Abstract
Necrotrophic fungal pathogens cause major losses to fruit, vegetable, and cereal crops annually and the economic impact is more than that of diseases caused by biotrophic pathogens. These pathogens are devastating because they kill as they colonize through production of cell wall-degrading enzymes and phytotoxins, obtaining nutrients for growth and reproduction from the dead plant cells. They explore a wide variety of virulence strategies and based on these the pathogens are classified into host-specific and broad host-range necrotrophs. Plants are equipped with an immune system as a defense mechanism while the necrotrophic fungal pathogenic arsenal suppresses the immune responses for disease manifestation. Plant defense response involves the interplay of signaling molecules which include various phytohormones like jasmonic acid, ethylene, salicylic and abscisic acid which also serve as regulators of the immune response. Coordination at the transcriptional level of genes for the production of defense molecules including antimicrobial phytoalexins and pathogenesis-related proteins by transcription factors such as WRKY33 and ERF which are responsive to the signaling molecules has been observed. The roles for several important transcription factors already unveiled through studies of mutants in the model plant, Arabidopsis thaliana and some of the information translatable to crop plants. The present chapter shows the interconnection between cell wall integrity and the action of signaling molecules in the expression of defense-related genes. Moreover, the epigenetic mechanism through DNA and histone modification is also discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Necrotrophic fungi
- Phytohormones
- Pathogenesis-related proteins
- Signaling molecules
- Transcription factors
- Epigenetic
1 Introduction
Infectious microbes found in the environment pose continuous threats to plant survival and productivity, leading to significant economic losses to major crops (Wilkinson et al. 2011). Plant microbial pathogens obtain nutrients from their host cells for growth and reproduction and the devastation caused by them relates to their feeding modes. They are divided into three classes: necrotrophs, biotrophs, and hemibiotrophs based on their methods of nutrient acquisition. The necrotrophs are the most damaging as they kill the host while colonizing and live on the contents of the dead cells. Biotrophs, on the other hand, maintain the viability of their host as they thrive on the nutrients from living cells. Hemibiotrophs display both modes starting at the early phase as biotrophs then switching to the necrotroph mode of nutrition. There is great variability in the duration of the biotrophic and necrotrophic phases among hemibiotrophs (Meinhardt et al. 2014).
The lifestyle and infection strategy differ between necrotrophs and biotrophs. The necrotrophic microbial pathogens which include bacterial, fungal, and oomycete species have notorious and aggressive pathogenesis strategies leading to extensive necrosis, tissue maceration, and rotting symptoms on the plant. Prior to or during colonization, the pathogens secrete disease causing agents which include phytotoxins and cell wall degrading enzymes (CWDEs) into host tissues. These toxic molecules and lytic enzymes kill the host cells and the decomposed plant tissues are consumed by the pathogens. In contrast, biotrophic pathogens, in general, do not produce toxins and secrete very limited amount of CWDEs (Oliver and Ipcho 2004). The present chapter shows the interconnection between cell wall integrity and the action of signaling molecules in the expression of defense-related genes. Moreover, the epigenetic mechanism through DNA and histone modification is also discussed.
2 Classification of Necrotrophs
2.1 Host-Specific Necrotrophs (HSNs)
Necrotrophs are classified into host-specific necrotrophs (HSNs) and broad host-range necrotrophs (BHNs). The HSN infects a single or few related plant species by producing a host-selective toxin (HST). The HST is a strain-specific effector required for pathogenicity and virulence on the natural hosts (Friesen et al. 2009). Cochliobolus and Alternaria spp. are examples of HSN that produce HSTs. C. victoriae produces victorin, which is responsible for the Victoria blight disease in oats (Wolpert et al. 2002). Similarly, A. alternata and A. brassicae also produce HSTs that define their host range (Thomma 2003; Mamgain et al. 2013).
2.2 Broad Host-Range Necrotrophs (BHNs)
The broad host range necrotrophs (BHNs) include fungal species belonging to the genera Monilia, Sclerotinia, and Botrytis. For instance, Botrytis cinerea can infect more than 200 hosts of dicotyledonous and monocotyledonous plant species widely distributed in diverse geographical locations. These include vegetables, ornamental plants, and important fruit species like kiwi-fruit and strawberry (Elad et al. 2004). Most BHNs infect aerial tissues, while some species from the genera Rhizoctonia, Fusarium, and Colletotrichum are soil borne necrotrophs that gain entry through roots. Their success is attributed to the extensive array of phytotoxic metabolites, CWDEs, and cell death elicitors which alone or together can interfere with the structural and biological functions found in common in the different plant families (Okubara and Paulitz 2005).
3 Fungal Necrotrophs Infection of Host Plant
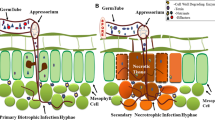
Infection of host plant by fungal necrotrophs in general follows a sequence of events as follows: conidial attachment, germination, penetration into the host, formation of primary lesion, lesion enlargement, and tissue maceration followed by sporulation (Fig. 1). Penetration may be achieved through either active or passive mechanisms. The active mechanisms include appressoria formation and enzymatic degradation. Passive mechanisms occur through an existing wound or infection sites as well as stomata. Lytic enzymes employed for initial penetration as well as toxic levels of reactive oxygen species (ROS) are involved in cellular dismantling to decompose the tissues. To ensure the progression of the disease, these fungi actively manipulate to suppress the host cell defense machinery (Prins et al. 2000). Among the necrotrophic fungal plant pathogens, the gray mold B. cinerea has been studied in great detail. Targeted mutagenesis of the genome sequence revealed the roles played by a variety of B. cinerea genes in the infection process. These include the genes required for signaling, penetration, killing the host cells, and decomposition of the plant tissues (van Kan 2006). The information generated is essential for developing knowledge-based strategies for disease control.
4 Molecular Mechanisms in Necrotrophic Fungi and Plant Interaction
The attack by the different types of microbial pathogens will trigger the plant immune response. This is a complex and multifaceted cellular reaction involving histological, physiological, biochemical, and molecular events, which work in concert to limit the pathogen spread and disease manifestation. Plant response in the early stages of necrotrophic fungal infection involves cell death, production of antimicrobial secondary metabolites, signaling molecules, including phytohormones and reactive oxygen species (ROS), callose deposition and the various cell wall modifications. Plants employ the same set of response to the different pathogens; however, the magnitude and rate of production of the different compounds vary in specific host–pathogen interactions and some of the responses may contribute to resistance. The necrotrophs employ various strategies for virulence and pathogenicity by counteracting the different processes and steps in the plant immune response pathways (Mengiste 2012). They may manipulate host phytohormone levels or synthesize their own hormone to disrupt defense signaling in the infected plants (Kazan and Lyons 2014). Some necrotrophs have mechanisms to detoxify host metabolites that interfere with virulence (Morrissey and Osbourn 1999). An effective plant immune system neutralizes the pathogenic arsenal of the infectious necrotroph or weakens its effect, curbing spread and development of disease symptom. Resistance traits against host-specific necrotrophs (HSNs) may be inherited; however, resistance to BHNs involves many contributing genes and pathways. Host genetic components and the processes at cellular and molecular levels that mediate and contribute to plant immune responses to necrotrophs are an important area of study for developing strategies for early diagnosis and disease management.
