Berger (1952) utilized ion-exchange resins to concentrate poliovirus, and shortly thereafter Kelly (1953) demonstrated the use of these same resins for the concentration of coxsackie viruses from sewage samples. This seminal research led to development of the first methods that relied on manipulation of virus surface charge for recovering viruses from large volumes of water. Virus adsorption to filter media followed by elution by a pH-adjusted solution (Wallis and Melnick 1967) became collectively known as VIRADEL (virus adsorption elution) method. Shortly after VIRADEL methods were introduced, alternative methods to concentrate viruses based on size exclusion (i.e. ultrafiltration based on nominal molecular weight cut-off) were investigated. Belfort et al. (1975) concentrated viruses from 5 L of water using hollow fiber membranes and from this initial report through the present day, numerous studies have shown hollow-fiber ultrafiltration is effective for the recovery of viruses from water (Fong and Lipp 2005; Gensberger and Kostić 2013; Hill et al. 2007).
At this point, two questions arise: (1) How are viruses in water related to viruses in food and (2) Why is it important? Simply stated, there is an intimate connection between food safety and water quality as water is used at almost every node in the food production process. Moreover, viruses are the primary cause of foodborne disease outbreaks (FBDO ) in the United States and in most other high-income countries (EFSA BIOHAZ Panel 2011; Lopman et al. 2003; Scallan et al. 2011). From an epidemiological perspective, human noroviruses (HuNoVs) are the most significant in foodborne transmission though other viruses of concern include hepatitis A virus (HAV), enteroviruses, and hepatitis E virus (HEV; Le Guyadar et al. 2008; Yugo and Meng 2013).
The most commonly reported connections between water quality and food safety are viruses in fresh produce and shellfish (i.e. bivalve mollusks such as oysters) (see Chaps. 6 and 7 in this book). For these foods, large volumes of water must be used during pre- and post-harvest processing. The ongoing conversation in shellfish production revolves around the relevance of using fecal indicator bacteria (Escherichia coli, Enterococcus spp.) as predictors of viral pathogen contamination when monitoring the quality of harvesting waters or shellfish meats (Richards et al. 2010). The issue arises because an indicator should correlate with the presence of pathogens; however, bacterial indicators have been repeatedly shown to correlate poorly with viruses (Field and Samadpour 2007). Fresh produce (i.e. fruits and vegetables) can become contaminated with waterborne bacteria and viruses during production via irrigation water, preparing and spraying pesticides, preventing dehydration, or during produce washing or cooling. The proposed agricultural water standards in the US FDA’s proposed rule for produce safety rely on E. coli to indicate sanitary quality of water (USFDA 2014). Thus, the same limitations would apply regarding the often observed absence of a relationship between bacterial indicators and human pathogenic viruses.
Human enteric viruses may be introduced into the water environment through various routes including discharge of sewage-contaminated water into food production settings (i.e. oyster harvesting waters and surface water used for irrigation), land application of municipal biosolids, wastewater, or septage with subsequent runoff, and groundwater contaminated by infiltrating surface water, faulty septic systems or leaking sanitary sewers (Gibson 2014). Overall, data are limited on virus occurrence in water used for food production even though the industry’s heavy reliance on water suggest it is a vehicle for dissemination of viruses in the food supply (Song et al. 2006; Stine et al. 2005).
There are several options for recovering viruses from water, although each method brings along its own set of challenges, primarily stemming from the low but significant levels of human enteric viruses that may be present in environmental water samples at sites located around the world (Dong et al. 2010; Gibson et al. 2012; Sinclair et al. 2009; World Health Organization 2009; Ye et al. 2012).
2 Virus Recovery Methods
Wallis et al. (1979) originally prescribed the ideal method for virus recovery as being able to: (1) process large volumes (>100 L) of various types of water in the least amount of time; (2) consistently concentrate most types of viruses present in water and wastewater, (3) be used easily and economically, and (4) recover viruses that are particle-associated or present in viral aggregates. In the subsections that follow, various methods are described for recovering and concentrating viruses from water.
2.1 VIRADEL
The first generation of VIRADEL methods was based on adsorption of viruses to electronegative (EN) membrane filters (Sobsey et al. 1973). However, because viruses have a net negative charge on their capsid surface at neutral pH, to facilitate virus adsorption to the negatively-charged filters, EN-based methods require cumbersome chemical modification of the water sample including acidification or addition of multivalent cations (Sobsey and Jones 1979). Shortly thereafter, Sobsey and Jones (1979) suggested that membrane filters with a more positive charge at the pH of natural waters and tap water (pH 5–9) would be more advantageous for virus recovery compared to EN-based methods. This initial investigation of electropositive (EP)-based membrane filter methods by Sobsey and Jones (1979) demonstrated a marked increase in virus recovery from tap water (e.g., <5 % recovery with EN filter vs. 64 % recovery with EP filter) and served as the basis for developing the standard method for virus recovery and concentration from large volumes of water (USEPA 2001).
Wallis et al. (1979) identified several factors influencing the adsorption of viruses to filter surfaces : filter surface charge and surface area resulting from the composition and design of the filter membrane; flow rate during sampling (i.e. high flow rate equates to less viruses adsorbed); ratio of pore diameter to virus diameter; pH; multivalent cations; and the presence of proteinaceous substances competing with viruses for adsorption. Filter surface charge plays a significant role in virus adsorption as demonstrated by the difference in viral recovery between EN- and EP-based methods. In addition, virus adsorption capacity decreases with decreasing filter surface area because there are a finite number of adsorption sites per unit area (Wallis et al. 1979). However, additions of acids (for pH manipulation) and salts (multivalent cations) can greatly increase filter adsorption capacity regardless of filter surface area; therefore, the virus adsorption capacity of filters is really never exceeded (Wallis et al. 1979). The impact of multivalent cations on filter charge is one of the primary reasons why EN-based methods outperform EP-based methods for the recovery of viruses from marine waters (Katayama et al. 2002; Lukasik et al. 2000). Overall, the net charge of the filter is one of the primary determining factors in virus adsorption.
Present day, researchers still use both EN- and EP-based methods for recovery of viruses from water though far more emphasis has been placed on EP-based methods. Table 10.1 summarizes the VIRADEL methods that are most commonly utilized for recovery of viruses, and Table 10.2 provides representative recovery efficiencies for each VIRADEL method by water type. For the EP-based methods, beef extract—glycine solutions at an alkaline pH is most often used for virus elution from the filter membrane (Table 10.1). However, other eluents have previously been used including 5× nutrient broth (Hill et al. 1974), tryptose phosphate broth (Farrah et al. 1976), and amino acids—arginine and lysine (Farrah and Bitton 1978). The type of eluent is important to note as organic and inorganic compounds in beef extract solutions are known to inhibit reverse transcription (RT) for cDNA synthesis from viral RNA as well as PCR amplification (Abbaszadegan et al. 1993). This is important because many waterborne viruses are not readily culturable requiring downstream detection methods that rely on RT and PCR technologies.
