Abstract
The description of the insulin–glucose metabolism has attracted much attention in the past decades, and several models based on physiology have been proposed. While these models provide a precious insight in the involved processes, they are seldom able to replicate and much less to predict the blood glucose (BG) value arising as a reaction of the metabolism of a specific patient to a given amount of insulin or food at a given time. Data-based models have proven to work better for prediction, but predicted and measured values tend to diverge strongly with increasing prediction horizon. Different approaches, for instance the use of vital signs, have been proposed to reduce the uncertainty, albeit with limited success. The key assumption hidden behind these methods is the existence of a single “correct” model disturbed by some stochastic phenomena. In this chapter, instead, we suggest using a different paradigm and to interpret uncertainty as an unknown part of the process. As a consequence, we are interested in models which yield a similar prediction performance for all measured data of a single patient, even if they do not yield a precise representation of any of them. This chapter summarizes two possible approaches to this end: interval models, which provide a suitable range; and probabilistic models, which provide the probability that the BG lies in predetermined ranges. Both approaches can be used in the framework of automated personalized insulin delivery, e.g., artificial pancreas or adaptive bolus calculators.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Blood Glucose
- Impulse Response
- Independent Component Analysis
- Gaussian Mixture Model
- Independent Component Analysis
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Glucose is the main energy source for the human body, but in excessive amounts it can be detrimental as well. In healthy persons, the correct glucose level is regulated by the insulin–glucose metabolism. In diabetes patients, this control system does not work properly any more, due, roughly speaking, either to lack of endogenous insulin production or to a resistance to insulin action [23]. In such cases, the therapy of choice consists in providing the necessary insulin externally. However, the delay times associated with this procedure and the danger of removing too much glucose from the blood circulation—with possibly lethal consequences—make the determination of the correct insulin quantity quite difficult and delicate.
Against this background, there has been a sustained interest in describing the glucose–insulin metabolism by physiological models of increasing complexity [2, 3, 25, 44] . A model resulting from the cooperation between the Universities of Padova and Virginia [12] has been accepted in 2008 by the Food and Drug Administration (FDA) as a substitute for preclinical trials for certain insulin treatments. The model was recently extended with new features [11].
And, of course, there has been a large number of attempts to use the model information to determine the right quantities of external insulin needed, so to say to replace the missing control action, to “close the loop.” Positive results have been reported for overnight operation [26], but also a reduction of variability for the daily use have been reported [33]. New and improved sensors have been introduced, faster insulin is entering the market, and all this nurtures the hope that more progress will come. Unfortunately, in spite of over 40 years of research [1], the results of this “artificial” or “virtual pancreas” are still not there where they should be and simple safety rules—e.g., avoiding insulin infusion during or near to hypoglycemia—seem to be able to offer the largest part of the benefits of closed-loop control in a much simpler way as well.
Against this background, it is natural to lean back for a moment and wonder whether we are asking the right questions. In this chapter, we are addressing this very issue by presenting two promising alternatives to modeling, namely interval models and probabilistic modeling techniques.
2 Alternatives for Modeling
The key assumption for modeling is the reproducibility of results. To some extent, of course, this is always true, for instance insulin does reduce the blood glucose (BG) concentration and carbohydrates (Carb) intake increases it. Many other effects are known but hard to quantify—e.g., some hormones increase it as well, muscular work reduces it—but there are many control loops in the human body which can lead to a complex response, e.g., in the case of prolonged physical work [21]. Other effects, like the circadian variation of sensitivities, are known as well [46].
Physiological models describe all these phenomena relying on deep understanding of physiology and frequently on very specific measurements, e.g., tracers [24, 42]. It is usually impossible to determine all parameters of such models with simple “external” measurements, e.g., taking only Carb and insulin administration and BG values. Indeed, it has been shown that the “external” behavior—the relation between insulin and Carb intake and BG—of these complex models can be approximated very well with very simple ones [17, 45] (see Fig. 1). This is the one reason why comparisons between the values predicted by physiological models and measurements are very seldom, exceptions being for instance [8, 47].