4.1 Pathogen-Triggered Immunity (PTI)
Pathogen associated molecular patterns (PAMPs) are signature-pattern molecules released from pathogens such as fragments of chitins from fungal cell wall when they infect plants. The microbial activity releases degradation products from constituents of plant cells such as fragments of plant cell wall release from the fungal CWDEs activities referred to as damage associated molecular patterns (DAMPs). Pathogen-triggered immunity (PTI) is a form of quantitative resistance that is activated upon recognition of PAMPs and DAMPs by pattern recognition receptors (PRRs). Membrane localized receptor-like kinases (RLKs), peptide receptors (Yamaguchi et al. 2010), and wall-associated kinase 1 (WAK1) as receptor of oligogalacturonides (OG) are examples of PRRs. Basal resistance and PTI responses to the corresponding pathogens diminished in the absence of PRRs (Mengiste 2012).
4.2 Roles of Reactive Oxygen Species and Nitric Oxide in Plant–Fungal Interaction
Attacks by biotrophs and necrotrophs trigger the production of ROS through the plant immune response pathways. The generation of ROS for regulating cell death is essential for resistance to biotrophs and hemibiotrophs through the hypersensitive response mechanism. It appears that the continuous generation of ROS facilitates cell death and promotes susceptibility to necrotrophs. However, early stimulation of ROS production could induce resistance mechanisms. Involvement of ROS in the activation of various immune responses plays a role in the plant resistance mechanism to both necrotrophs and biotrophs. In this context, the kinetics of the oxidative burst are more important than the absolute levels of ROS being produced as an indicator of resistance in plant microbial pathogen interaction (Mengiste 2012).
The ROS serve as the virulence factor for some necrotrophs. The pathogen induces generation of ROS once infection is established, resulting in plant cell death, which offers significant advantages to the pathogen as this increases susceptibility of the plant to the fungal attack (Govrin and Levine 2000). The fungal secreted enzyme superoxide dismutase may play a role in stimulating oxidative burst by B. cinerae from the time of cuticle penetration. Deletion of the gene encoding the enzyme reduced the virulence of the fungal pathogen on its different host plants (van Kan 2006). Over-accumulation of ROS resulting from interference with the breakdown of chlorophyll led to increased susceptibility to some necrotrophs (Kariola et al. 2005). Disease progression is promoted through a large disturbance of the redox status in the infected tissues (Lyon et al. 2004). Furthermore, the pathogenicities of B. cinerea and S. sclerotiorum correlated with the level of hydroxyl (OH−) radicals and hydrogen peroxide (H2O2) produced. At advanced stages of infection, it was found that Botrytis could trigger significant changes in the antioxidant system of the peroxisomes, causing the collapse of the protective mechanism (Kuzniak and Sklodowska 2005). The type 3 metallothionein, MT3-A and MT3-B expression was induced in leaf tissues of oil palm artificially inoculated with the necrotrophic fungi, Ganoderma boninense. Since one of the important roles of metallothionein is in scavenging ROS, MT3-A and MT3-B may be produced in response to the generation of ROS in the infected oil palm tissues. Thus, the oil palm type 3 metallothioneins could potentially be suitable biomarkers for the oxidative stress due to G. boninense infection (Fahimeh et al. 2011).
Nitric oxide is produced in response to both biotic and abiotic stresses and evidence is accumulating to suggest its significant role in the plant–pathogen interaction and signaling (Crawford and Guo 2005; Mur et al. 2013). Nitric oxide concentration influences the synthesis of cellulose an essential component of plant cell wall (Correa-Aragunde et al. 2008), alleviate cadmium and aluminum stress through alteration of cell wall composition (Xiong et al. 2009; Zhang et al. 2011). Plant hormones, including auxins and cytokinins, are involved in regulating the production and signaling by nitric oxide. Auxins work synergistically with nitric oxide in various developmental and morphological plant responses, whereas nitric oxide’s interaction with cytokinins is complex and the outcome appeared to be dependent on tissue type and plant species (Freschi 2013; Nafisi et al. 2015).
4.3 Fungal Toxins and Phytotoxic Proteins as Necrotrophic Effectors
Toxins and phytotoxic proteins produce by necrotrophs are critical for their virulence strategies. Together they are referred to as necrotrophic effectors. Phytotoxins such as HC-toxins, victorin, AM-toxin, ergotamine, and sirodesmin PL are produced by plant-pathogenic fungi through the activity of polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs) (Vleeshouwers and Oliver 2014). The effectors target specific host proteins and cellular processes. For example, Fumonisin B1 and AAL toxin inhibit ceramide synthesis in the plant host that consequently interferes with sphingolipid metabolism (Abbas et al. 1994). However, the interaction of the necrotrophic effectors may not directly bind to the dominant host susceptible gene, even though Hmr interacted with the necrotrophic effector. It was the first disease resistance gene cloned (Walton 2006).
HSTs are secreted as virulence factors by many necrotrophs. Specific host target proteins conferred susceptibility to the pathogen. Periconia circinata produces HST causing milo disease in sorghum. Susceptibility to the pathogen is conditioned by the Pc gene which encodes the nucleotide-binding site leucine-rich repeat (NBS-LRR) protein (Nagy et al. 2007). The wheat tan spot caused by Pyrenophora tritici-repentis is mediated by ToxA toxins and Tsn 1, the wheat toxin sensitivity gene that encodes the R-like protein (Faris et al. 2010). However, it was suggested that the toxin does not physically interact with the R-protein, but rather the protein functions indirectly for toxin sensitivity and pathogen susceptibility. As effectors, the HSTs suppress host defenses while, serving as determinants of host responses. Resistance to HSN emulates effector-triggered immunity (ETI). It involves a single gene-encoded protein not affected by the toxin or can detoxify the HSTs (Wolpert et al. 2002). For example, HC-toxin can be detoxified by the carbonyl reductase encoded by the maize HM1 gene. This blocked the inhibition of histone deacetylases (HDACs) by the toxin, thus conferring race-specific resistance (Johal and Briggs 1992).
BHNs produce nonspecific toxins that most probably target host proteins. BHNs secrete necrosis and ethylene-inducing proteins (NEPs) causing cell death in dicotyledonous (Staats et al. 2007). NEPs serve as the virulence factors for the BHNs. The Fusarium oxysporum secreted necrosis and ethylene-inducing protein (NEP1) that cause plant cell death. NEP1-like fungal proteins (NLPs) are small conserved molecules that induce HR-like cell death. NLPs only exhibit conserved toxicity to dicot plants across a wide range of taxa (Ottmann et al. 2009). The response triggered by both HSTs and NEPs mimics the plant immune response (Qutob et al. 2006; Wolpert et al. 2002). The toxins behave like virulence promoting cytolytic toxins that function by interfering with integrity of the plasma membrane (Ottmann et al. 2009). The disruption once detected by the host plants leads to the activation of defenses.
