Abstract
Strong evidence suggests a genetic susceptibility to suicidal behavior, including familial heritability and common occurrence in twins. In the recent advent of scientific research, the genome-wide association study (GWAS), an alternative to the candidate-gene approach, is widely utilized to examine hundreds of thousands of SNPs by high-throughput genotyping technologies. In addition to the candidate-gene approach, the GWAS approach has recently been employed to study the determinants of suicidal behavior. Several recent findings have demonstrated that some SNPs and genes are closely associated with suicidal behavior. This chapter addresses recent molecular genetic studies in suicidal behavior. First, we surveyed the SNPs and genes identified as genetic markers that are correlated and associated with suicidal behavior in the candidate-gene association studies. Next, we reviewed the SNPs and genes that have been suggested as contributors to suicidal behavior in the GWAS studies. Finally, we summarized the limitations and future directions. Future research with independent replication in large sample sizes is needed to confirm the role of the SNPs and genes identified in the candidate-gene and GWAS studies in suicidal behavior.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Bipolar disorder
- Genome-wide association study
- Major depressive disorder
- Genomics
- Single nucleotide polymorphisms
- Schizophrenia
- Suicidal behavior
1 Introduction
Numerous studies over the last three decades have reported that abnormalities in the functioning of the central serotonergic system are linked to the pathogenesis of suicidal behavior (Ryding et al. 2008). To date, most of the molecular genetic studies focused on the serotonergic pathway as the basis of established biological correlations of suicidal behavior, and thus, the candidate genes were primarily related to the serotonergic system (Bondy et al. 2006). Genetic association studies using genetic variants such as single nucleotide polymorphisms (SNPs) have shown that genes contribute to suicide risks and have suggested several genes such as serotonin transporter related to suicidal behavior, but not all reports support these findings (Tsai et al. 2011).
In addition, evidence from other research designs (such as adoption, family, geographical, immigrant, surname, and twin studies of suicide) suggested genetic contributions to suicide risk (Baldessarini and Hennen 2004). Further, the contribution of additive genetic factors is estimated to be 30–50 % for suicidal behavior including ideation, plans, and attempts (Voracek and Loibl 2007). Twin and family studies also documented a higher concordance rate for suicide in monozygotic than dizygotic twins (24.1 % vs. 2.8 %) and nearly fivefold greater relative risk of suicidal acts in the relatives of individuals who die by suicide, even after adjusting for psychiatric disorder.
2 Candidate-Gene Association Studies
Genes that code for proteins involved in regulating serotonergic neurotransmission have been major candidate genes for the association studies of suicidal behavior. Among them, genes for serotonergic receptors, serotonin transporter, tryptophan hydroxylase, and other monoaminergic systems have received the most research attention.
2.1 Brain-Derived Neurotrophic Factor Pathway
The protein encoded by the brain-derived neurotrophic factor (BDNF) gene is a member of the neurotrophin family and plays an important role in neuronal survival and brain plasticity (Dwivedi 2010). In the past decade, evidence is accumulating to suggest BDNF and its relevant genes such as the neurotrophic tyrosine kinase receptor type 2 (NTRK2) and nerve growth factor receptor (NGFR) genes in the pathogenesis of suicidal behavior and major depression from clinical studies and postmortem studies. The functional Val66Met (rs6265) SNP in BDNF has attracted much attention in suicide research. However, evidence for assessing the risk of BDNF Val66Met in suicidal behavior is currently controversial. BDNF Val66Met has been reported to predispose to suicidal behavior in Japanese, Taiwanese, Korean, and Italian populations (Tsai et al. 2011). On the contrary, this association with suicide behavior has not been replicated in Taiwanese, Slovenian, Caucasian, and German studies (Tsai et al. 2011). Regarding the conflicting findings in these studies, a meta-analysis may help to elucidate the role of BDNF Val66Met in suicidal behavior. Zai et al. (2012) performed the meta-analysis of BDNF Val66Met in suicidal behavior using data from 12 studies and suggested that BDNF Val66Met is involved in suicidality.
In addition to the aforementioned findings, a report failed to find an association between the BDNF SNPs and suicidal behavior but found the associations of SNPs in NTRK2 with suicide attempt in depressed German patients, which were also replicated in the African American population (Kohli et al. 2010). McGregor et al. (2007) also investigated three SNPs in NGFR for an association with suicide behavior in childhood-onset mood disorder, and the results did not support an association of the NGFR SNPs with suicide risk.
