Abstract
Classification of brain images is frequently done using kernel based methods, such as the support vector machine. These lend themselves to improvement via multiple kernel learning, where a number of different kernels are linearly combined to integrate different sources of information and increase accuracy. Previous applications made use of a small number of kernels representing different image modalities or kernel functions. Here, the kernels instead represent 83 anatomically meaningful brain regions. To find the optimal combination of kernels and perform classification, we use a Gaussian Process framework to infer the maximum likelihood weights. The resulting formulation successfully combines voxel level features with prior anatomical knowledge. This gives an improvement in classification accuracy of MRI images of Alzheimer’s disease patients and healthy controls from the ADNI database to almost 88 %, compared to less than 86 % using a single kernel representing the whole brain. Moreover, interpretability of the classifier is also improved, as the optimal kernel weights are sparse and give an indication of the importance of each brain region in separating the two groups.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Machine learning methods have become increasingly common in the analysis of brain image data, both for computer aided diagnosis (CAD) of disease and in a more exploratory fashion to discover biomarkers that can be informative about disease processes. For Alzheimer’s disease (AD), grey matter (GM) density maps obtained from structural MRI images are used as sources of data in the classification. However the actual features derived from the image can take two forms: as the intensities of MRI voxels themselves [1], or as aggregations of all GM voxels within different anatomical regions. The regions can be defined by an atlas [2] or can themselves be generated from voxel level data [3]. There is a trade-off between these methods. Regional level features reduce the data dimensionality and can introduce prior information relevant to the classification problem, but also eliminate fine detail that may be informative about disease state. Voxel level data can introduce noise by including uninformative brain regions and results in a very high dimensional problem. The different feature extraction methods are compared and discussed in depth in [4].
Our proposed method combines the strengths of these two approaches. It uses both voxel level features and atlas derived regions, and automatically gives less weight to voxels within less relevant regions. This is done using multiple kernel learning (MKL), a method that can be applied to any kernel based classifier, such as the support vector machine (SVM) or Gaussian Process (GP). These use a linear combination of kernels, where the kernels can be derived from different data modalities [2, 5] or kernel functions [6]. Conversely, in our approach each kernel represents the voxel level data within a different anatomical region to produce anatomical regional kernels (ARKs). This takes a similar approach to [7, 8] presented a related method using hierarchical groups of regional features. Although the work was developed from our previous use of MKL, and is presented as a specific case of MKL, it is related to other families of methods. Specifically, it can be seen as a way to incorporate explicit spatial regularisation into the classifier. A number of other methods have been developed to do this specifically for three dimensional medical image data. Spatial smoothness and sparsity can be enforced with a joint \(\ell 1\) and total variation penalty [9]. Alternative a smoothness penalty is derived from the image voxel neighbourhood structure, which can be built into a kernel function for use with an SVM or other kernel method [10] or used directly as a term in the objective function [11].
Our method and [11] can also both be interpreted as a variant of automatic relevance determination (ARD) [12, 13], a Bayesian method of automatic feature selection. Our method, however, operates at the regional level in the kernel space, rather than at the voxel level in the input space. Our approach builds on the existence of a brain atlas in a custom groupwise template. We explain how this was achieved, and how MKL is performed within a GP framework.
We apply this method to a large population of AD and control subjects from the ADNI study. In terms of classification accuracy, our method outperforms a single kernel with voxel level features by a susbtantial margin, and a single kernel with regional features by a smaller amount. We also introduce a new method to assess the quality of a classifier that exploit the probabilistic predictions made by GPs. Finally, we show that the optimal kernel weights in the MKL formulation are informative about which regions are affected by AD.
2 Materials and Methods
2.1 Image and Biomarker Data
All data were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) databaseFootnote 1. The MRI images were T1 weighted structural scans from a mixture of 1.5T and 3T scanners. All were subjected to quality control and automatically corrected for spatial distortion caused by gradient nonlinearity and B1 field inhomogeneity and downloaded from the ADNI database. Subjects were classified as healthy control (HC), AD or mild cognitive impairment by neuropsychological and clinical testing at the time of the baseline scan, and only HC and AD subjects were used. For the classification experiments, a further quality control step was taken which removed 16 subjects with registration errors, leaving a final total of 627 subjects. Their demographics are given in Table 1.