4.4 Downstream Defense Responses
4.4.1 Pathogenesis-Related Proteins
The biosynthesis and accumulation of inducible pathogenesis-related (PR) proteins is one of the main biochemical defense responses of plants to infection by pathogens. These proteins are of low molecular mass in the range of 6–43 kDa and strongly resistant to proteolytic cleavage and low pH values which enables survival in the harsh environment such as the vacuolar compartment or the intercellular spaces (van Loon 1985). PRs are found in almost all organs, including roots, flowers, and stems with maximum abundance levels in the leaves (van Loon and Van Strien 1999). To date, there are 17 families of structurally and functionally unrelated PR-proteins where they are numbered sequentially based on the time of discovery (van Loon et al. 2006). The PR-proteins are strongly induced by infections. Their expression and accumulation is strongly linked with the type of pathogen and the type of plant tissues being infected. They are produced at the infection site, surrounding tissues and systemic tissues further away from the infected site. Production in uninfected parts of the plants will protect the affected plants from further infection. Acidic PR proteins are found in the intercellular space, whereas the basic forms are transported to the vacuole by C-terminal signals (van Loon et al. 2006). Proteins which have sequence homology with PR proteins, but are constitutively expressed in some tissues or during specific developmental stages are referred to as PR-like proteins (Linthorst et al. 1990).
Several families of PR proteins, including PR-1, PR-2 proteins (β-1,3-glucanases), PR-3 and PR-4 proteins (chitinases), thaumatin-like proteins and osmotins (PR-5), peroxidases (PR-9), defensins (PR-12), and thionin (PR-13) exhibit antifungal activities which were mostly demonstrated through in vitro studies suggesting their involvement in plant defense. Since both β-1,3-glucans and chitins are the main constituents of fungal cell wall, co-induction for a coordinated expression of β-1,3-glucanase and chitinase following infection has been reported in many plant species. Thaumatin-like proteins and osmotins are believed to be involved in creating transmembrane pores, while, during resistance responses, peroxidases have been linked with accumulation of phenolics in plant cell walls. Induction of peroxidase activities in infected plants is associated with phenolics-mediated resistance against necrotrophs (van Loon et al. 2006).
Functional characterization to demonstrate the antifungal properties of PR-proteins, either using overexpressed recombinant proteins or transgenic systems has been done. For example, rice overexpressing thaumatin-like protein demonstrated increased resistance to Rhizoctonia solani (Datta et al. 1999), while thaumatin-like protein, chitinase, and glucanase were stably expressed in wheat for resistance against the devastating pathogen, Fusarium graminearum (Anand et al. 2003). There is interest in using PR-proteins as natural substances to protect plants from pathogen infection; however, it should be realized that some PR-proteins cause allergic reactions in humans (Hoffmann-Sommergruber 2002). The PR-proteins are also being used to develop biomarkers for defense response of specific plant pathogen and tissue interaction.
4.4.2 Phytoalexins
Phytoalexins are antimicrobial secondary metabolites that are of low molecular weight, synthesized and accumulated in plants when exposed to microorganisms. For example, in response to necrotrophic fungal pathogens, camalexin is produced in Arabidopsis, zealexin in maize, scopoletin in tobacco, and resveratrol in grapevine (Ahuja et al. 2012). Phytoalexins play a role in disease resistance (Dixon 2001) and most of the evidence has been obtained based on the studies done on camalexin, an indole derivative of tryptophan which is one of the most widely studied phytoalexins. It was initially isolated from the Camelina sativa leaves infected by Alternaria brassica (Browne et al. 1991). In Arabidopsis, it is a major phytoalexin, where pad2, pad3, bos2, bos4, and esa1 mutants which are defective in camalexin synthesis and accumulation showed susceptibility towards the different pathogens (Glazebrook et al. 1997; Veronese et al. 2004). Induced synthesis of camalexin occurs at the site of infection by necrotrophs. It exhibits antimicrobial activity similar to the systemic fungicide thiabedazole. High concentrations of camalexin induce ion leakage and inhibit proline uptake resulting in the disruption of pathogen membrane (Pedras et al. 2006).
The necrotrophic fungus Alternaria alternata causes major damages in Nicotiana species. It infects mature tobacco leaves; however, young leaves are highly resistant to the pathogen. Scopoletin, a phenolic coumarin, which produces strong blue fluorescence under ultraviolet light, is synthesized through the phenylpropanoid pathway (Kai et al. 2008). These secondary metabolites can be isolated from Nicotiana and several other plant species (Gnonlonfin et al. 2012). In both in vitro and in vivo conditions in tobacco, scopoletin exhibited strong antifungal activity against A. alternata and it is recognized as an important phytoalexins against this pathogen. A. alternata induced blue fluorescence in N. attenuata leaves due to production of scopoletin and scopolin. The young wild tobacco leaves produces more scopoletin in response to attack by A. alternata and the synthesis was dependent on jasmonic acid signaling. Thus, the higher level of accumulated scopoletin was the main contribution to the resistance against the A. alternata in young leaves (Sun et al. 2014). Similarly, in the oil palm it was observed that the tolerant progenies produced higher percentages of defense-related secondary metabolites such as sterols and tocopherols than the susceptible progenies when treated with the necrotrophic fungal pathogen, G. boninense (Nusaibah et al. 2011).
Grapevine is susceptible to B. cinerea and produces resveratrol (an antifungal phytoalexins) as the result of its defense machinery. Zheng et al. (2011) showed that resveratrol impedes germination of the fungal conidium, thus preventing it from penetrating the host cell wall. The researchers also carried out gene-expression profiling using LongSAGE (long serial analysis of gene expression) libraries constructed from B. cinerea conidia treated in vitro with resveratrol in comparison to non-treated germlings in order to determine the influence of the phytoalexins on transcriptional regulation in B. cinerea germlings. Functional categorization of differentially expressed genes demonstrated that primary metabolism of the germinating conidia was strongly affected by resveratrol treatment, while concomitant induction of putative metabolic pathway genes for secretion of virulence-effector that disrupt the plant barriers and detoxify the resveratrol phytoalexin was observed. It was believed that the huge amount of virulence-effector being produced is the key to the eventual success of the pathogen.
4.5 Roles of Phytohormones
Plant hormone biosynthesis and signaling are important in modulating plant response to necrotrophic pathogens. The interactive roles of jasmonic acid (JA), ethylene (ET), and salicylic acids (SA) in defense signaling and plant immune response are well established, however, in recent years, the important contributions of other phytohormones such as abscisic acid (ABA), gibberellic acid (GA), auxins, cytokinins, and brassinosteroids in regulating plant defense response either alone or in conjunction with JA, ET, or SA have been discovered (Robert-Seilaniantz et al. 2011). Resistance and susceptibility to pathogens are affected by pathological conditions created through changes in plant hormonal homeostasis. JA and ET works in synergy in defense against necrotrophs, in direct antagonism to SA-mediated defense.
Infection with certain pathogens or wounding, induce the synthesis of JA and ET followed by activation of their signaling pathways. Necrotrophic fungal infection promotes ET production in the host plant. Ethylene biosynthesis involves conversion of S-adenosyl-l-methionine to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase and ACC is converted to ET by ACC oxidase. There are overwhelming data implicating JA and ET in resistance to necrotrophs. For example, Arabidopsis mutants affecting genes involved in JA and ET biosynthesis or signaling such as fad3/fad7/fad8, ein2, ein3, and eil1 mutants are more susceptible to infection by necrotrophs (Stintzi et al. 2001; Pre et al. 2008). The molecular components of ET response pathways and factors involved in their regulation contribute to plant immune responses to necrotrophic infection. Many components of the ET-response pathway such as EIN3 and EIL1 are known to have a defense function against necrotrophs where loss of function mutations led to disease manifestations (Alonso et al. 2003; van Wees et al. 2003).