2.2 Dopaminergic System
Genes in the dopaminergic system could be candidates for suicidal behavior study due to the fact that the striatal dopaminergic activity is related to impulsivity, a character of suicidal behavior. In a Japanese report, Suda et al. (2009) implicated possible involvement of the TaqIA and − 141C insertion/deletion variants of the dopaminergic D2 receptor (DRD2) gene in the biological susceptibility to suicidal behavior. Furthermore, this finding is in line with an earlier report that linked the DRD2 − 141C insertion/deletion variant with attempted suicide in German alcoholics (Johann et al. 2005).
Catechol-O-methyltransferase (COMT), the postsynaptic enzyme that metabolizes released dopamine, is a critical enzyme in the metabolic degradation of dopamine in the prefrontal cortex. Kia-Keating et al. (2007) conducted a meta-analysis using six studies and demonstrated evidence of a modest significant association between the COMT Val158Met SNP and suicidal behavior. In addition, two following studies identified the role of COMT Val158Met in suicidal behavior among the male subjects in Korean (Lee and Kim 2011) and Caucasian (Pivac et al. 2011) populations. These findings are in line with a report associating COMT Val158Met with attempted suicide in Caucasian alcoholics (Nedic et al. 2011).
Monoamine oxidase (MAO) is a mitochondrial outer membrane enzyme that degrades biogenic amines, including neurotransmitters such as dopamine. The MAOA gene contains a functional variable number tandem repeat (or VNTR) in the upstream regulatory region (MAOA-uVNTR). Hung et al. (2012) indicated that there was no association between MAOA-uVNTR and suicidal behaviors in the meta-analysis using data from seven case-control association studies. However, Antypa et al. (2013) found that the MAOA rs909525 SNP was associated with suicidality after examining the following markers: MAOA rs909525, MAOA rs6323, MAOA rs2064070, and MAOB rs1799836.
2.3 Hypothalamic–Pituitary–Adrenal (HPA) Axis
The HPA axis is a neuronal and endocrine system that regulates the body’s response to stress. Dysregulation of the HPA axis may be related to the risk of suicidal behavior owing to the fact that stress plays a major role in the various pathophysiological processes associated with suicidal behavior (Pompili et al. 2010). De Luca et al. (2010) tested the HPA axis-related genes including the corticotrophin-releasing hormone (CRH), the corticotrophin-releasing hormone receptor 1 (CRHR1), CRHR2, CRH binding protein (CRHBP), and melanocortin 2 receptor (MC2R) genes in a cohort of schizophrenia subjects with attempted suicide. The genotype analyses yielded a significant association between CRHBP and suicide attempt. They also demonstrated a significant interaction between CRHR1 and CRHBP in influencing suicide attempt and the severity of suicidal behavior, suggesting that SNPs in the HPA-axis-related genes could be associated with suicidal behavior in schizophrenia. Wasserman et al. (2008) also reported CRHR1 rs4792887 for suicidality in depressed males exposed to low stress. In addition, Roy et al. (2012) showed an interaction between CRHBP and childhood trauma on suicidal behavior. Moreover, there was an additive effect with the FK506 binding protein 5 (FKPB5) gene.
The protein encoded by FKBP5 is a member of the immunophilin protein family, which plays a role in causing subsensitivity of the glucocorticoid receptor. In a study with seven FKBP5 SNPs in families with bipolar offspring, Willour et al. (2009) suggested that FKBP5 may influence attempted suicide and number of depressive episodes in the bipolar subjects. In addition, Supriyanto et al. (2011) reported that the haplotypes (comprised of rs3800373 and rs1360780) in FKBP5 were associated with completed suicide in the Japanese population. Furthermore, Roy et al. (2012) showed that an interaction between FKBP5 and childhood trauma may increase the risk for attempting suicide.
2.4 Serotonergic Receptors
Suicide genetic studies mostly investigated the 5-hydroxytryptamine receptor 1A (HTR1A) and 5-hydroxytryptamine receptor 2A (HTR2A) genes that encode two of the serotonin receptors, which have opposing functions in a variety of cellular and behavioral processes.