After filtration with EN-based methods the filter is rinsed with a dilute acid followed by elution with alkaline buffer (pH > 9.0) containing either high salt concentration and surfactant (Hamza et al. 2009) or proteinaceous substances such as skimmed milk (Wyn-Jones et al. 2011). De Keuckelaere et al. (2013) compared four methods, two EN-based, one EP-based, and one using tangential flow filtration for recovery of five types of human enteric viruses from four irrigation water sources and processing water from a fresh cut lettuce processing plant. The investigators concluded the EN-based method including virus elution with high salt concentration buffer provided the highest virus recoveries across the range of water types tested. However, De Keuckelaere et al. (2013) reported much lower recovery efficiency for three of the four methods evaluated as compared to previously published reports.
A novel filter-less method was reported by Calgua et al. (2008) where 5–10 L volumes of seawater or freshwater amended with artificial sea salts are flocculated with 1 % (w/v) skimmed milk solution at pH 3.5. Samples are stirred for 8 h, the floc is allowed to settle for another 8 h, and then centrifuged. Mean virus recoveries from freshwater were in the range of 40–50 % (Calgua et al. 2013). The EN-based methods and skimmed milk method may be considered advantageous when compared to EP-based methods and ultrafiltration based on the volume of water needed. For instance, only 2–10 L (Fong et al. 2005; Lee and Kim 2008; Hamza et al. 2011) of a given water sample can be concentrated to detect viruses in the former methods; however, given the same water sample, hundreds of liters may need to be concentrated by EP-based and ultrafiltration to detect the viruses present. Simultaneous concentration of PCR inhibitors and particulates during large volume filtration is speculated to offset the benefits of concentrating more viruses (Albinana-Gimenez et al. 2009; De Keuckelaere et al. 2013), but why small sample volumes appear to provide the same limits of virus detection has not been systematically investigated. Confounder effects , such as correlation between the method selected for sampling and virus concentrations in the waters being sampled, or between sampling method and the use of nested PCR for virus detection, are possible considerations.
The EP-based filtration method described by Sobsey and Jones (1979) served as the basis for USEPA to prescribe using the positively-charged 1MDS filter for virus concentration from water for the Information Collection Rule (USEPA 2001). Although effective (Table 10.2), the 1MDS method has been criticized for being cost prohibitive for routine monitoring purposes—each filter cartridge costs approximately $200–300 (Cashdollar and Wymer 2013). In 2010, USEPA published Method 1615, a revised method for detecting enteroviruses and noroviruses in water that permits virus concentration by the positively charged NanoCeram® filter, which cost roughly 20 % ($40–60) of the cost of the 1MDS filter and is equally as effective for these two viruses (Fout et al. 2010). The Nanoceram filter does not appear to be as effective for concentrating adenoviruses (Gibbons et al. 2010; Ikner et al. 2011; Pang et al. 2012; McMinn 2013).
Glass wool filtration is another effective EP-based method (Table 10.1). Oiled sodocalcic glass wool is washed in series with 1 N HCl, distilled water, 1 N NaOH, and a final wash with distilled water until a neutral pH is achieved. The washed glass wool can be stored in sterile phosphate buffered saline for up to 1 week at 4 °C. Filters are made by packing washed glass wool tightly into column housings with size and fittings appropriate for the virus sampling plan at hand (Vilagines et al. 1993; Millen et al. 2012). Filter construction configurations are highly flexible for specific projects and can be used to filter large volumes of water (Gibson et al. 2012; Lambertini et al. 2008; Vilagines et al. 1997). Similar to other EP-based filter cartridges, glass wool requires the use of an alkaline eluent such as a beef extract—glycine solution. Glass wool filters are a fraction of the cost—$4.40 per filter (not including labor)—of NanoCeram® and 1MDS filters (Lambertini et al. 2008) and are demonstrated to achieve similar virus recovery efficiencies (Table 10.2). The primary disadvantage of glass wool filters is that they are not commercially available, lending to the perception that hand-packing in the laboratory may lead to filter-to-filter variability in virus recovery efficiency (Cashdollar and Wymer 2013). Even then, the United Kingdom Environment Agency utilizes the glass wool filtration technique in standard methods for the recovery and concentration of viruses in groundwater (Environment Agency 2000).
As described in USEPA Method 1615, the basic setup and steps for recovering viruses from tap water using an EP-based cartridge filtration method are shown in Figs. 10.1 and 10.2, respectively. Overall, when using EP filters such as NanoCeram®, 1MDS, or glass wool, the steps taken to recover viruses from a given water sample will depend on the need to neutralize chlorine residual disinfectant, adjust pH to the optimal level for virus adsorption, and add a pre-filter for highly turbid samples.
Virus Adsorption—Elution (VIRADEL) filtration method procedure. aIf the water sample has a disinfectant present based on measured chlorine residual, then it must be dechlorinated with 2 % sodium thiosulfate using the optional injector module (Fig. 10.1) prior to filtration. bIf the water sample has a pH >9.0 or >8.0 for NanoCeram® and 1MDS Zetapor® filter cartridges, respectively, then the pH must be adjusted with 0.12 M HCl until it measures 6.5–7.5. c If the water sample turbidity is >20 NTU or >50 NTU for NanoCeram® and 1MDS Zetapor® filter cartridges, respectively, then a 10-μm polypropylene prefilter cartridge should be added prior to filtration but after dechlorination and pH adjustment. dThe volume of water filtered depends on the water source and flow rate (Fout et al. 2010) eVarious secondary concentration steps may be applied as described in Sect. 2.3 of this Chapter.