2.2 Image Processing
Groupwise Registration. As our method defines features at the voxel level, it was necessary to transfer images into a common space. All native space images were rigidly and then affinely registered to a randomly chosen image, coalescing the registered images to update the template after each round of registrations. This was then followed by ten rounds of nonrigid registration to produce a final template in the groupwise space. All registrations were performed using the Niftyreg package [14].
Image Segmentation. All images were segmented into GM, white matter (WM), cerebrospinal fluid (CSF), and non-brain tissues components using the new segment module of SPM12 with the cleanup option set to maximum. A brain mask generated from the original structural image was then applied to the GM segmentations to further exclude any non-brain material.
Image Parcellation. The native space images were also anatomically parcellated into 83 regions. This was done with a novel label fusion algorithm [15] in a multi-atlas label propagation scheme. A library of 30 atlases manually labelled with 83 anatomical regions was used as a basis for the parcellation [16].
Atlas Construction. Unlike in other approaches using anatomical regions, features were defined at the level of the voxel rather than regions, requiring that all images share a common space. As kernels were constructed from the voxels within anatomical regions common across subjects, the parcellation defining the region was also required to be in the common space. However, our initial parcellations were in the native spaces of each subject. To combine these initial parcellations in the groupwise space, the following procedure was used. First, all the parcellations were warped into the groupwise space, using the parameters from the native space of each image to the final groupwise template. Care was taken to preserve the integer labels in the parcellations during resampling. Finally to combine the individual parcellations, a consensus atlas was produced by majority voting among the set of N parcellations X to assign a single label l to each voxel \(v_{i}\) of the groupwise space \(\mathbf {\Omega }\):
The pipeline to construct the atlas is summarised graphically in Fig. 1.
2.3 Gaussian Process Classification
Gaussian processes (GPs) provide a Bayesian, kernelised framework for solving both regression and classification problems. As an in depth explanation of GPs is beyond the scope of this paper, we refer the reader to [13] for a more theoretical treatment. Briefly, however, a GP (essentially a multivariate Gaussian) forms the prior on the value of a latent function f. For binary classification, the value of the latent function is linked to the probability of being in class \(y,y\in \left\{ {-1, +1}\right\} \) by a sigmoidal function. The GP is parameterised by a mean function \(\mu (\mathbf {x})\) and a covariance kernel function \(k(\mathbf {x},\mathbf {x'})\) where \(\mathbf {x}\) is a feature vector, whose elements represent voxel values in this case.
For classification, the non-Gaussian link function means that the posterior is also non-Gaussian so an approximation made be used. We make use of expectation propagation (EP) [17] as an approximation.
The GP prior is a function not only of the data but also of any hyperparameters \(\theta \) that specify the form of the prior. We trained the GP by tuning the values of these hyperparameters to maximise the log likelihood of the training data, which can be done with standard gradient based optimisation algorithms. Once the hyperparameters have been set, predictions on unseen data are made by integrating across this optimised prior.
2.4 Gaussian Processes as Multimodal Kernel Methods
As Eq. 2 implies, GP classification belongs to the family of kernel methods. Hence a positive sum of valid kernels is a valid kernel, and a valid kernel multiplied by a positive scalar is also a valid kernel. The covariance between the ith and jth subject, \(\mathbf {K}_{ij}\), is a kernel function k of the feature vectors for the ith and jth subject \(\mathbf {x}_{i}\) and \(\mathbf {x}_{j}\) and hyperparameters \(\theta \). For ARKs, the final kernel \(\mathbf {K}\) is the weighted sum of 83 linear subkernels, each of which in turn is the dot product between the voxels within a particular anatomical region of the ith and jth image. These regions are defined using masks for each label derived from the groupwise atlas. This is illustrated in Fig. 2.
The covariance hyperparameters are the weights of the subkernels \(\alpha \) and bias term \(\beta \), so the final kernel value \(\mathbf {K}\) is given by
where r indexes regions 1 to 83 and \(\beta \) is a bias term. There are thus 84 covariance hyperparameters: \(\theta _{cov}=(\alpha _{1},\alpha _{2},\ldots ,\alpha _{83},\beta )\). All the above calculations are carried out within the GPML toolkitFootnote 2, modified to take precomputed kernel matrices.