HTR1A C-1019G (rs6295) has attracted considerable interest in suicidal behavior research because this SNP influences serotonergic neurotransmission. Angles et al. (2012) performed a meta-analysis of a possible correlation between HTR1A C-1019G and suicidal behavior with 4 studies and found no association. Similarly, González-Castro et al. (2013) performed another meta-analysis with nine studies and confirmed no association.
There are many genetic association studies of the HTR2A gene with suicidal behavior. Li et al. (2006) have made a good summary of these reports with a meta-analysis of 73 association studies for HTR2A and suicidal behavior. They detected a significant association between HTR2A A-1438G and suicidal behavior. However, they failed to find a significant association of HTR2A T102C with suicidal behavior.
In addition, a meta-analysis utilizing seven studies demonstrated no significant association between the G861C (rs6296) SNP in the 5-hydroxytryptamine receptor 1B (HTR1B) gene and suicidal behavior (Kia-Keating et al. 2007).
2.5 Serotonin Transporter
The solute carrier family 6 member 4 (SLC6A4) gene codes for the serotonin transporter. Following serotonin release, the serotonin transporter is the major site of serotonin reuptake into the presynaptic neuron. By regulating the reuptake of the released serotonin, the serotonin transporter is central to fine-tuning the serotonergic neurotransmission.
Among the genetic variants in SLC6A4, a 44-base pair (bp) insertion–deletion in the promoter region (serotonin-transporter-linked polymorphic region; 5-HTTLPR) polymorphism and a 17-bp VNTR in the second intron of SLC6A4 have been extensively studied for an association with suicidal behavior. Li and He (2007) reviewed 39 studies that examined the association between the functional 5-HTTLPR polymorphism and suicidal behavior in groups with psychiatric diagnoses. Their report suggested a significant association (P = 0.0068) with S allele as the risk allele for suicidal behavior. For the SLC6A4 VNTR polymorphism, they demonstrated no significant association for both the 10-repeat allele and 12-repeat allele, either in allelic or genotypic analysis.
2.6 Tryptophan Hydroxylase
The encoded proteins by the tryptophan hydroxylase 1 (TPH1) and tryptophan hydroxylase 2 (TPH2) genes are the rate-limiting enzymes in the biosynthesis of serotonin. In humans, as well as in other mammals, there are two isoforms of TPH, referred to as TPH1 and TPH2.
Numerous studies have tested for an association between the genetic variants in TPH1 and suicidal behavior but produced contradictory results. Li and He (2006) reported a meta-analysis with published 22 studies. They demonstrated a significant overall association between suicidal behavior and the TPH1 A218C/A779C SNPs, suggesting the involvement of TPH1 in the pathogenesis of suicidal behavior. In contrast, a following meta-analysis is conflicting and does not support TPH1 A218C/A779C being a susceptibility locus for suicidal behavior (Saetre et al. 2010). However, a recent meta-analysis with 37 studies provided evidence that TPH1 A218C/A779C may be a risk factor to manifest suicidal behavior at the clinical level (González-Castro et al. 2014).
The TPH2 gene has attracted much attention for its role in suicidal behavior pathophysiology since its identification. Several studies have tested TPH2 in recent years for associations with suicidal behavior with positive findings, although some studies found negative findings (Tsai et al. 2011). A meta-analysis of these studies may help to clarify the role of TPH2 in suicidal behavior. González-Castro et al. (2014) performed a meta-analysis with 37 studies and could not find an association with suicidal behavior with regard to the TPH2 SNPs (including G-703T, A-473T, and G19918A).
2.7 Gene–Gene Interaction
Because suicidal behavior may be related to multiple genes, several studies have investigated gene–gene interaction in suicidal behavior. Studies have tested the gene–gene interaction between MAOA and COMT in suicide attempt history in families with at least one member having bipolar disorder (De Luca et al. 2005) and in patients with schizophrenia (De Luca et al. 2006); however, these studies found no additive effect in conferring suicidal behavior risk. Another report showed no association among the HTR2C and MAOA intergenic haplotype combination and suicide attempt in bipolar patients (De Luca et al. 2008).
Moreover, Murphy et al. (2011) identified a three-locus gene–gene interaction (including HTR1B rs6296, SLC1A2 rs4755404, and NTRK2 rs1659400) as a significant predictor of suicidal behavior after controlling for possible confounders such as age, gender, alcohol abuse, and schizophrenia. They also found four SNPs (including SLC1A2 rs4755404, SLC1A3 rs2269272, HTR1B rs6296, and NTRK2 rs1659400), which showed evidence of association with suicidal behavior compared to a non-attempter control group.