2.2 Hollow Fiber Ultrafiltration
Hollow fiber ultrafiltration (HFUF) for virus concentration and recovery from water was initially investigated by Belfort et al. (1975). There was a renewed interest in this method a decade ago for its potential to concentrate not only viruses but also other classes of microorganisms. Ultrafiltration methods rely on size exclusion for virus concentration rather than adsorption and hence difficulties associated with adsorbing viruses to various filter media are avoided. Ultrafilters are rated by molecular weight cut-off (MWCO) ranging from 1 to 300 kilodaltons (kDa), corresponding to a nominal pore size between 5 and 35 nm (Pall Corporation 2015). There are many different shapes of ultrafilter membranes available including spiral-wound, flat sheets, flat discs, and hollow fibers; the latter is the most commonly used shape for waterborne virus concentration. The shape of the filter impacts the membrane surface area available for filtration. Most hollow fiber membrane filters used for recovery of viruses range from 1.3 to 2.5 m2 (Table 10.2). Ultrafiltration membranes are constructed from a variety of polymers such as polysulfone, polyacrylonitrile, and cellulose triacetate and can be used in two different modes; tangential (cross) flow (TF) and direct (dead-end) flow (DE).
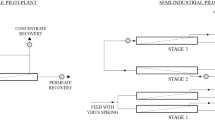
In TF filtration , a portion of the water (i.e. “feed” water) recirculates back to a central reservoir while the remaining portion passes through the membrane. The feed water is continuously recirculated until only a concentrate remains. Conversely, direct flow filtration pushes water through the membrane without recirculation, thus increasing the likelihood of membrane fouling, or the loss of membrane permeability that occurs due to accumulation of aquatic substances on or inside the membrane (Huang et al. 2008). During TF-HFUF, an applied pressure serves to force a portion of the feed water through the membrane to the filtrate side while viruses are concentrated in the retentate as opposed to being filtered out (Fig. 10.3). In TF, the retained components do not build up at the surface of the membrane but instead are swept along by the cross directional flow, dramatically reducing fouling and decreasing the tendency for microbes to adhere to the filter surfaces (Huang et al. 2008; Morales-Morales et al. 2003).
In DE filtration , water is not recirculated but flows directly through the filter, retaining viruses and other particulates greater in size than the MWCO. Ultrafilters are attached directly to a pressurized water source (e.g. drinking water distribution system), or in unpressurized systems (e.g. irrigation canal), a peristaltic pump is used. Input pressure less than 20 lb/in2 is recommended (Mull and Hill 2012; Smith and Hill 2009). Dead-end HFUF may be advantageous in field settings where set-up of TF-HFUF equipment can be cumbersome and filter clogging is not problematic as in low turbidity water.
Belfort et al. (1975, 1983, 1985) initially investigated ultrafiltration for recovery of viruses in water in the mid-1970s to 1980s and interest was renewed in the 1990s with the potential ultrafiltration afforded for recovering multiple classes of microorganisms (pathogenic viruses, bacteria, and protozoa) with a single sampling method. Kfir et al. (1995) first demonstrated the simultaneous recovery of viruses and protozoa from 10 L water samples using 50 kDa MWCO ultrafiltration disc membranes. Moreover, the study showed that UF provided higher, more consistent recovery of enteric viruses (e.g., 63–100 % with an average of 82 %) when compared to traditional VIRADEL methods.
Additional studies by Morales-Morales et al. (2003) and Hill et al. (2005) switched the focus to hollow fiber ultrafilters operated in tangential flow for the recovery and concentration of viruses in water. In Morales-Morales et al. (2003), two different hollow fiber ultrafilters with 50 kDa MWCOs were evaluated to recover microorganisms in 2 and 10 L water samples (tap water, groundwater, and surface water). Reported recoveries for PP7 and T1 bacteriophage were 31–74 % for 10 L surface water samples (Morales-Morales et al. 2003). Similarly, Hill et al. (2005) described a TF-HFUF method for the simultaneous recovery of microbes in tap water using commercially available dialysis filters with hollow fiber membranes (Table 10.3). The process was optimized through addition of (1) sample amendments such as sodium polyphosphate (NaPP); (2) an alternative membrane blocking procedure (e.g., NaPP solution as opposed to calf serum as described in Morales-Morales et al. (2003)); and (3) a backwashing step with a solution containing Tween 80 (Hill et al. 2005). Polyphosphates increase the negative surface charge of microbes suspended in water, creating surface charge repulsion. Tween 80 serves as a surfactant for reducing hydrophobic interactions between the filter surface and microbes, reducing adsorption of microbes to the hollow fiber membrane surface. Average recovery efficiencies for viruses using the optimized TF-HFUF method were 91 and 49 % for MS2 bacteriophage and echovirus 1, respectively (Hill et al. 2005).
Shortly thereafter, Hill et al. (2007) published another study on the concentration of viruses from 100 L samples of tap water. This study is the foundation for the development and optimization of additional TF-HFUF methods for concentration of viruses from 100 L water samples (Polaczyk et al. 2008; Hill et al. 2009; Francy et al. 2009; Gibson and Schwab 2011; Rhodes et al. 2011; Liu et al. 2012; Wu et al. 2013). Based on these studies, the basic steps for recovering viruses and other classes of microbes from water using HFUF are shown in Fig. 10.4, and the configuration for TF-HFUF is shown in Fig. 10.5a.
Hollow fiber ultrafiltration (HFUF) method steps. UF ultrafilter. aThe ultrafilter may be blocked to reduce the potential for virus adsorption to the membranes; however, the need for blocking may depend on the brand of ultrafilter and water type (Liu et al. 2012). Blocking procedures may include (1) overnight incubation with 5 % calf serum (Hill et al. 2005); (2) recirculation of 500 ml 5 % calf serum for 5 min through the UF; (3) recirculation of 500 ml of blocking solution containing 0.055 % Tween 80, 0.001 % Antifoam A, and 0.1 % NaPP through the ultrafilter for 5 min (USEPA and CDC 2011); or (4) filtration of 1 L 0.1 % NaPP through the UF at 1.7 L/min with no back pressure (Hill et al. 2005). bSample may be amended with sodium polyphosphate (NaPP) to a final concentration of 0.01 % prior to beginning filtration. cFollowing filtration, the ultrafilter can be eluted, or back flushed in the case of DE-HFUF, to capture any viruses that may be bound to the membrane. Elution buffers are recirculated within the TF-HFUF system and may include a solution containing 0.001 % Tween 80 or a solution containing 0.01 % Tween 80, 0.01 % NaPP, or 0.0001 % Y-30 antifoam emulsion. On the other hand, backflushing buffers are pumped through the permeate/filtrate port and may include solutions containing slightly higher concentrations of components found in the elution buffers used in TF-HFUF, such as 0.5 % Tween 80, 0.01 % NaPP, and 0.001 % Y-30 antifoam emulsion. dVarious secondary concentration steps may be applied to the filter concentrate as described in Sect. 2.3 of this Chapter.