The fad3/fad7/fad8 triple mutant lacking in JA which showed enhanced susceptibility to Pythium mastophorum (Vijayan et al. 1998) led to the discovery of the functional role of JA in defense response. A common occurrence in plants following injury is peroxidation of free linolenic acid catalyzed by 13-lipoxygenases, then allene oxide synthase-mediated epoxide formation followed by cyclization by allene oxide cyclase (AOC), and finally three rounds of β-oxidation to produce JA (Wasternack and Hause 2013). JA and its derivatives are well accepted as key regulators of defense mechanism against fungal pathogens. CORONATINE INSENSITIVE1 (COI1), an F-box protein is the JA-receptor essential for most JA-dependent responses. It was implicated in B. cinerea and A. brassicicola induced gene expression and resistance to necrotrophic pathogens (van Wees et al. 2003; AbuQamar et al. 2006). The strong resistance to B. cinerea and A. brassicicola displayed by Arabidopsis rst1 mutant was dependent on COI1 (Mang et al. 2009). The increase in the levels of JA-biosynthesis and JA responsive gene expression and elevated cuticular lipids were the phenotypes displayed by rst1.
The extensive interactions among the plant hormones enable fine-tuning of host immune responses towards specific pathogen. The Arabidopsis dde2/ein2/pad4/sid2 quadruple mutant with defects affecting essential components of the different phytohormone signaling pathways was studied in order to evaluate the contributions of JA, ET and SA to immunity against A. brassicicola. Interestingly, it was found that JA, ET, and SA signaling components contribute positively to immunity against A. brassicicola (Tsuda et al. 2009). Such information on the complex interaction is valuable and it could not be derived from single-mutant analysis.
4.5.1 Intervention of Phytohormone Balance and Function by Necrotrophs
Manipulation of host phytohormone biosynthesis and/or signaling pathways for defense is often employed by most pathogens. Many of these pathogens can synthesize the different phytohormones affecting the balance and functions of the endogenous plant hormones. Some pathogens manipulate the phytohormone signaling pathways essential for physiological and/or developmental features, for example stomatal opening and senescence in order to ease their entry and disease symptom development in the hosts (Melotto et al. 2008). Molecules that resemble phytohormones or phytohormone signaling components in structure, function, or both, referred to as “phytohormone mimics,” are also used by pathogens to trick the hosts.
SA and JA modulate downstream defenses when attack by pathogens. To counter the effects, the pathogens often manipulate phytohormone cross talk to promote disease development. It is well known that the pathogens exploit the SA and JA cross talk when infecting plants (Thaler et al. 2012). For example, B. cinerea secretes β-(1,3)(1,6)-d-glucan as an exopolysaccharide (EPS) effector molecule which contributes to disease development by exploiting the SA-JA antagonism. Non-expressor of pathogenesis-related gene1 (NPR1) is a master regulator of SA signaling. Enhanced susceptibility to B. cinerea was observed in tomato plants pretreated with the EPS. It was suggested that EPS acts by activation of the SA pathway through NRP1, JA signaling was then suppressed (El Oirdi et al. 2011).
4.5.2 Cell Wall, Cuticles, and Phytohormone Interplay
The plant cell wall and cuticle serve as a physical barrier for inhibiting infection by necrotrophs. The severity of disease in the host is closely related to the susceptibility of the cell wall to degradation by CWDEs. Cellulose and pectins are attacked by the fungal enzymes to facilitate penetration. Disruption of cell wall integrity upregulates the biosynthetic machinery of the cell wall to maintain its integrity. It provides PAMPs/DAMPs and signals for recognition by the host. The cell wall is equipped with an integrity sensory system for activating the intracellular signaling cascades to induce the multitude of host defense responses. Phytohormones influence cell wall composition and structure. This affects the ability of necrotrophs to digest the cell wall and the cell wall capacity in generating DAMPs which will be detected by the cell wall integrity sensory systems. When the host sensed the degradation of cell walls by the invading pathogen through DAMP, the signaling cascades involving phytohormones cross talks leading to induced or increased expression of defense related genes are activated.
Cuticle serves as a platform for activating immune responses. It facilitates the host in sensing and signaling early irritations by the invading necrotrophic pathogen. Altered cuticle development and composition involving cuticular lipid and cutin polymer profile can enhance resistance to B. cinerea through faster recognition of fungal elicitors, better diffusion of defense signals to the infected site, and faster oxidative burst to counteract the pathogen virulence (L’Haridon et al. 2011; Mang et al. 2009).
Short fragments of OGs are released through the degradation of pectin homogalacturonan in plant cell wall by the polygalacturonase enzyme secreted by necrotrophs during infection. Mechanical damage can also induce the expression of endogenous polygalacturonases (PG) that is involved in the production of OGs (Orozco-Cardenas and Ryan 1999). OGs are DAMPs effective in eliciting defense responses (Cervone et al. 1989). A multitude of defense responses in plants, including accumulation of antimicrobial secondary metabolites or phytoalexins (Davis et al. 1986), expression of PR genes, including chitinase and glucanase (Broekaert and Pneumas 1988), deposition of callose, generation of ROS (Ridley et al. 2001; Denoux et al. 2008) and nitric oxide are induced by OGs. The extent of pectin methylation effects PG hydrolysis and the length of OGs produced and this influence host defense (De Lorenzo et al. 2001; Lionetti et al. 2007). It was found that OGs with a degree of polymerization between 10 and 15 are optimal for inducing the plant defense responses (Ferrari et al. 2013).
The cell wall-phytohormone homeostasis is a highly integrated system that is important in plant–necrotrophic pathogen interaction (Nafisi et al. 2014). Resistance can be produced through mutation of the member of the CesA family of cellulose synthase subunits. It was shown that the mutation led to an increase in the level of endogenous ABA and expression of ABA-responsive genes (Hernandez-Blanco et al. 2007). Further, cev1 mutant, which is cellulose deficient elicits ET and JA signaling which results in improved resistance to pathogens (Ellis et al. 2002a, b). Auxin has an antagonistic effect on OGs and vice versa. It was found that auxin can abolish the effects of improved resistance of tobacco plants to B. cinerea conferred by a fungal endo-PG, which most probably through the constitutive production of OGs (Ferrari et al. 2008). The catalytic activity of fungal PGs can be inhibited by polygalacturonase inhibiting proteins (PGIPs) expressed in plants (Sicilia et al. 2005). PGIPs can limit the degradation of homogalacturonan by fungal PG, hence reducing the generation of the elicitor-active OGs (Ferrari et al. 2013). Transgenic tobacco overexpressing a grapevine PGIP (Vvpgip1) had a higher level of IAA and higher content of lignins and a decreased level of xyloglucan endotransglycosylase activity (Alexandersson et al. 2011). It was revealed through detailed cell wall analysis that overexpression of Vvpgip1 leads to constitutive compositional changes. Thus, suggesting that PGIP-induced initial changes in the cell wall. This leads to altered auxin accumulation and stress responses, resulting in additional cell wall structural and compositional changes.