Souza et al. (2011) evaluated whether genetic variants in HTR3A and HTR3B were susceptibility SNPs for suicidal behavior in Caucasian subjects with schizophrenia. Although HTR3A and HTR3B may not play a major role in the susceptibility for suicidal behavior, there were two nominally significant gene–gene interactions.
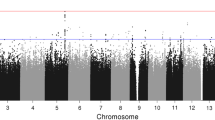
3 Genome-Wide Association Study
The genome-wide association study (GWAS) is an alternative to the candidate-gene approach (Christensen and Murray 2007). Unlike the candidate-gene approach, there is no a priori hypothesis about the involved genes in the GWAS studies, which examine common genetic variations (500,000 to 2 million SNPs) across the entire human genome in an attempt to identify genetic associations with observable traits by using high-throughput genotyping technologies.
3.1 GWAS by Perlis and Colleagues
In a GWAS study, Perlis et al. (2010) examined the association between common genome-wide variation and lifetime suicide attempts in bipolar I and II disorder as well as major depressive disorder. Strongest evidence of association for suicide attempt in bipolar disorder was observed for the rs1466846 SNP without known gene within 400 kb. In major depression, strongest evidence of association was shown for the rs2576377 SNP in the ABI family member 3 binding protein (ABI3BP) gene. Replication samples did not provide further support for these two SNPs. However, they conducted a meta-analysis incorporating all available mood disorder subjects (N = 8737) and identified ten SNPs in four loci, including the sorbin and SH3 domain containing 1 (SORBS1) and protein kinase C epsilon (PRKCE) genes.
The SORBS1 gene has been indicated in insulin signaling (Perlis et al. 2010). Knockout mice for the PRKCE gene have been shown to exhibit reduced anxiety behavior. In addition, a study found differences in expression of multiple protein kinases, including the one encoded by the PRKCE gene, in individuals with depression relative to control subjects.
3.2 GWAS by Willour and Colleagues
Willour et al. (2012) conducted a GWAS study that compared the genotypes of 1201 bipolar subjects with a history of suicide attempts to the genotypes of 1497 bipolar subjects without a history of suicide attempts. Genotyping was performed using the Affymetrix 6.0 array. Their analysis produced an association with the rs300774 SNP at the threshold of genome-wide significance (P = 5.07 × 10(−8)). The associated rs300774 SNP is within a large linkage disequilibrium block that includes the SH3 and SYLF domain containing 1 (SH3YL1), acid phosphatase 1 (ACP1), and family with sequence similarity 150 member B (FAM150B) genes.
Expression in the ACP1 gene was significantly elevated in bipolar subjects who have completed suicide (Willour et al. 2012). Furthermore, the protein encoded by the ACP1 gene is a tyrosine phosphatase that influences the Wnt signaling pathway regulated by lithium, indicating ACP1 as a functional candidate for involvement in suicidal behavior. Little is known about the potential functional contribution in the brain for SH3YL1 and FAM150B.
3.3 GWAS by Perroud and Colleagues
Suicidal ideation or suicidal plans that emerge during the course of antidepressant treatment have received considerable public attention. A small subset of patients with major depressive disorder develops this uncommon adverse event. Perroud et al. (2012) conducted a GWAS study to identify SNPs involved in increasing suicidal ideation during antidepressant treatment, where the subjects (n = 706) were either treated with escitalopram (n = 394) or nortriptyline (n = 312). They utilized high-quality Illumina Human610-quad chip genotyping data. There were 244 subjects who experienced an increase in suicidal ideation during follow-up.
Perroud et al. (2012) revealed that the rs11143230 SNP (P = 8.28 × 10(−7)) in the guanine deaminase (GDA) gene was significantly associated with increasing suicidality. The GDA gene encodes an enzyme responsible for the hydrolytic deamination of guanine and found to be differentially expressed in thalami from patients with schizophrenia.
3.4 Limitations in GWAS
With respect to the aforementioned GWAS studies, there were several limitations. First, the small size of the sample does not allow drawing definite conclusions. Small sample sizes can result in none of the findings reaching genome-wide significance due to insufficient statistical power (Spencer et al. 2009). In future work, independent replications in large sample sizes are needed to confirm the role of the polymorphisms found in these GWAS studies.