DE-HFUF methods gained attention due to the relative ease of field deployment compared to the TF-HFUF setup requirements. Olzewski et al. (2005) initially compared DE-HFUF to tangential flow ultrafilter cassettes (i.e. not hollow fiber dialyzers) and demonstrated similar recovery efficiencies (>65 %). However, high turbidity samples caused membrane fouling (or clogging) of the DE-HFUF (Olzewski et al. 2005). Smith and Hill (2009) demonstrated the recovery of diverse microbes including viruses from 100 L tap water and from mock surface water samples (i.e. tap water spiked with surface water to obtain a turbidity of 5 NTU) using DE-HFUF. The reported recovery efficiencies using DE-HFUF were similar to TF-HFUF (Table 10.4), making DE-HFUF an acceptable option for field-based sampling efforts. Additionally, Smith and Hill (2009) determined that Asahi Kasei REXEED 25S ultrafilters were better suited for use in DE-HFUF set-ups based on their hydraulic performance (i.e. ability to maintain an acceptable permeate flow rate at consistent, low system pressures) as compared to Exeltra Plus 210, F200NR, and REXEED 21S filters (Table 10.3).
Mull and Hill (2012) also investigated DE-HFUF for recovery of viruses from medium (approx. 50 NTU) to high (approx. 100 NTU) turbidity surface waters. There was a decrease in virus recovery with increasing turbidity levels; however, the recovery rates were still deemed acceptable (Table 10.4). Aside from differences in filtration configurations, DE-HFUF differs from TF-HFUF in the back flushing step. For DE-HFUF, an elution buffer is passed back through the permeate/filtrate port to recover viruses as opposed to recirculation of an elution buffer as done in TF-HFUF. The configuration for DE-HFUF based on Smith and Hill (2009) is shown in Fig. 10.5b and the basic steps are outlined in Fig. 10.4.
2.3 Secondary Concentration
Even though HFUF and VIRADEL methods can concentrate viruses from water by 200- and >4000-fold, respectively, downstream methods for detection of viruses often require a secondary concentration and purification step after the initial concentration step. This additional step is needed since organic compounds (mostly humic acids) and metallic ions are often adsorbed or concentrated along with the vi ruses during primary concentration. These compounds can have inhibitory effects on downstream detection methods such as RT-PCR and real time qPCR. The methods most commonly used for secondary concentration and purification can be roughly categorized into one of the following groups: flocculation (organic or chemical), polymer-based hydroextraction, and centrifugal ultrafiltration.
One common method for secondary concentration is organic flocculation of the filter eluate (eluent + viruses) obtained during VIRADEL methods. This concept was originally proposed by Katzenelson et al. (1976) and involves the acidification of the eluate to pH 3.5 to cause flocculation of proteins, thus trapping any viruses present. The suspended floc is then centrifuged to a pellet followed by resuspension in 30 ml of sodium phosphate buffer. Organic flocculation is the method currently prescribed by USEPA Method 1615 for the concentration and processing of waterborne viruses (Fout et al. 2010). Additional flocculation methods include lanthanum-based chemical flocculation (Zhang et al. 2013) and celite (diatomaceous earth) concentration (Dahling and Wright 1986; Fout et al. 2003; McMinn et al. 2012, 2013).
Other effective methods for secondary concentration of viruses from water include polyethylene glycol (PEG) hydroextraction (Ramia and Sattar 1979; Lewis and Metcalf 1988; Schwab et al. 1996; Huang et al. 2000; Lambertini et al. 2008; Lee et al. 2011) and centrifugal ultrafiltration devices (e.g., Centricon Plus-70) (Hill et al. 2007; Gibson and Schwab 2011; Ikner et al. 2011). Hydroextraction with PEG has been paired with both VIRADEL and HFUF primary concentration methods, whereas centrifugal filtration devices have primarily been used in conjunction with HFUF. As with the primary methods for recovery of viruses, all secondary concentration steps have both advantages and disadvantages including cost, reproducibility, and ease of use, etc.
3 Method Selection: What Is Important?
Despite differences in the details of elution and secondary concentration steps for various virus filters, the costs for reagents and disposables are about the same; approximately $5.00–10.00 per sample with the exception of secondary concentration by centrifugal UF devices which adds $30 or more per sample. Processing times, however, can differ greatly among the elution and secondary concentration procedures and depend also on the laboratory equipment and personnel available for processing filters in batches. Time necessary for clean-up and equipment sterilization must also be considered.
What, then, are the criteria for selecting a filtration method for virus sampling ? Several criteria are important. (1) If in addition to viruses, concentrating bacteria and protozoa is required, ultrafiltration and glass wool filtration are good choices because there is sufficient evidence to indicate that these methods effectively concentrate a variety of microbial targets. (2) Sample flow rate can be important when there are many sample sites, long travel times between sites, and limited field sampling personnel. The Nanoceram® filter at flow rate of 10 L/min is advantageous in that respect, requiring one third less time than DE-HFUF at 2.9 L/min. (3) For water samples with pH exceeding 7.5, if the standard approach of decreasing pH inline during sampling is not workable, glass wool filtration should be avoided. However, Nanoceram® filters are reported to be effective at pH values of up to pH 9.0. (4) Tangential flow ultrafiltration is not always practical in field settings, although the requirement for large sample volumes to be transported to the laboratory, might also be impractical. (5) DE-HFUF is prone to clogging and virus recovery has been shown to decrease with increases in turbidity. (6) Lastly, intangible factors such as experience and confidence with equipment and technical procedures should be considered when making a decision on selection of virus filter.
Regardless of filtration method, recovery controls are essential to demonstrate the chosen method does, in fact, concentrate the target microbes from the water being sampled. The general approach is to transport a test water volume (e.g., 10–20 L) to the laboratory, seed it with quantified target, filter and process the water, quantify the target in the final concentrate, and calculate the percent of target recovered. Another approach, more representative of field settings and typical for large sample volumes (hundreds of liters), is to filter the sample at the field site with a target volume minus 10 L, transport the remaining 10 L back to the laboratory, seed it with the target microbes, and finish filtration with the same filter from the field (Fout et al. 2010). A companion volume of unseeded test water should also be filtered in order to quantify indigenous target microbes present in the water. This quantity is then subtracted from the numerator of the recovery calculation.