4.6 Transcription Factors
The outcome of plant–pathogen interactions is strongly influenced by the transcriptional control of gene expression by the specific transcription factors. The transcription factors specifically bind to cis-acting elements in the promoters of genes involved in immune response pathways of plants, enabling coordinated and precise timing of their expression in response to infection by necrotrophs. These include genes involved in stress signaling, cell death, cell wall dynamics, and biosynthesis of phytohormones and secondary metabolites. Since resistance and susceptibility of host are dependent on the speed and level of expression of the immune response pathway genes, transcription factors have great impact on plant defense. Certain families of plant transcription factors are involved in plant defense to microbial pathogens; for example, different transcription factor families, including ERFs, WRKY, MYB, zinc-finger, and HD-ZIP, are induced in response to B. cinerea infection (AbuQamar et al. 2006). WRKY33, MYB, ERF1 and ERF104, ZFAR1, AS1, and HD-ZIP homeodomain (Smith et al. 2014) are important for resistance to necrotrophic fungi.
The WRKYs are plant specific transcription factors believed to be the intermediate signaling components of the various PTI responses (Asai et al. 2002). WRKY33 and WRKY70 are required for responses to B. cinerea, suggesting a potential role in plant resistance (AbuQamar et al. 2006; Zheng et al. 2006). Arabidopsis WRKY33 was shown to be a major regulator of immune responses to necrotrophic fungi based on analysis of loss-of-function mutants and interactions with other PTI pathway components. It is involved in regulating expression of camalexin (Qiu et al. 2008; Mao et al. 2011), an antimicrobial secondary metabolite secreted by the plants at the early stage of infection and the autophagy gene ATG18a (Lai et al. 2011a). Arabidopsis AtWRKY33 interacts with the Sigma Factor interacting proteins 1 and 2 (SIB1 and SIB2) whose production are induced by the necrotrophic fungus B. cinerea. Deletion mutants (SIB1 and SIB2) showed a decrease in plant resistance supporting their role as AtWRKY33 activators (Lai et al. 2011b). From 16 canola (Brassica napus) BnWRKY genes assayed, 13 BnWRKY were responsive to hormonal treatment as well as Sclerotinia sclerotiorum and Alternaria brassicae, the fungal pathogen causing stem rot and Alternaria black spot, respectively. This suggests that in response to hormonal stimuli and fungal pathogen, a large proportion of BnWRKY proteins play a role in the transcriptional regulation of defense-related genes (Yang et al. 2009).
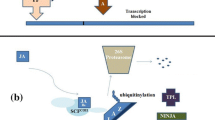
Many transcription factors regulate transcription of genes in the JA and ET pathways and affecting resistance to necrotrophs. Selective binding of the MYB-related gene ASYMMETRIC LEAVES 1 (AS1) to the promoters of JA-regulated genes (Nurmberg et al. 2007) suppressed inducible resistance against B. cinerea while the loss of AS1 function, enhanced resistance in Arabidopsis to B. cinerea. MYB46 suppresses resistance to B. cinerea through transcriptional reprogramming of genes for cell wall proteins and enzymes (Ramirez et al. 2011). Many MYB proteins regulate the expression of genes involved in the production of secondary metabolites. For example, MYB51 regulates the transcriptional activation of genes for the biosynthesis of indole glucosinolate (Clay et al. 2009), a tryptophan-derived secondary metabolite that contributes to resistance to necrotrophic fungi (Bednarek et al. 2009; Kliebenstein et al. 2005; Sanchez-Vallet et al. 2010). BOTRYTIS SUSCEPTIBLE 1 (BOS1) that encodes an R2R3 MYB was shown to restrict necrosis caused by necrotrophic pathogen. BOS1 and BOTYRYTIS SUSCEPTIBL E1 INTERACTOR (BOI), a ligase that restricts cell death may integrate plant response to various signals caused by stress factors.
The necrotrophic fungus Gaeumannomyces graminis var. tritici (Ggt) causes a devastating root disease in wheat. At least six wheat defense-related genes showed significant upregulated expression in the TiMYB2R-1 transgenic wheat lines. It was shown that enhanced resistance to the disease which is known as take-all was achieved in the TiMYB2R-1-overexpressing wheat lines (Liu et al. 2013).
ERF belongs to AP2/ERF superfamily of transcription factors which is responsive to ET. ERF binds to the ethylene-responsive element (ERE), also known as the GCC-box (AGCCGCC) a common promoter element found in ethylene-inducible defense genes. In Arabidopsis, a subgroup known as octadecanoid-responsive Arabidopsis AP2/ERF (ORA) is rapidly induced by JA. Many of the ORA transcription factors play important roles in disease resistance involving JA and ethylene signaling pathways. Infection with certain pathogens or wounding induces the synthesis of JA and ethylene followed by activation of their signaling pathways. The expression of ORAs is activated by JA and or related oxylipins via COI1 which serves as the central regulator of all JA-dependent responses (Garcia-Marcos et al. 2013)
ORA59 is involved in increasing expression of several defense genes such as PDF1.2 and ChiB and β-glucosidase 2 (BGL2) and its overexpression results in increased resistance against the necrotrophic fungus B. cinerea. The ORA37 transcription factor is different from the other ORAs because it contains an ERF-associated amphiphilic repression (EAR) motif at its C-terminal region that functions as a transcriptional repressor (Otha et al. 2001). The induction of a subset of JA- and ethylene-responsive genes was significantly lower in plants overexpressing ORA37 including the defense genes PDF1.2, HEL, and ChiB when treated with JA or ET (Atallah 2005).
Rhizoctonia solani is a fungal pathogen that has led to significant economic losses to diverse range of crops including cereals, canola, and legumes. Root specific expression of a Medicago truncatula ERF transcription factor MtERF1-1 in Medicago enhanced resistance to R. solani and Phytophthora medicaginis without adversely affecting symbiosis with Rhizobia (Anderson et al. 2010). The defense regulatory mechanism of ERF transcription factors may involve interaction with other transcription factors, for example, the interaction observed between bZIP transcription factor and ERF72 for regulating expression of a PR-1 type protein from tobacco (Büttner and Singh 1997; Alves et al. 2013). The MYC2 has been well characterized where in Arabidopsis it was shown to differentially regulate the expression of JA-responsive pathogen defense and wound response (VSP) genes (Dombrecht et al. 2007). Expression of PDF1.2, ChiB, and HEL are regulated by JA and ET-signaling pathways during necrotrophic fungal infection. Their induced expression is negatively regulated by AtMYC2 indirectly via negative regulation of expression of ERF1 transcription factors. We have summarized the various transcription factors involved in regulating defense response in tabular form (Table 1).
4.7 Chromatin and Histone Modifications
Alterations in the expression of genes due to processes not associated with changes in the underlying DNA sequence are referred to as epigenetics. The processes include DNA methylation, histone modification, and nucleosome remodeling which alters the dynamics of the chromatin, a highly compacted organized complex of DNA and histone proteins. Histone modifications alter the net charge of nucleosomes and these affect DNA histone and histone–histone interactions at the inter- or intra-nucleosome level.
The chromatin states in eukaryotic nuclei are influenced by the different combinations of posttranslational modifications of the histone proteins. The lysine residues of the histones in a eukaryotic chromatin can be covalently modified through acetylation, methylation, and mono-ubiquitination, resulting in an epigenome which plays an important role in the fine-tuning of transcription process. Acetylation of the lysine residues on histone 3 and histone 4 is often linked with active transcription. Histone acetyltransferases (HATs) catalyze the addition of an acetyl group to multiple lysine residues and this is a dynamic process where changes occur over time. The opposing action is catalyzed by HDACs. The antagonistic action between the two families of enzymes maintained the homeostatic balance of histone acetylation for proper cellular function and differentiation (Jeon et al. 2014).