In addition, most of the loci found in the GWAS studies are not immediately informative because they are noncoding variants, which have incomplete annotation and unknown mechanisms (Ward and Kellis 2012). Therefore, extensive experimental work is needed to uncover the molecular mechanisms responsible for suicidal behavior. Moreover, the GWAS studies did not replicate across studies or populations, causing us to question the validity of novel associations, especially when the loci found are noncoding (Nebert et al. 2008).
4 Future Perspective
Future research may benefit from examining gene–gene interactions with novel computational techniques such as generalized multifactor dimensionality reduction (Lin et al. 2009). It is essential to address these interactions in order to describe complex traits in genetics. Epistasis analysis for gene–gene interactions has been advocated for deciphering complex mechanisms, particularly when each involved factor only demonstrates a minor marginal effect. Association studies based on individual SNPs or haplotypes, using a locus-by-locus or region-by-region approach, may overlook associations that can only be found when gene–gene interactions are investigated (Lane et al. 2012). To assess gene–gene interactions, there are many promising methods available, including regression models, multifactor dimensionality reduction, generalized multifactor dimensionality reduction, Bayesian approaches, and artificial neural network algorithms.
In addition, epigenetic approaches, which are modulated by environmental factors, should be considered to obtain clinically meaningful prediction of suicide owing to the fact that suicide may involve effects of the environment, genes, and their interaction. Guintivano et al. (2014) conducted an epigenome-wide association study using Illumina Infinium Human Methylation (HM) 450 BeadChip and generated a prediction model for suicidal behaviors with the rs7208505 SNP in the spindle and kinetochore-associated complex subunit 2 (SKA2) gene. However, their results were limited by the small size of the cohort. Future epigenetic studies with a much larger cohort of patients are needed to better assess sensitivity and specificity of the proposed predictive models and biomarkers.
Other approaches such as transcriptomics should also be employed to identify and prioritize biomarkers of relevance to suicidality. Le-Niculescu et al. (2013) investigated whole-genome gene expression profiling in the blood samples using Affymetrix HG-U133 Plus 2.0 GeneChips and found four biomarkers (including the spermidine/spermine N1–acetyltransferase 1 (SAT1), phosphatase and tensin homolog (PTEN), myristoylated alanine-rich protein kinase C substrate (MARCKS), and mitogen-activated protein kinase kinase kinase 3 (MAP3K3) genes) to be predictive of suicidality in bipolar disorder and psychosis. However, their results should be interpreted with caution because of small sample sizes, and future transcriptome-based studies with large sample sizes should be carried out to further evaluate their findings.
Furthermore, studies in future work can examine the contributions of genetic markers by whole-genome sequencing (Ng and Kirkness 2010) or exome sequencing (Bamshad et al. 2011). It has been suggested that gene variants with relatively large effects on drug efficacy or side effects are rare rather than common ones because of additive effects of significant loci (Tucker et al. 2009). Rare variants can only be discovered through whole-exome and whole-genome sequencing or family-based studies, instead of GWAS studies. Whole-genome sequencing represents a new era of scientific research and provides the most comprehensive collection of an individual’s genetic variation owing to the reduced cost and the increased throughput of next-generation sequencing technologies. Exome sequencing, which selectively sequences the nucleotides of protein-coding exons in an individual, has recently been introduced as an alternative and efficient approach for Mendelian disorders and common diseases (Bamshad et al. 2011). In summary, combining whole-genome approaches with novel computational tools may potentially lead to a better understanding on suicide.