In addition, when recovery is measured by qPCR, a third companion volume of unseeded test water should be filtered and processed identically as the seeded test water. The target microbial seed is then added to the final concentrate created from this third volume and quantified by qPCR . This quantified value is used as the denominator for calculating virus recovery. This step is crucial for accuracy because filtration and the downstream processing steps can affect qPCR inhibition or other water constituents that shift quantification cycle (Cq) values (Borchardt et al. 2013). Lambertini et al. (2008) observed that inhibition during qPCR was detected in the beef extract eluent from the sample spiked with microbes processed by glass wool filtration; however, qPCR inhibition was not detected in the mock beef extract eluent spiked with microbes indicating that constituents in the water concentrated during filtration contained inhibitors. In this instance, qPCR inhibition underestimated the quantity of microorganisms spiked in the water sample, resulting in an overestimate of virus recovery. Using six viruses, Calgua et al. (2013) demonstrated that percent recoveries varied depending on which step, and consequently which matrix, in the concentration process the viral seeds were quantified.
Ideally, a recovery control would be performed for each sample, but that is impractical in terms of cost and time. Water matrix type (i.e., the combined attributes of turbidity, pH, dissolved organic matter, inorganic compounds, divalent cations, etc.) is an important determinant of recovery for both VIRADEL and HFUF methods (Hill et al. 2007; Lambertini et al. 2008; Mull and Hill 2012; Wu et al. 2013). Insofar as the waters to be sampled are known to vary in their matrices, for example by location, season, or aquifer type, the number of recovery controls can be planned to correspond with these various matrices. However, this number of recovery controls can be impractical as well and ultimately the number of controls is decided in the context of the sampling plan goals and whether potentially false-negative samples can be tolerated. At a minimum, several recovery controls should be performed during the sampling period just to ensure the procedures in the laboratory are working, particularly if there is a change in reagents. For example, beef extract lots are reported to contribute to variation in virus recovery rates (Fout et al. 2010). Regardless of the number of recovery controls, these data should be reported along with the virus results for the unknown field samples.
4 Advantages of Virus Sampling
Human enteric viruses are the primary cause of foodborne disease outbreaks, and the food commodity most often implicated is fresh produce (i.e. leafy greens and berries) (Koopmans and Duizer 2004; Scallan et al. 2011). The common assumption is that fresh produce is contaminated with viruses at the point of preparation or service, instead of during production, harvest, and post-harvest steps (Berger et al. 2010; Hall et al. 2012). However, a limited number of studies are available on occurrence of viruses in agricultural water sources (van Zyl et al. 2006; Cheong et al. 2009; Kokkinos et al. 2012; Pachepsky et al. 2011). This knowledge gap stems, in part, from the regulatory emphasis of agricultural water standards that focus on the control and detection of bacteria, not viruses. In the US, the proposed rule for produce safety and standards for the sanitary quality of agricultural water sources is based on Escherichia coli density exceedance criteria established by the US EPA for recreational waters (USEPA 2012). These criteria were previously adopted by the California Leafy Greens Marketing Agreement (LGMA) and several other state produce safety programs (LGMA 2013; USFDA 2014).
The variety of agricultural water sources—groundwater wells, ponds, rivers, streams, irrigation ditches, municipal water, reclaimed (treated wastewater) water, and irrigated liquid manure, combined with the diversity of potential fecal contamination sources, makes it difficult to rely on a single indicator, like E. coli, to reflect the fate, transport, and occurrence of all potential pathogens in agricultural water resources. Several criteria define an effective indicator: (1) rapidly measurable, (2) representative of the pathogens of concern, and (3) fate and transport characteristics similar to pathogens (Brookes et al. 2005). Unfortunately, E. coli meets only one criterion, rapid measurement. Rarely is E. coli fully concordant with pathogenic virus levels (Payment and Franco 1993; Harwood et al. 2005; Payment and Locas 2011). An E. coli positive sample suggests the presence of fecal material and indirectly viruses. However, an E. coli-negative sample does not necessarily mean viruses are absent. Sampling for viruses provides the definitive advantage of knowing whether viruses are present without the risk of making incorrect inferences based on E. coli.
Virus sampling methods offer several other advantages for ensuring the sanitary quality of foods: (1) Methods capable of filtering large sample volumes (e.g., 1000 L) can help account for any spatial and temporal variability of viruses in the water source; (2) Concentrated samples can be stored frozen and analyzed later if necessary; (3) Continuous sampling instead of grab sampling is possible from low turbidity waters using cartridge filter methods like glass wool or hollow fiber ultrafilters; and (4) Some methods are effective in concentrating multiple classes of pathogens (e.g., protozoa, bacteria, and viruses) in one step.
5 Summary and Conclusions
Based on what has been presented here, it is clear that no single method may universally be recognized as superior or suitable for all monitoring scenarios or contamination events. The efficiency, performance consistency, robustness, complexity of the method, and cost are all factors that must be considered when selecting the appropriate method. Additionally, regardless of the method, performance characteristics must be continuously monitored to ensure recovery efficiency and consistency over time. Finally, there is evidence that applying sampling methods targeted for recovery of viruses and other microorganisms would be advantageous (i.e. when compared to traditional 100 ml grab sample methods for detection of fecal indicator bacteria) in the assessment of the sanitary quality of agricultural water sources.