3 Results
We performed binary classification of all subjects as HC or AD. To generate results, we use a leave-one-out cross validation (LOOCV) across the entire set of 627 subjects. For the ARK formulation described above, the feature vectors \(\mathbf {x}\) consist of voxel level data. For the purposes of comparison to existing methods, we also deploy two more conventional methods related to those introduced, representing opposite ends of the tradeoff between detail and use of prior anatomical information discussed in the introduction:
-
‘voxels’ method: This again uses voxel level data for the whole brain. However this is just used with a single kernel for the whole brain and no use of the atlas or anatomical prior information.
-
‘regions’ method: In place of voxel GM densities, this method takes the total GM volumes of each region as its features. These are normalised by the intra-cranial volume to control for variability in head size. The resulting feature vectors, of much lower dimensionality than either ARK or the voxels methods, are then used to build the single kernel.
3.1 Binary Accuracy
We compare the three methods by thresholding predicted probabilities at 0.5 and comparing to ground truth labels for HC or AD status. The resulting sensitivity, specificity and accuracy are shown in Table 2. We also show the area under the ROC curve (AUC), and a p-value for difference in accuracy with McNemar’s test. The ARK formulation displays a greater accuracy and AUC then both competing methods. While the advantage over the voxels method is substantial, we do not quite have enough subjects and thus statistical power to show that it or the smaller advantage over regions is significant. We can, however visualise the effect of the ARK formulation across all the individual predictions.
3.2 Individual Predictions
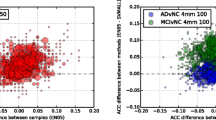
Figure 3 shows the difference in predicted p(AD) between ARKs and each competing method for all subjects. Results are colour-coded so AD subjects are shown in red and HC ones in blue, and sorted by the value of the p(AD) for the competing method. Hence blue (HC) subjects will be represented by a line extending left from the baseline, and red (AD) subjects by a line extending right, if ARKs improve the baseline classification. The plots also show how most subjects are correctly classified: The AD subjects mostly occupied the right hand side of the plots (\(p(AD) > 0.5\)) and the HC ones the left side of the plots.
3.3 Interpretation of Hyperparameters
The optimised weights \(\alpha \) tell us about the importance of the corresponding regions in the classification, and hence in AD. For each of the 627 sets of \(\alpha \), we normalise \(\alpha \) so they represent a fraction of the total weight, than average each normalised weight across all folds. Only 14 regions have weights of more than 1 % of the total, shown in Fig. 3. These include temporal lobe regions frequently implicated in AD in studies such as [18], as well as the GM tissue adjacent to the temporal horn of the left lateral ventricle, which will be very sensitive to expansion of the horn. However, other structures much more widely distributed across the brain are also important in the classification, suggesting that atrophy may more quite widespread. The largest weight value is given to the right nucleus accumbens, and the left nucleus accumbens and right caudate are also given large weights, which may be a result of recently identified AD related atrophy in deeper structures [19] (Fig. 4).
4 Discussion
Our results show that ARKs successfully combine voxel level data with prior anatomical knowledge, offering a substantial accuracy improvement compared to voxel level data alone, and also offer a smaller improvement over features based on predefined regions. We are also able to show the improvements ARKs bring to individual subjects. Moreover, the kernel weights enhance model interpretability by showing new regions which may be involved in the AD process. The chief disadvantage of ARKs is speed of classifier training, due to the high dimensionality of the data and the large number of hyperparameters; however this is largely compensated for by the use of modified software that uses precomputed (sub)kernel matrices.
The method is quite general, and could also be applied to any type of training data where low level features and a parcellation are available in a common space, for example voxelwise cortical thickness data and a cortical atlas.