Indeed, in general, physiological models are not able to provide a personalized description.
As the comparison makes clear, the measurements have a high degree of complexity not reflected in the physiological model, the main reason being the many unmodeled effects, e.g., related to the emotional state, which may affect very strongly the BG values, and which cannot be captured by the model because the critical quantities, in this case the concentration of some hormones, are not known.
There are several ways to cope with this problem. On one side, the attempt can be made to find additional measurements to extend the model, e.g., vital signs—acceleration, heart frequency, body temperature, and so on. Similar techniques have proved very useful in the industrial framework, e.g., to detect changes in machines [13], but have never really succeeded in the case of diabetes treatment.
Another approach, related to another chapter of this book (see “Empirical Representation of Blood Glucose Variability in a Compartmental Model” by S. Patek et al.) consists essentially in estimating a “corrective” Carb input to explain the difference between measured and computed values. While this method cannot be used in real time, it allows to study the effect of some changes in therapy, e.g., different amounts of insulin.
If we are interested in obtaining models which are sufficiently simple to allow their use and parameter estimation in real time, it might be better to look for other approaches. A very efficient adaptive model was developed by [5] which relies on a simple hypothesis, a so-called ARX model, and determines the parameters continuously, concentrating on the most critical BG ranges. Figure 2 shows the performance of such a model as predictor.
45 min ahead prediction of glucose for two different patients from [6]
In this chapter, however, we suggest two different approaches. The key idea is not to get rid of uncertainty, assuming one particular value to be true, but to design models valid for the whole region, implicitly assuming that a full range of values are possible and in some sense true. One possible approach to this end are interval models, i.e., models which compute an output range and not a single value. The other alternative is using a probabilistic approach. Indeed, the exact BG value is not really important in itself, the clinician is more interested in keeping it inside the usual (“euglycemic”) range and preventing to reach a dangerous one, e.g., hypoglycemia. Markov jump model can help in describing the physiology the way it really is—i.e., to some extent random.
Both models can be used for automated insulin delivery as well. In the case of interval models, the problem can be stated in terms of a min/max problem such that the carbohydrate amount optimizes the cost functions for all cases of uncertainty. In the probabilistic case, it becomes the minimization of the probability that, under the action of insulin and meals, the BG leaves the good range to reach a dangerous one.
The further sections of this chapter are organized as follows: Before coming to the topic of interval models itself, a review about empirical continuous time transfer function models is given in Sect. 3 and the differences between possible model structures are discussed. The models presented in that section are a special form of control-oriented data-based models for describing the input–output relationship of the glucose metabolism that have proven quite powerful in the recent past (see e.g., [37, 38]). One of those presented model structures is then further used for the interval modeling introduced in Sect. 4. After a quick overview about the topic of interval modeling in general, some details about the methods used here for deriving interval models from data are given in Sect. 4.1. In following Sect. 4.1, results for the interval modeling are shown, both for simulated and for real patient data. The subsequent Sect. 5 then describes a probabilistic framework that can be used for predicting changes from one BG range to another. It starts in Sect. 5.1 with an overview about Gaussian mixture models that have been used in this context. Section 5.2 gives some details about the used model structure and the methodology of predicting transitions in the BG range, whereas actual prediction results for real patient data are presented in the following Sect. 5.3. The chapter finished with some final conclusions and discussion given in Sect. 6.
3 Model Structures
A model consists of a mathematical structure and of parameters. Thus the first step in modeling consists of fixing the model structure, and thereafter the parameters have to be tuned to get the best correspondence between measured and computed values. Of course, every model represents a simplification of the real system, and not every model structure is able to capture the behavior of the system under observation in a sufficiently general way. This is especially true in the case of a simplified model we are interested in.