References
Abbaszadegan M, Huber MS, Gerba CP, Pepper IL (1993) Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl Environ Microbiol 59:1318–1324
Albinana-Gimenez N, Clemente-Casares P, Cagua B, Huguet JM, Courtois S, Girones R (2009) Comparison of methods for concentrating human adenoviruses, polyomaviruses JC and noroviruses in source waters and drinking water using quantitative PCR. J Virol Methods 158:104–109
Belfort G, Rotem Y, Katzenelson E (1975) Virus concentration using hollow fiber membranes. Water Res 9:79–85
Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, Hand P, Frankel G (2010) Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol 12:2385–2397
Borchardt MA, Kieke BA Jr, Spencer SK (2013) Ranking filter methods for concentrating pathogens in lake water. Appl Environ Microbiol 79:5418–5419
Brookes JD, Hipsey MR, Burch MD, Regel RH, Linden LG, Ferguson CM, Antenucci JP (2005) Relative value of surrogate indicators for detecting pathogens in lakes and reservoirs. Environ Sci Technol 39:8614–8621
Calgua B, Mengewein A, Grunert A, Bofill-Mas S, Clemente-Casares P, Hundesa A, Wyn-Jones AP, López-Pila JM, Girones R (2008) Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. J Virol Methods 153:79–83
Calgua B, Fumian T, Rusiñol M, Rodriguez-Manzano J, Mbayed VA, Bofill-Mas S, Miagostovich M, Girones R (2013) Detection and quantification of classic and emerging viruses by skimmed-milk flocculation and PCR in river water from two geographical areas. Water Res 47:2797–2810
Cashdollar JL, Dahling DR (2006) Evaluation of a method to re-use electropositive cartridge filters for concentrating viruses from tap and river water. J Virol Methods 132:13–17
Cashdollar JL, Wymer L (2013) Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J Appl Microbiol 115:1–11
Cheong S, Lee C, Song SW, Choi WC, Lee CH, Kim SJ (2009) Enteric viruses in raw vegetables and groundwater used for irrigation in South Korea. Appl Environ Microbiol 75:7745–7751
Dahling DR, Wright BA (1986) Recovery of viruses from water by a modified flocculation procedure for second-step concentration. Appl Environ Microbiol 51:1326–1331
De Keuckelaere A, Baert L, Duarte A, Stals A, Uyttendaele M (2013) Evaluation of viral concentration methods from irrigation and processing water. J Virol Methods 187:294–303
Dong Y, Kim J, Lewis GD (2010) Evaluation of methodology for detection of human adenoviruses in wastewater, drinking water, stream water, and recreational waters. J Appl Microbiol 108:800–809
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) (2011) Scientific opinion on an update on the present knowledge on the occurrence and control of foodborne viruses. EFSA J 9:2190–2196
Environment Agency (2000) Optimization of a new method for detection of viruses in groundwater, Report no. NC/99/40. Environment Agency, National Groundwater and Contaminated Land Centre, West Midlands
Farrah SR, Bitton G (1978) Elution of poliovirus adsorbed to membrane filters. Appl Environ Microbiol 36:982–984
Farrah SR, Gerba CP, Wallis C, Melnick JL (1976) Concentration of viruses from large volumes of tap water using pleated membrane filters. Appl Environ Microbiol 31:221–226
Field KG, Samadpour M (2007) Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41:3517–3538
Fong T, Lipp EK (2005) Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev 69:357–371
Fong T-T, Griffin DW, Lipp EK (2005) Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl Environ Microbiol 71:2070–2078
Fout GS, Martinson BC, Moyer MWN, Dahling DR (2003) A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl Environ Microbiol 69:3158–3164
Fout GS, Brinkman NE, Cashdollar JL, Griffin SM, McMinn BR, Rhodes ER, Varughese EA, Karim MR, Grimm AC, Spencer SK, Borchardt MA (2010) Method 1615: measurement of enterovirus and norovirus occurrence in water by culture and RT-qPCR, publication no EPA/ 600/R-10/181. US Environmental Protection Agency, Cincinnati
Francy DS, Bushon RN, Brady AMG, Bertke EE, Kephart CM, Likirdopulos CA, Mailot BE, Schaefer FW, Lindquist HDA (2009) Comparison of traditional and molecular analytical methods for detecting biological agents in raw and drinking water following ultrafiltration. J Appl Microbiol 107:1479–1491
Francy DS, Stelzer EA, Brady AMG, Huitger C, Bushon RN, Ip HS, Ware MW, Villegas EN, Gallardo V, Lindquist HDA (2013) Comparison of filters for concentrating microbial indicators and pathogens in lake water samples. Appl Environ Microbiol 79:1342–1352
Gensberger ET, Kostić T (2013) Novel tools for environmental virology. Curr Opin Virol 3:61–68
Gibbons CD, Rodríguez RA, Tallon L, Sobsey MD (2010) Evaluation of positively charged alumina nanofiber cartridge filters for the primary concentration of noroviruses, adenoviruses and male-specific coliphages from seawater. J Appl Microbiol 109:635–641
Gibson KE (2014) Viral pathogens in water: occurrence, public health impact, and available control strategies. Curr Opin Virol 4:50–57
Gibson KE, Schwab KJ (2011) Tangential-flow ultrafiltration with integrated inhibition detection for recovery of surrogates and human pathogens from large-volume source water and finished drinking water. Appl Environ Microbiol 77:385–391
Gibson KE, Schwab KJ, Spencer SK, Borchardt MA (2012) Measuring and mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res 46:4281–4291
Gilgen M, Germann D, Lüthy J, Hübner P (1997) Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int J Food Microbiol 37:189–199
Grabow WOK., Taylor MB (1993) New methods for virological analysis of drinking water supplies. In: Proceeding (Vol. 1): Biennial conference and exhibition of the Water Institute of Southern Africa, Durban. Johannesburg, Water Institute of Southern Africa, pp. 259–264, 24–27 May 1993
Hall AJ, Eisenbart VG, Etingüe AL, Gould LH, Lopman BA, Parashar UD (2012) Epidemiology of foodborne norovirus outbreaks, United States, 2001-2008. Emerg Infect Dis 18:1566–1573
Hamza IA, Jurzik L, Stang A, Sure K, Uberla K, Wilhelm M (2009) Detection of human viruses in rivers of a densely-populated area in Germany using virus adsorption elution method optimized for PCR analyses. Water Res 43:2657–2668
Hamza IA, Jurzik L, Überla K, Wilhelm M (2011) Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res 45:1358–1368
Haramoto E, Katayama H, Utagawa E, Ohgaki S (2009) Recovery of human norovirus from water by virus concentration methods. J Virol Methods 160:206–209
Haramoto E, Kitajima M, Kishida N, Katayama H, Asami M, Akiba M (2012) Occurrence of viruses and protozoa in drinking water sources of Japan and their relationship to indicator microorganisms. Food Environ Virol 4:93–101
Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB (2005) Validity of the indicator organism paradigm for the pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170
Hill WF, Akin EW, Benton WH, Mayhew CJ, Metcalf TG (1974) Recovery of poliovirus from turbid estuarine water on microporous filters by use of Celite. App Microbiol 27:506–512
Hill VR, Polaczyk AL, Hahn D, Narayanan J, Cromeans TL, Roberts JM, Amburgey JE (2005) Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl Environ Microbiol 71:6878–6884
Hill V, Kahler A, Jothikumar N, Johnson T (2007) Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl Environ Microbiol 73:4218–4225
Hill VR, Polaczyk AL, Kahler AM, Cromeans TL, Hahn D, Amburgey JE (2009) Comparison of hollow-fiber ultrafiltration to the USEPA VIRADEL technique and USEPA method 1623. J Environ Qual 38:822–825
Hill V, Mull B, Jothikumar N, Ferdinand K, Vinjé J (2010) Detection of GI and GII noroviruses in ground water using ultrafiltration and TaqMan real-time RT-PCR. Food Environ Virol 2:218–224
Holowecky PM, James RR, Lorch DP, Straka SE, Lindquist HDA (2009) Evaluation of ultrafiltration cartridges for water filtration apparatus. J Appl Microbiol 106:738–747
Huang PW, Laborde D, Land VR, Matson DO, Smith AW, Jiang X (2000) Concentration and detection of caliciviruses in water samples by reverse transcription-PCR. Appl Environ Microbiol 66:4383–4388
Huang H, Young TA, Jacangelo JG (2008) Unified membrane fouling index for low pressure membrane filtration of natural waters: Principles and methodology. Environ Sci Technol 42:714–720
Ikner LA, Soto-Beltran M, Bright KR (2011) New method using a positively charged microporous filter and ultrafiltration for concentration of viruses from tap water. Appl Environ Microbiol 77:3500–3506
Karim MR, Rhodes ER, Brinkman N, Wymer L, Fout GS (2009) New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl Environ Microbiol 75:2393–2399
Katayama H, Shimasaki A, Ohgaki S (2002) Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Appl Environ Microbiol 68:1033–1039
Katzenelson E, Fattal B, Hostovesky T (1976) Organic flocculation: an efficient second-step concentration method for the detection of viruses in tap water. Appl Environ Microbiol 32:632–639
Kelly SM (1953) Detection and occurrence of Coxsackie virus in sewage. Am J Public Health Nations Health 43:1532–1538
Kfir R, Hilner C, du Preez M, Bateman B (1995) Studies evaluating the applicability of utilizing the same concentration techniques for the detection of protozoan parasites and viruses in water. Water Sci Technol 31:417–423
Knappett PSK, Layton A, McKay LD, Williams D, Mailloux BJ, Huq MR, Alam MJ, Matin Ahmed K, Akita Y, Serre ML, Sayler GS, van Geen A (2011) Efficacy of hollow-fiber ultrafiltration for microbial sampling in groundwater. Ground Water 49:53–65
Kokkinos P, Kozyra I, Lazic S, Bouwknegt M, Rutjes S, Willems K, Moloney R et al (2012) Harmonized investigation of the occurrence of human enteric viruses in the leafy green vegetable supply chain in three European countries. Food Environ Virol 4:179–191
Koopmans M, Duizer E (2004) Foodborne viruses: an emerging problem. Int J Food Microbiol 90:23–41
Lambertini E, Spencer SK, Bertz PD, Loge FJ, Kieke BA, Borchardt MA (2008) Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl Environ Microbiol 74:2990–2996
Lambertini E, Spencer SK, Kieke BA, Loge FJ, Borchardt MA (2011) Virus contamination from operation and maintenance events in small drinking water distribution systems. J Water Health 9:799–812
Le Guyadar FS, Le Saux JC, Ambert-Balay K, Krol J, Serais O, Parnaudeau S, Giraudon H, Delmas G, Pommepuy M, Pothier P, Atmar RL (2008) Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J Clin Microbiol 46:4011–4017
Lee C, Kim S-J (2008) The genetic diversity of human noroviruses detected in river water in Korea. Water Res 42:4477–4484
Lee H, Kim M, Paik SY, Lee CH, Jheong WH, Kim J, Ko G (2011) Evaluation of electropositive filtration for recovering norovirus in water. J Water Health 9:27–36
Lewis GD, Metcalf TG (1988) Polyethylene glycol precipitation for recovery of pathogenic viruses, including Hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl Environ Microbiol 54:1983–1988
LGMA (2013) Commodity specific guidelines for the production and handling of lettuce and leafy greens. Accessed online 3/20/14 at: http://www.caleafygreens.ca.gov/food-safety-practices#downloads
Liu P, Hill VR, Hahn D, Johnson TB, Pan Y, Jothikumar N, Moe CL (2012) Hollow-fiber ultrafiltration for simultaneous recovery of viruses, bacteria and parasites from reclaimed water. J Microbiol Methods 88:155–161
LoGrippo G, Berger B (1952) Use of ion exchange resins in partial purification and concentration of poliomyelitis virus. J Lab Clin Med 39:970–973
Lopman BA, Reacher MH, Van Duijnhoven Y, Hanon F, Brown D, Koopmans M (2003) Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg Infect Dis 9:90–96
Lukasik J, Scott TM, Andryshak D, Farrah SR (2000) Influence of salts on virus adsorption to microporous filters. App Environ Microbiol 66:2914–2920
McMinn BR (2013) Optimization of adenovirus 40 and 41 recovery from tap water using small disk filters. J Virol Methods 193:284–290
McMinn BR, Cashdollar JL, Grimm AC, Fout GS (2012) Evaluation of the celite secondary concentration procedure and an alternate elution buffer for the recovery of enteric adenoviruses 40 and 41. J Virol Methods 179:423–428
Millen HT, Gonnering JC, Berg RK, Spencer SK, Jokela WE, Pearce JM, Borchardt JS, Borchardt MA (2012) Glass wool filters for concentrating waterborne viruses and agricultural zoonotic pathogens. J Vis Exp 61, e3930. doi:10.3791/3930
Morales-Morales H, Vidal G, Olszewski J, Rock CM, Dasgupta D, Oshima KH, Smith GB (2003) Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl Environ Microbiol 69:4098–4102
Mull B, Hill VR (2012) Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J Microbiol Methods 91:429–433
Olzewski J, Winona L, Oshima KH (2005) Comparison of 2 ultrafiltration systems for the concentration of seeded viruses from environmental waters. Can J Microbiol 51:295–303
Pachepsky Y, Shelton DR, McLain JET, Patel J, Mandrell RE (2011) Irrigation waters as a source of pathogenic microorganisms in produce: a review. Adv Agron 113:73–138
Pall Corporation (2015) Selection guide: separation products for centrifugal and tangential flow filtration. Accessed online 7/15/2015 at: http://www.pall.com/main/laboratory/literature-library-details.page?id=7046
Pang XL, Lee BE, Pabbaraju K, Gabos S, Craik S, Payment P, Neumann N (2012) Pre-analytical and analytical procedures for the detection of enteric viruses and enterovirus in water samples. J Virol Methods 184:77–83