References
Angles MR, Ocaña DB, Medellín BC et al (2012) No association between the HTR1A gene and suicidal behavior: a meta-analysis. Rev Bras Psiquiatr 34(1):38–42
Antypa N, Giegling I, Calati R et al (2013) MAOA and MAOB polymorphisms and anger-related traits in suicidal participants and controls. Eur Arch Psychiatry Clin Neurosci 263(5):393–403
Baldessarini RJ, Hennen J (2004) Genetics of suicide: an overview. Harv Rev Psychiatry 12:1–13
Bamshad MJ, Ng SB, Bigham AW et al (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12:745–755
Bondy B, Buettner A, Zill P (2006) Genetics of suicide. Mol Psychiatry 11:336–351
Christensen K, Murray JC (2007) What genome-wide association studies can do for medicine. N Engl J Med 356(11):1094–1097
De Luca V, Tharmalingam S, Sicard T et al (2005) Gene–gene interaction between MAOA and COMT in suicidal behavior. Neurosci Lett 383:151–154
De Luca V, Tharmalingam S, Müller DJ et al (2006) Gene–gene interaction between MAOA and COMT in suicidal behavior: analysis in schizophrenia. Brain Res 1097:26–30
De Luca V, Tharmaligam S, Strauss J et al (2008) 5-HT2C receptor and MAO-A interaction analysis: no association with suicidal behaviour in bipolar patients. Eur Arch Psychiatry Clin Neurosci 258:428–433
De Luca V, Tharmalingam S, Zai C et al (2010) Association of HPA axis genes with suicidal behaviour in schizophrenia. J Psychopharmacol 24:677–682
Dwivedi Y (2010) Brain-derived neurotrophic factor and suicide pathogenesis. Ann Med 42:87–96
González-Castro TB, Tovilla-Zárate CA, Juárez-Rojop I et al (2013) Association of 5HTR1A gene variants with suicidal behavior: case-control study and updated meta-analysis. J Psychiatr Res 47(11):1665–1672
González-Castro TB, Juárez-Rojop I, López-Narváez ML et al (2014) Association of TPH-1 and TPH-2 gene polymorphisms with suicidal behavior: a systematic review and meta-analysis. BMC Psychiatry 14:196
Guintivano J, Brown T, Newcomer A et al (2014) Identification and replication of a combined epigenetic and genetic biomarker predicting suicide and suicidal behaviors. Am J Psychiatry 171(12):1287–1296
Hung CF, Lung FW, Hung TH et al (2012) Monoamine oxidase A gene polymorphism and suicide: an association study and meta-analysis. J Affect Disord 136(3):643–649
Johann M, Putzhammer A, Eichhammer P et al (2005) Association of the − 141C Del variant of the dopamine D2 receptor (DRD2) with positive family history and suicidality in German alcoholics. Am J Med Genet B Neuropsychiatr Genet 132B:46–49
Kia-Keating BM, Glatt SJ, Tsuang MT (2007) Meta-analyses suggest association between COMT, but not HTR1B, alleles, and suicidal behavior. Am J Med Genet B Neuropsychiatr Genet 144B:1048–1053
Kohli MA, Salyakina D, Pfennig A et al (2010) Association of genetic variants in the neurotrophic receptor-encoding gene NTRK2 and a lifetime history of suicide attempts in depressed patients. Arch Gen Psychiatry 67:348–359
Lane HY, Tsai GE, Lin E (2012) Assessing gene-gene interactions in pharmacogenomics. Mol Diagn Ther 16:15–27
Lee HY, Kim YK (2011) Gender effect of catechol-O-methyltransferase Val158Met polymorphism on suicidal behavior. Neuropsychobiology 63(3):177–182
Le-Niculescu H, Levey DF, Ayalew M et al (2013) Discovery and validation of blood biomarkers for suicidality. Mol Psychiatry 18(12):1249–1264
Li D, He L (2006) Further clarification of the contribution of the tryptophan hydroxylase (TPH) gene to suicidal behavior using systematic allelic and genotypic meta-analyses. Hum Genet 119:233–240
Li D, He L (2007) Meta-analysis supports association between serotonin transporter (5-HTT) and suicidal behavior. Mol Psychiatry 12:47–54
Li D, Duan Y, He L (2006) Association study of serotonin 2A receptor (5-HT2A) gene with schizophrenia and suicidal behavior using systematic meta-analysis. Biochem Biophys Res Commun 340:1006–1015
Lin E, Hong CJ, Hwang JP et al (2009) Gene-gene interactions of the brain-derived neurotrophic-factor and neurotrophic tyrosine kinase receptor 2 genes in geriatric depression. Rejuvenation Res 12:387–393
McGregor S, Strauss J, Bulgin N et al (2007) p75(NTR) gene and suicide attempts in young adults with a history of childhood-onset mood disorder. Am J Med Genet B Neuropsychiatr Genet 144B:696–700