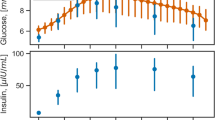
Table 1 summarizes previously proposed model structures to describe the blood glucose dynamics where BG(s), Carb(s), and I(s) correspond to the blood glucose concentration, ingested meal carbohydrates, and subcutaneously injected insulin bolus, respectively, all transformed into the Laplace domain. The table also lists the number of parameters which need to be estimated from data. All models thus use the same amount of information. A typical dataset which could be used for parameter estimation is shown in Fig. 3 (data from the DIAdvisor project [14]).
Illustration of the relevant measurement data for a specific patient, CHU0102 (data from the DIAdvisor project [14])
It is immediately visible that the model inputs are impulse-shaped quantities which are zero most of the time. That is because subcutaneous insulin injections are discrete events and meal ingestions, regardless of the quantity and time it takes to actually consume the food, are commonly treated as discrete events also. Therefore, for model analysis, the impulse responses are of great interest whereas step responses, commonly used in control engineering, do not provide sufficient information. A step response would actually mean that food or insulin is added to the metabolism in a continuous way over an extended period of time. While this might be true for the case of insulin, it is definitely an unrealistic case for food. In all given references, continuous insulin delivery is not considered for modeling.
From a physiological point of view, the impulse response gives precise information on the effect of one gram of carbohydrate and one unit of insulin, respectively, on the blood glucose concentration.
The impulse responses for both inputs of model structure 1 in Table 1 are shown in Fig. 4 for various selections of the parameters \(T_1\) and \(T_2\). It is immediately clear that the parameters \(K_1\) and \(K_2\) correspond to the steady-state change in BG. Furthermore, the parameters \(T_1\) and \(T_2\) are time constants which determine the time it takes until this steady state is reached. From this perspective, the physiological interpretation of the model parameters is straightforward.
Impulse response of model structure 1 from Table 1 using \(K_1=10\), \(K_2=-8\)
The impulse responses for both inputs of model structure 2 in Table 1 are shown in Fig. 5 for various selections of the parameters \(T_1\) and \(T_2\). Compared to the impulse responses in Fig. 4, model structure 2 involves a time delay determined by the parameters \(\tau _1\) and \(\tau _2\). Furthermore, after this delay, the impulse response shows a discontinuity (see the magnified plots in Fig. 5) which hardly appears in real subjects. However, the overall responses of model structures 1 and 2 are similar.
Impulse response of model structure 2 from Table 1 using \(K_1=10\), \(\tau _1=10\), \(K_2=-8\), \(\tau _2=5\)
The impulse responses for model structure 3 in Table 1 are not shown explicitly since they are very similar to those of model structure 2 shown in Fig. 5, except for the time delay which is zero in this case (\(\tau _1=0\), \(\tau _2=0\)).
Model structure 4 of Table 1 gives a significantly different impulse response than the models discussed above, see Fig. 6. Since there is no integrating behavior, the impulse responses return to the steady state of zero. Physiologically, the assumption is that even in the absence of insulin, glucose will be removed from the circulation after meal ingestion. This is fundamentally different than the assumptions in the previous structures, where glucose will not be removed until insulin is supplied. As it can be seen in the experimental results in the corresponding papers, both model assumptions can lead to a good approximation of real data. Note also that model parameters are not directly visible from the impulse response.
Impulse response of model structure 4 from Table 1 using \(K_1=100\), \(K_2=-80\)
Finally, the impulse response related to the carbohydrate input from model structure 5 in Table 1 is shown in Fig. 7. The response related to the insulin input is the same as for model structure 2 and can be seen in Fig. 5. The main difference in this structure is the assumption of two different time constants \(T_1\), \(T_2\), both for the carbohydrate dynamics. Therefore, there is more flexibility in the response compared to the case of using only one value for both constants (\(T_1=T_2\)). However, it is questionable whether distinctive values for \(T_1\) and \(T_2\) can really be identified from clinical data [29].