4.7.1 Fungal Toxins as HDAC Inhibitors
The toxins produced by the necrotrophic fungal pathogens C. carbonum and A. brassicicola inhibit HDACs. Thus, the toxins are targeting chromatin modifying enzymes possibly to interfere with the transcriptional activity of immune response genes. The HC-toxin is an HST secreted by C. carbonum. The toxin specifically inhibits maize HDACs, but does not have an effect on HATs. It was proposed that the HC-toxin helps the pathogen to establish compatibility with maize via the interference of HDACs (Brosch et al. 1995). A. brassicicola produces depudecin as a virulence factor which is also a HDAC inhibitor (Kwon et al. 1998; Matsumoto et al. 1992). Induced HDAC19 expression in A. thaliana infected with A. brassicicola correlates with changes in expression of JA and ET-regulated genes (Zhou et al. 2005). Thus, it is possible that plant HDACs are conserved regulators of defense-related genes in plants and inhibition of their activities enable the pathogens to disrupt the defense response transcriptional programs of host plants (Jeon et al. 2014).
4.7.2 Epigenetic Control of Defense Priming
It has been hypothesized that chromatin modification primes the defense genes for more rapid and robust activation (Bruce et al. 2007). This led to the concept of epigenetic control of defense priming. SET DOMAIN GROUP 8 (SDG8)-mediated methylation of histone H3 lysine 6 in the promoters of JA-inducible defense genes accompanied the induction of expression of these genes in response to fungal infection (Berr et al. 2010). This enables a long-lasting priming of JA-dependent defense genes against future infection by the necrotrophs. Priming of the WRKY transcription factor 29 gene (WRKY29) with benzothiadiazole is associated with the trimethylation and acetylation of specific lysine of the histone proteins in the WRKY29 gene promoter (Jaskiewicz et al. 2011). This allows the activation of the expression of the WRKY29 gene when the plants were confronted with further stress stimulus. Thus, defense priming essentially involves chromatin modification of promoters of defense genes before true activation of these genes. The modifications in the chromatin could loosen histone–DNA interaction to allow access to the open chromatin structure of the various transcription co-activators, effector proteins, and chromatin remodeling factors (Alvarez-Venegas 2013).
5 Conclusions and Future Prospects
The mechanisms of infection, including secretion of the phytotoxins and cell wall degrading enzymes and strategies in mimicking components of plant defense by pathogen and the series of plant immune response pathways to counteract the effects are well studied for model crops involving certain pathogens. The information needs to be translated to other crops of economic importance and peculiarities of specific pathogen plant interaction must be understood. It is important to have an early intervention strategy to protect the plant following initial plant–pathogen interaction due to the notorious nature of the necrotrophic fungal pathogens. Often the control measures taken were too late to save the plant as when the physical symptoms are recognized, the toxins and reactive oxygen species had killed the cells in the important tissues and organs essential for the plants survival. Thus, work on predisposing factors and strategies for early disease diagnosis before the appearance of symptoms needs to be developed. For these purposes early inducible defense response, including components of the signaling cascades that have been discussed and early defense response secondary metabolites such as the phytoalexins are valuable targets and their roles in inhibiting fungal ingress is a key area of research to focus on for producing disease resistant plants. It is clear that the plant immune response is complex, involving the interplay of various phytohormones, cell wall components, and production of different antimicrobial molecules playing their roles at the different infection phases from pathogen entry to disease development. This is the result of the transcriptional reprogramming that occurs in response to the different signaling molecules. Mechanisms of transcriptional controls addressed through systems approach via omics platform which had enabled identification of the key pathways of pathogenic attack and plant defense response should be emphasized. This would lead to the identification of different compounds and proteins with functional and regulatory roles covering both transcriptional and translational processes. Chromatin modifications that fine-tune transcriptional regulation of immune responses through histone modification are also being targeted by the pathogen to overcome the plant’s immune response. Epigenetic control of defense priming enables the plants to be more prepared for recurrent attack and should be an important area of research for a holistic and effective approach in sustaining crop survival and productivity.
References
Abbas HK, Tanaka T, Duke SO, Porter JK, Wray EM, Hodges L, Sessions AE, Wang E, Merrill AHJ, Riley RT (1994) Fumonisin and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol 106:1085–1093
AbuQamar S, Chen X, Dahwan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J 48:28–44
Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17:73–90
Alexandersson E, Becker JV, Jacobson D, Nguema-Ona E, Steyn C, Denby KJ, Vivier MA (2011) Constitutive expression of a grapevine polygalacturonase inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Res Notes 4:493
Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci U S A 100:2992–2997
Alvarez-Venegas R (2013) Chromatin modifications and plant immunity in Phaseolus vulgaris L. J Plant Biochem Physiol 1:3
Alves MS, Dadalto SP, Gonçalves AB, De Souza GB, Barros VA, Fietto LG (2013) Plant bZIP transcription factors responsive to pathogens: a review. Int J Mol Sci 14:7815–7828
Anand A, Zhou T, Trick HN, Gill BS, Bockus WW, Muthukrishnan S (2003) Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot 54:1101–1111
Anderson JP, Lichtenzveig J, Gleason C, Oliver RP, Singh KB (2010) The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia. Plant Physiol 154:861–873
Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983
Atallah M (2005) Jasmonate-responsive AP2-domain transcription factors in Arabidopsis. Ph.D. thesis, Leiden University, Leiden, The Netherlands
Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze-Lefert P (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323:101–106
Berr A, McCallum EJ, Alioua A, Heintz D, Heitz T et al (2010) Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol 154:1403–1414
Broekaert WF, Pneumas WJ (1988) Pectic polysaccharides elicit chitinase accumulation in tobacco. Physiol Plant 74:740–744
Brosch G, Ransom R, Lechner T, Walton JD, Loidl P (1995) Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7:1941–1950
Browne LM, Conn KL, Ayert WA, Tewari JP (1991) The camalexins: new phytoalexins produced in the leaves of Camelina sativa (cruciferae). Tetrahedron 47:3909–3914
Bruce TJA, Matthes MC, Napier JA, Pickett JA (2007) Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173:603–608
Büttner M, Singh K (1997) Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci U S A 94:5961–5966
Cervone F, Hahn MG, Delorenzo G, Darvill A, Albersheim P (1989) Host-pathogen interactions. A plant protein converts a fungal pathogenesis factor into an elicitor of plant defense responses. Plant Physiol 90:542–548
Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323:95–101
Correa-Aragunde N, Lombardo C, Lamattina L (2008) Nitric oxide: an active nitrogen molecule that modulates cellulose synthesis in tomato roots. New Phytol 179:386–396
Crawford NM, Guo FQ (2005) New insights into nitric oxide metabolism and regulatory functions. Trends Plant Sci 10:195–200
Datta K, Velazhahan R, Oliva N, Ona I, Mew T, Khush GS, Muthukrishnan S, Datta SK (1999) Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet 98:1138–1145
Davis KR, Darvill AG, Albersheim P, Dell A (1986) Host–pathogen interactions. XXIX. Oligogalacturonides released from sodium polypectate by endopolygalacturonic acid lyase are elicitors of phytoalexins in soybean. Plant Physiol 80:568–577
De Lorenzo G, D’Ovidio R, Cervone F (2001) The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39:313–335
Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J (2008) Activation of defense response pathways by OGS and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 3:423–445
Dixon RA (2001) Natural products and plant disease resistance. Nature 411:843–847
Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, Kazan K (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245
El Oirdi M, El Rahman TA, Rigano L, El Hadrami A, Rodriguez MC, Daayf F, Vojnov A, Bouarab K (2011) Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23:2405–2421
Elad Y, Williamson B, Tudzynski P, Delen N (2004) Botrytis: biology, pathology and control. Kluwer, Dordrecht, p 416
Ellis C, Karafyllidis I, Turner JG (2002a) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant Microbe Interact 15:1025–1030
Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002b) The Arabidopsis mutant Cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14:1557–1566
Fahimeh A, Siti Nor Akmar A, Alireza K, Faridah A, Umi KY, Pei Pei C (2011) Differential expression of oil palm pathology genes during interactions with Ganoderma boninense and Trichoderma harzianum. J Plant Physiol 168:1106–1113
Faris JD, Zhang Z, Lu H, Lu S, Reddy L, Cloutier S, Fellers JP, Meinhardt SW, Rasmussen JB, Xu SS, Oliver RP, Simons KJ, Friesen TL (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci U S A 107:13544–13549
Ferrari S, Galletti R, Pontiggia D, Manfredini C, Lionetti V, Bellincampi D, Cervone F, De Lorenzo G (2008) Transgenic expression of a fungal endopolygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol 146:669–681
Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, Lorenzo GD (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci 4:49
Freschi L (2013) Nitric oxide and phytohormone interactions: current status and perspectives. Front Plant Sci 4:398
Friesen TL, Chu CG, Liu ZH, Xu SS, Halley S, Faris JD (2009) Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor Appl Genet 118:1489–1497
Garcia-Marcos A, Pacheco R, Manzano A, Aguilar E, Tenllado F (2013) Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair Potato virus X-Potato virus Y and tomato spotted wilt virus. J Virol 87:5769–5783
Glazebrook J, Zook M, Mert F, Kagan I, Rogers EE, Crute IR, Holub EB, Hammerschmidt R, Ausubel FM (1997) Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146:381–392
Gnonlonfin GJB, Sanni A, Brimer L (2012) Review scopoletin—a coumarin phytoalexin with medicinal properties. Crit Rev Plant Sci 31:47–56
Govrin EM, Levine A (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10:751–757
Hernandez-Blanco C, Feng DX, Hu J, Sanchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, Anderson LK, Somerville S, Marco Y, Molina A (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19:890–903
Hoffmann-Sommergruber K (2002) Pathogenesis-related (PR)-proteins identified as allergens. Biochem Soc Trans 30:930–935
Jaskiewicz M, Conrath U, Peterhänsel C (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12:50–55
Jeon J, Kwon S, Lee YW (2014) Histone acetylation in fungal pathogens of plants. Plant Pathol J 30:1–9
Johal GS, Briggs SP (1992) Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258:985–987
Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguchi H, Sakata K, Shimizu B (2008) Scopoletin is biosynthesized via orthohydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J 55:989–999
Kariola T, Brader G, Li J, Palva ET (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17:282–294
Kazan K, Lyons R (2014) Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26:2285–2309
Kliebenstein DJ, Rowe HC, Denby KJ (2005) Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J 44:25–36
Kuzniak E, Sklodowska M (2005) Fungal pathogen-induced changes in the antioxidant systems of leaf peroxisomes from infected tomato plants. Planta 222:192–200
Kwon HJ, Owa T, Hassig CA, Shimada J, Schreiber SL (1998) Depudecin induces morphological reversion of transformed fibroblasts via the inhibition of histone deacetylase. Proc Natl Acad Sci U S A 95:3356–3361
L’Haridon F, Besson-Bard A, Binda M, Serrano M, Abou-Mansour E, Balet F, Schoonbeek HJ, Hess S, Mir R, Léon J, Lamotte O, Métraux JP (2011) A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog 7, e1002148
Lai Z, Wang F, Zheng Z, Fan B, Chen Z (2011a) A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J Cell Mol Biol 66:953–968
Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z (2011b) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23:3824–3841
Linthorst HJ, van Loon LC, van Rossum CM, Mayer A, Bol JF, van Roekel JS, Meulenhoff EJ, Cornelissen BJ (1990) Analysis of acidic and basic chitinases from tobacco and petunia and their constitutive expression in transgenic tobacco. Mol Plant Microbe Interact 3:252–258
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Liu X, Yang L, Zhou X, Zhou M, Lu Y, Ma L, Ma H, Zhang Z (2013) Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. J Exp Bot 64:2243–2253
Lyon GD, Goodman BA, Williamson B (2004) Botrytis cinerea perturbs redox processes as an attack strategy in plants. In: Elad Y, Williamson B, Tudzynski P, Delen N (eds) Botrytis: biology, pathology and control. Kluwer, Dordrecht, pp 119–141
Mamgain A, Roychowdhury R, Tah J (2013) Alternaria pathogenicity and its strategic controls. Res J Biol 1:1–9
Mang HG, Laluk KA, Parsons EP, Kosma DK, Cooper BR, Park HC, AbuQamar S, Boccongelli C, Miyazaki S, Consiglio F, Chilosi G, Bohnert HJ, Bressan RA, Mengiste T, Jenks MA (2009) The Arabidopsis RESURRECTION1 gene regulates a novel antagonistic interaction in plant defense to biotrophs and necrotrophs. Plant Physiol 151:290–305
Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23:1639–1653
Matsumoto M, Matsutani S, Sugita K, Yoshida H, Hayashi F, Terui Y, Nakai H, Uotani N, Kawamura Y, Matsumoto K, Shoji J, Yoshida T (1992) Depudecin: a novel compound inducing the flat phenotype of NIH3T3 cells doubly transformed by ras- and src-oncogene, produced by Alternaria brassicicola. J Antibiot 45:879–885
Meinhardt LW, Costa GGL, Thomazella DPT, Teixeira PJPL, Carazzolle MF, Schuster SC, Carlson JE, Guiltinan MJ, Mieczkowski P, Farmer A, Ramaraj T, Crozier J, Davis RE, Shao J, Melnick RL, Pereira GAG, Bailey BA (2014) Genome and secretome analysis of the hemibiotrophic fungal pathogen, Moniliophthora roreri, which causes frosty pod rot disease of cacao: mechanisms of the biotrophic and necrotrophic phases. BMC Genomics 15:164
Melotto M, Underwood HW, He SY (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol 46:101–122
Mengiste T (2012) Plant immunity to necrotrophs. Annu Rev Phytopathol 50:267–294