Murphy TM, Ryan M, Foster T et al (2011) Risk and protective genetic variants in suicidal behaviour: association with SLC1A2, SLC1A3, 5-HTR1B & NTRK2 polymorphisms. Behav Brain Funct 7:22
Nebert DW, Zhang G, Vesell ES (2008) From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev 40(2):187–224
Nedic G, Nikolac M, Sviglin KN et al (2011) Association study of a functional catechol-O-methyltransferase (COMT) Val108/158Met polymorphism and suicide attempts in patients with alcohol dependence. Int J Neuropsychopharmacol 14(3):377–388
Ng PC, Kirkness EF (2010) Whole genome sequencing. Methods Mol Biol 628:215–226
Perlis RH, Huang J, Purcell S et al (2010) Genome-wide association study of suicide attempts in mood disorder patients. Am J Psychiatry 167(12):1499–1507
Perroud N, Uher R, Ng MY et al (2012) Genome-wide association study of increasing suicidal ideation during antidepressant treatment in the GENDEP project. Pharmacogenomics J 12(1):68–77
Pivac N, Pregelj P, Nikolac M et al (2011) The association between catechol-O-methyl-transferase Val108/158Met polymorphism and suicide. Genes Brain Behav 10(5):565–569
Pompili M, Serafini G, Innamorati M et al (2010) The hypothalamic–pituitary–adrenal axis and serotonin abnormalities: a selective overview for the implications of suicide prevention. Eur Arch Psychiatry Clin Neurosci 260(8):583–600
Roy A, Hodgkinson CA, Deluca V et al (2012) Two HPA axis genes, CRHBP and FKBP5, interact with childhood trauma to increase the risk for suicidal behavior. J Psychiatr Res 46(1):72–79
Ryding E, Lindström M, Träskman-Bendz L (2008) The role of dopamine and serotonin in suicidal behaviour and aggression. Prog Brain Res 172:307–315
Saetre P, Lundmark P, Wang A et al (2010) The tryptophan hydroxylase 1 (TPH1) gene, schizophrenia susceptibility, and suicidal behavior: a multi-centre case-control study and meta-analysis. Am J Med Genet B Neuropsychiatr Genet 153B(2):387–396
Souza RP, De Luca V, Manchia M et al (2011) Are serotonin 3A and 3B receptor genes associated with suicidal behavior in schizophrenia subjects? Neurosci Lett 489(3):137–141
Spencer CC, Su Z, Donnelly P et al (2009) Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet 5(5):e1000477
Suda A, Kawanishi C, Kishida I et al (2009) Dopamine D2 receptor gene polymorphisms are associated with suicide attempt in the Japanese population. Neuropsychobiology 59:130–134
Supriyanto I, Sasada T, Fukutake M et al (2011) Association of FKBP5 gene haplotypes with completed suicide in the Japanese population. Prog Neuropsychopharmacol Biol Psychiatry 35(1):252–256
Tsai SJ, Hong CJ, Liou YJ (2011) Recent molecular genetic studies and methodological issues in suicide research. Prog Neuropsychopharmacol Biol Psychiatry 35(4):809–817
Tucker T, Marra M, Friedman JM (2009) Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet 85:142–154
Voracek M, Loibl LM (2007) Genetics of suicide: a systematic review of twin studies. Wien Klin Wochenschr 119:463–475
Ward LD, Kellis M (2012) Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol 30(11):1095–1106
Wasserman D, Sokolowski M, Rozanov V et al (2008) The CRHR1 gene: a marker for suicidality in depressed males exposed to low stress. Genes Brain Behav 7:14–19
Willour VL, Chen H, Toolan J et al (2009) Family-based association of FKBP5 in bipolar disorder. Mol Psychiatry 14:261–268
Willour VL, Seifuddin F, Mahon PB et al (2012) A genome-wide association study of attempted suicide. Mol Psychiatry 17(4):433–444
Zai CC, Manchia M, De Luca V et al (2012) The brain-derived neurotrophic factor gene in suicidal behaviour: a meta-analysis. Int J Neuropsychopharmacol 15(8):1037–1042
Acknowledgments
The authors extend their sincere thanks to Vita Genomics, Inc. and SBIR grants from the Department of Economic Affairs in Taiwan for funding this research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Lin, E., Tsai, SJ. (2016). Genetics and Suicide. In: Courtet, P. (eds) Understanding Suicide. Springer, Cham. https://doi.org/10.1007/978-3-319-26282-6_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-26282-6_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-26280-2
Online ISBN: 978-3-319-26282-6
eBook Packages: MedicineMedicine (R0)