References
Klöppel, S., Stonnington, C.M., Chu, C., Draganski, B., Scahill, R.I., Rohrer, J.D., Fox, N.C., Jack, C.R., Ashburner, J., Frackowiak, R.S.J.: Automatic classification of MR scans in Alzheimerss disease. Brain: J. Neurol. 131(Pt 3), 681–689 (2008)
Zhang, D., Wang, Y., Zhou, L., Yuan, H., Shen, D.: Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage 55(3), 856–867 (2011)
Fan, Y., Shen, D., Gur, R.C., Gur, R.E., Davatzikos, C.: COMPARE: classification of morphological patterns using adaptive regional elements. IEEE Trans. Med. Imaging 26(1), 93–105 (2007)
Cuingnet, R., Gerardin, E., Tessieras, J., Auzias, G., Lehricy, S., Habert, M.O., Chupin, M., Benali, H., Colliot, O.: Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. NeuroImage 56(2), 766–781 (2011)
Young, J., Modat, M., Cardoso, M.J., Mendelson, A., Cash, D., Ourselin, S.: Accurate multimodal probabilistic prediction of conversion to Alzheimer’s disease in patients with mild cognitive impairment. NeuroImage: Clin. 2, 735–745 (2013)
Hinrichs, C., Singh, V., Xu, G., Johnson, S.C.: Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage 55(2), 574–589 (2011)
Chu, C., Bandettini, P., Ashburner, J., Marquand, A., Kloeppel, S.: Classification of neurodegenerative diseases using Gaussian process classification with automatic feature determination. In: 2010 First Workshop on Brain Decoding: Pattern Recognition Challenges in Neuroimaging (WBD), pp. 17–20. IEEE (2010)
Liu, F., Zhou, L., Shen, C., Yin, J.: Multiple kernel learning in the primal for multi-modal Alzheimer’s disease classification (2013). arXiv e-print 1310.0890
Gramfort, A., Thirion, B., Varoquaux, G.: Identifying predictive regions from fMRI with TV-L1 prior. In: Proceedings of the 2013 International Workshop on Pattern Recognition in Neuroimaging. PRNI 2013, pp. 17–20. IEEE Computer Society, Washington, DC (2013)
Cuingnet, R., Glaunès, J.A., Chupin, M., Benali, H., Colliot, O.: Spatial and anatomical regularization of SVM: a general framework for neuroimaging data. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 682–696 (2013)
Sabuncu, M.R., Leemput, K.V.: The relevance voxel machine (RVoxM): a self-tuning Bayesian model for informative image-based prediction. IEEE Trans. Med. Imaging 31(12), 2290–2306 (2012)
Neal, R.M.: Bayesian Learning for Neural Networks. Springer, New York (1996)
Rasmussen, C.E., Williams, C.K.I.: Gaussian Processes for Machine Learning. MIT Press, Cambridge (2006)
Modat, M., Ridgway, G.R., Taylor, Z.A., Lehmann, M., Barnes, J., Hawkes, D.J., Fox, N.C., Ourselin, S.: Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 98(3), 278–284 (2010)
Cardoso, M., Modat, M., Ourselin, S., Keihaninejad, S., Cash, D.: Multi-STEPS: multi-label similarity and truth estimation for propagated segmentations. In: 2012 IEEE Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA), pp. 153–158 (2012)
Gousias, I.S., Rueckert, D., Heckemann, R.A., Dyet, L.E., Boardman, J.P., Edwards, A.D., Hammers, A.: Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. NeuroImage 40(2), 672–684 (2008)
Minka, T.: Expectation propagation for approximate bayesian inference. In: Proceedings of the Seventeenth Conference Annual Conference on Uncertainty in Artificial Intelligence (UAI 2001), pp. 362–369. Morgan Kaufmann, San Francisco (2001)
Braak, H., Braak, E.: Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 16(3), 271–278 (1995)
Madsen, S., Ho, A., Hua, X., Saharan, P., Toga Jr, A., Jack, C., Weiner, M., Thompson, P.: 3D maps localize caudate nucleus atrophy in 400 Alzheimers disease, mild cognitive impairment, and healthy elderly subjects. Neurobiol. Aging 31(8), 1312–1325 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Young, J., Mendelson, A., Cardoso, M.J., Modat, M., Ashburner, J., Ourselin, S. (2015). Improving MRI Brain Image Classification with Anatomical Regional Kernels. In: Bhatia, K., Lombaert, H. (eds) Machine Learning Meets Medical Imaging. MLMMI 2015. Lecture Notes in Computer Science(), vol 9487. Springer, Cham. https://doi.org/10.1007/978-3-319-27929-9_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-27929-9_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27928-2
Online ISBN: 978-3-319-27929-9
eBook Packages: Computer ScienceComputer Science (R0)