Morrissey JP, Osbourn AE (1999) Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol Mol Biol Rev 63:708–724
Mur LA, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, Hall MA, Harren FJ, Hebelstrup KH, Gupta KJ (2013) Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants 5:pls052
Nafisi M, Stranne M, Zhang L, van Kan JAL, Sakuragi Y (2014) The endo-arabinanase BcAra1 is a novel host-specific virulence factor of the necrotic fungal phytopathogen Botrytis cinerea. Mol Plant Microbe Interact 27:781–792
Nafisi M, Fimognari L, Sakuragi Y (2015) Interplays between the cell wall and phytohormones in interaction between plants and necrotrophic pathogens. Phytochemistry 112:63–71
Nagy ED, Lee TC, Ramakrishna W, Xu Z, Klein PE, SanMiguel P, Cheng CP, Li J, Devos KM, Schertz K, Dunkle L, Bennetzen JL (2007) Fine mapping of the Pc locus of Sorghum bicolor, a gene controlling the reaction to a fungal pathogen and its host-selective toxin. Theor Appl Genet 114:961–970
Nurmberg PL, Knox KA, Yun BW, Morris PC, Shafiei R, Hudson A, Loake GJ (2007) The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc Natl Acad Sci U S A 104:18795–18800
Nusaibah SA, Siti Nor Akmar A, Mohamad Pauzi Z, Idris AS, Sariah M (2011) Detection of phytosterols in Ganoderma boninense-infected oil palm seedlings through GC-MS analysis. J Oil Palm Res 23:1069–1077
Okubara PA, Paulitz TC (2005) Root defense responses to fungal pathogens: a molecular perspective. Plant Soil 274:215–226
Oliver RP, Ipcho SV (2004) Arabidopsis pathology breathes new life into the necrotrophs-vs-biotrophs classification of fungal pathogens. Mol Plant Pathol 5:347–352
Orozco-Cardenas M, Ryan CA (1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci U S A 96:6553–6655
Otha M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13:1959–1968
Ottmann C, Luberacki B, Kufner I, Koch W, Brunner F, Weyand M, Mattinen L, Pirhonen M, Anderluh G, Seitz HU, Nurnberger T, Oecking C (2009) A common toxin fold mediates microbial attack and plant defense. Proc Natl Acad Sci U S A 106:10359–10364
Pedras MSC, Sarwar MG, Suchy M, Adio AM (2006) The phytoalexins from cauliflower, caulilexins A, B and C: isolation, structure determination, syntheses and antifungal activity. Phytochemistry 67:1503–1509
Pre M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147:1347–1357
Prins TW, Tudzynski P, Tiedemann AV, Tudzynski B, Ten Have A, Hansem ME, Tenberge K, van Kan JAL (2000) Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In: Kronstad JW (ed) Fungal pathology. Kluwer, Dordrecht, pp 33–64
Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, Brodersen P, Grasser KD, Mattsson O, Glazebrook J, Mundy J, Petersen M (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27:2214–2221
Qutob D, Kemmerling B, Brunner F, Kufner I, Engelhardt S, Gust AA, Luberacki B, Seitz HU, Stahl D, Rauhut T, Glawischnig E, Schween G, Lacombe B, Watanabe N, Lam E, Schlichting R, Scheel D, Nau K, Dodt G, Hubert D, Gijzen M, Nürnberger T (2006) Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18:3721–3744
Ramirez V, Agorio A, Coego A, Garcia-Andrade J, Hernandez MJ, Balaguer B, Ouwerkerk PB, Zarra I, Vera P (2011) MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol 155:1920–1935
Ridley BL, O’Neill MA, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57:929–967
Robert-Seilaniantz A, Grant M, Jones JD (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49:317–343
Sanchez-Vallet A, Ramos B, Bednarek P, Lopez G, Pislewska-Bednarek M, Schulze-Lefert P, Molina A (2010) Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J 63:115–127
Sicilia F, Fernandez-Recio J, Caprari C, De Lorenzo G, Tsernoglou D, Cervone F, Federici L (2005) The polygalacturonase-inhibiting protein PGIP2 of Phaseolus vulgaris has evolved a mixed mode of inhibition of endopolygalacturonase PG1 of Botrytis cinerea. Plant Physiol 139:1380–1388
Smith JE, Mengesha B, Tang H, Mengiste T, Bluhm BH (2014) Resistance to Botrytis cinerea in Solanum lycopersicoides involves widespread transcriptional reprogramming. BMC Genomics 15:334
Staats M, van Baarlen P, Schouten A, van Kan JA, Bakker FT (2007) Positive selection in phytotoxic protein-encoding genes of Botrytis species. Fungal Genet Biol 44:52–63
Stintzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci U S A 98:12837–12842
Sun H, Wang L, Zhang B, Ma J, Hettenhausen C, Cao G, Sun G, Wu J, Wu J (2014) Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signaling. J Exp Bot 65:4305–4315
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17:260–270
Thomma BPHJ (2003) Alternaria spp.: from general saprophyte to specific parasite. Mol Plant Pathol 4:225–236
Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5, e1000772
Van Kan JA (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci 11:247–253
van Loon LC (1985) Pathogenesis-related proteins. Plant Mol Biol 4:111–116
van Loon LC, van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
van Wees S, Chang HS, Zhu T, Glazebrook J (2003) Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol 132:606–617
Veronese P, Chen X, Bluhm B, Salmeron J, Dietrich R, Mengiste T (2004) The BOS loci of Arabidopsis are required for resistance to Botrytis cinerea infection. Plant J 40:558–574
Vijayan P, Shockey J, Levesque CA, Cook RJ, Browse J (1998) A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci U S A 95:7209–7214
Vleeshouwers VGAA, Oliver RP (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Interact 27:196–206
Walton JD (2006) HC-toxin. Phytochemistry 67:1406–1413
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review. Ann Bot 111:1021–1058
Wilkinson K, Grant WP, Green LE, Hunter S, Jeger MJ, Lowe P, Medley GF, Mills P, Phillipson J, Poppy GM, Waage J (2011) Infectious diseases of animals and plants: an interdisciplinary approach. Phil Trans R Soc B 366:1933–1942
Wolpert TJ, Dunkle LD, Ciuffetti LM (2002) Host-selective toxins and avirulence determinants: what’s in a name. Annu Rev Phytopathol 40:251–285
Xiong J, An L, Lu H, Zhu C (2009) Exogenous nitric oxide enhances cadmium tolerance of rice by increasing pectin and hemicellulose contents in root cell wall. Planta 230:755–765
Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22:508–522
Yang B, Jiang Y, Muhammad HR, Deyholos MK, Kav NNV (2009) Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L) in response to fungal pathogens and hormone treatments. BMC Plant Biol 9:68
Zhang ZY, Wang HH, Wang XM, Bi YR (2011) Nitric oxide enhances aluminum tolerance by affecting cell wall polysaccharides in rice roots. Plant Cell Rep 30:1701–1711
Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48:592–605
Zheng C, Choquer M, Zhang B, Ge H, Hu S, Ma H, Chen S (2011) LongSAGE gene-expression profiling of Botrytis cinerea germination suppressed by resveratrol, the major grapevine phytoalexin. Fungal Biol 115:815–832
Zhou C, Zhang L, Duan J, Miki B, Wu K (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17:1196–1204
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Abdullah, S.N.A., Akhtar, M.S. (2016). Plant and Necrotrophic Fungal Pathogen Interaction: Mechanism and Mode of Action. In: Hakeem, K., Akhtar, M., Abdullah, S. (eds) Plant, Soil and Microbes. Springer, Cham. https://doi.org/10.1007/978-3-319-27455-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-27455-3_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27453-9
Online ISBN: 978-3-319-27455-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)