Payment P, Franco E (1993) Clostridium perfringens and somatic coliphages as indicators of the efficiency of drinking water treatment for viruses and protozoan cysts. Appl Environ Microbiol 59:2418–2424
Payment P, Locas A (2011) Pathogens in water: values and limits of correlation with microbial indicators. Groundwater 49:4–11
Polaczyk A, Narayanan J, Cromeans T, Hahn D, Roberts J, Amburgey J, Hill V (2008) Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J Microbiol Methods 73:92–99
Rajal VB, McSwain BS, Thompson DE, Leutenegger CM, Kildare BJ, Wuertz S (2007) Validation of hollow fiber ultrafiltration and real-time PCR using bacteriophage PP7 as surrogate for the quantification of viruses from water samples. Water Res 41:1411–1422
Ramia S, Sattar SA (1979) Second-step concentration of viruses in drinking water and surface waters using polyethylene glycol hydroextraction. Can J Microbiol 25:587–592
Rhodes ER, Hamilton DW, See MJ, Wymer L (2011) Evaluation of hollow-fiber ultrafiltration primary concentration of pathogens and secondary concentration of viruses from water. J Virol Methods 176:38–45
Richards GP, McLeod C, Le Guyadar FS (2010) Processing strategies to inactivate enteric viruses in shellfish. Food Environ Virol 2:183–193
Rose JB, Singh SN, Gerba CP, Kelley LM (1984) Comparison of microporous filters for concentration of viruses from wastewater. Appl Environ Microbiol 47:989–992
Scallan E, Hoekstra R, Angulo F, Tauxe R (2011) Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis 17:7–15
Schwab KS, De Leon R, Sobsey MD (1996) Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcription PCR. Appl Environ Microbiol 62:2086–2094
Shields PA, Ling TF, Tjatha V, Shah DO, Farrah SR (1986) Comparison of positively charged membrane filters and their use in concentrating bacteriophages in water. Water Res 20:145–151
Sinclair RG, Jones EL, Gerba CP (2009) Viruses in recreational water-borne disease outbreaks: a review. J Appl Microbiol 107:1769–1780
Smith CM, Hill VR (2009) Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl Environ Microbiol 75:5284–5289
Sobsey MD, Glass JS (1980) Poliovirus concentration from tap water with electropositive adsorbent filters. Appl Environ Microbiol 40:201–210
Sobsey MD, Jones BL (1979) Concentration of poliovirus from tap water using positively charged microporous filters. Appl Environ Microbiol 37:588–595
Sobsey MD, Wallis C, Henderson M, Melnick JL (1973) Concentration of enteroviruses from large volumes of water. Appl Microbiol 26:529–534
Song I, Stine SW, Choi CY, Gerba CP (2006) Comparison of crop contamination by microorganisms during subsurface drip and furrow irrigation. J Environ Qual 132:1243–1248
Stine SW, Song IH, Choi CY, Gerba CP (2005) Application of microbial risk assessment to the development of standards for enteric pathogens in water used to irrigate fresh produce. J Food Protect 68:913–918
USEPA (2001) Concentration and processing of waterborne viruses by positive charge 1MDS cartridge filters and organic flocculation. In: Manual of methods for virology, EPA/600/4-84/013. Accessed online 2/14/14 at http://www.epa.gov/nerlcwww/documents/chapt14.pdf
USEPA, CDC (2011) Comparison of ultrafiltration techniques for recovering biothreat agents in water, EPA 600/R-11/103. Accessed online 2/20/14 at http://cfpub.epa.gov/si/si_public_record_report.cfm?address=nhsrc/&dirEntryId=238310
USEPA (2012) Recreational water quality criteria, EPA 820-F-12-058. Accessed online 3/1/14 at http://water.epa.gov/scitech/swguidance/standards/criteria/health/recreation/
USFDA (2014) FSMA Facts: more on the proposed agricultural water standards – FDA’s proposed rule for produce safety. Accessed online 3/16/14 at http://www.fda.gov/downloads/Food/GuidanceRegulation/FSMA/UCM360242.pdf
van Zyl WB, Page NA, Grabow WOK, Steele AD, Taylor MB (2006) Molecular epidemiology of group A rotaviruses in water sources and selected raw vegetables in southern Africa. Appl Environ Microbiol 72:4554–4560
Victoria M, Guimarães F, Fumian T, Ferreira F, Vieira C, Leite JP, Miagostovich M (2009) Evaluation of an adsorption-elution method for detection of astrovirus and norovirus in environmental waters. J Virol Methods 156:73–76
Vilagines P, Sarrette B, Husson G, Vilagines R (1993) Glass wool for virus concentration at ambient water pH level. Water Sci Technol 27:299–306
Vilagines P, Sarrette B, Champsaur H, Hugues B (1997) Round robin investigation of glass wool method for poliovirus recovery from drinking water and sea water. Water Sci Technol 35:445–449
Wallis C, Melnick JL (1967) Concentration of enteroviruses on membrane filters. J Virol 1:472–477
Wallis C, Melnick JL, Gerba CP (1979) Concentration of viruses from water by membrane chromatography. Annu Rev Microbiol 33:413–437
Winona LJ, Ommani AW, Olszewski J, Nuzzo JB, Oshima KH (2001) 2001, Efficient and predictable recovery of viruses from water by small scale ultrafiltration systems. Can J Microbiol 47:1033–1041
World Health Organization (2009) Outbreaks of waterborne diseases, Fact Sheet 1.1. Accessed online 3/20/14 at http://www.euro.who.int/__data/assets/pdf_file/0009/96885/1.1.-Outbreaks-of-waterborne-diseases-EDITED_layout_V03.pdf
Wu J, Simmons IO, Sobsey MD (2013) Uncertainty analysis of the recovery of hollow-fiber ultrafiltration for multiple microbe classes from water: a bayesian approach. J Microbiol Methods 93:161–167
Wyn-Jones AP, Carducci A, Cook N, D’Agostino M, Divizia M, Fleischer J, Gatzer C, Gawler A, Girones R, Höller C, de Roda Husman AM, Kay D, Kozyra I, López-Pila J, Muscillo M, São José Nascimento M, Papageorgiou G, Rutjes S, Sellwood J, Szewzyk R, Wyer M (2011) Surveillance of adenoviruses and noroviruses in European recreational waters. Water Res 45:1025–1038
Ye XY, Ming X, Zhang YL, Xiao WQ, Huang XN, Cao YG, Gu KD (2012) Real-time PCR detection of enteric viruses in source water and treated drinking water in Wuhan, China. Curr Microbiol 65:244–253
Yugo DM, Meng X (2013) Hepatitis E virus: foodborne, waterborne, and zoonotic transmission. Int J Environ Res Public Health 10:4507–4533
Zhang Y, Riley LK, Lin M, Purdy GA, Hu Z (2013) Development of a virus concentration method using lanthanumbased chemical flocculation coupled with modified membrane filtration procedures. J Virol Methods 190:41–48
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Gibson, K.E., Borchardt, M.A. (2016). Methods for Virus Recovery in Water. In: Goyal, S., Cannon, J. (eds) Viruses in Foods. Food Microbiology and Food Safety(). Springer, Cham. https://doi.org/10.1007/978-3-319-30723-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-30723-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-30721-3
Online ISBN: 978-3-319-30723-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)