Impulse response of model structure 5 from Table 1 using \(K_1=100\), \(T_2=20\), \(\tau _1=10\)
4 Interval Models
There are essentially two ways to characterize interval models: one viewpoint is to design the model such that all possible realizations of the uncertainties will be considered and the true system output will be within the computed model output all the time, minimizing at the same time the conservativeness. The other viewpoint is to allow a certain amount or points to lie outside of the computed interval and thereby obtaining a much smaller interval [35]. The first approach is treated in another chapter of this book (see “Physiology-Based Interval Models: A Framework for Glucose Prediction Under Intra-Patient Variability” by J. Bondia and J. Vehi) while we focus on the latter here.
We will choose the model structure 1 from Table 1
because it contains only four parameters to be estimated and does not lead to a discontinuity in the impulse response. The generalized description of this model (assuming only one input for simplicity of notation) is a continuous time process model of the form
Modeling a specific patient means to assign values to the variables \(a_i\), \(b_i\) in (2). This is an identification problem [34, 43] and can be tackled in two ways. By transforming (2) into an equivalent discrete-time formulation, all the available tools of discrete-time system identification can be applied. However, such a transformation might introduce additional parameters and the initial (physiological) interpretation of the continuous time parameters gets lost [20]. On the opposite, there are continuous time identification methods which directly estimate the parameters in (2) without any need for transformation [19]. The benefits of such a direct estimation in the context of the human glucose insulin system have been treated in [10, 30]. In this contribution, we will thus also focus on a direct continuous time estimation method.
4.1 Continuous Time System Identification
Beginning first without taking into account the uncertainty, estimates of the model parameters can be found by minimizing a quadratic criterion of the form
where \(\varepsilon \) denotes the difference between measurement and model output,
N is the total number of available measurements and the vector \(\theta \) contains all parameters. Various optimization techniques exist for actually minimizing the criterion (3) (see e.g., [43]).
Now, considering uncertainty in the system to be modeled we assume two separate datasets of the same system with different underlying dynamics. The cost function which is minimized now is the sum of two similar terms as above, again taking into account the deviation between model output and measurement. Additionally, we will introduce a third term which penalize the standard deviation of the estimated model parameters for the first dataset compared to the second dataset (5). For example, if this third term is not present, the model parameters for both datasets will be estimated independently and might deviate to a great extent from each other. By introducing and weighting (using the weighting matrix \(\varPi \) which has the tuning parameters in the main diagonal) the third term, a compromise between a good model fit of the individual datasets and compact parameter sets can be obtained.
The function \(\bar{\sigma }\) denotes a vector standard deviation operator which determines component-wise the standard deviation of the model parameters of the parameter vector \(\theta \).
Minimization can be done with a Gauss–Newton algorithm [43]. Thanks to the definition of the cost and the model structure, gradient vector and Hessian matrix can be determined analytically [30]. Nevertheless, the optimization is nonlinear and iterative and thus care has to be taken to avoid improper starting values for the parameters. A generalization of the cost function to \(N_{exp}\) datasets and the corresponding gradient vectors and the Hessian was derived in [30].
As a result of the continuous time system identification, there is one parameter vector per experiment. The next step is to extract parameter intervals which then define the model output interval. Considering again the model structure 1 from Table 1, there are four model parameters which means the parameter vector has the form
Denoting with superscripts the corresponding dataset number, at the end of the identification \(N_{exp}\) estimates \(K_1^1, K_1^2,\ldots K_1^{N_{exp}}\) are available and their maximum value is \(K_1^{max}\) and minimum value \(K_1^{min}\). The computation of the interval model output is then
Note that \(K_2\) describes the effect of insulin and is thus negative. In the following subsection, we will present results when applying the interval model estimation to short data segments representing the breakfast period. The starting point of the data is breakfast time (around 8:00 in the morning) and data end points just before lunch were taken (around 12:30).
4.2 Interval Model Results
Figure 8 shows a typical result obtained with the methodology described above for three independent measurement sets of a single patient. The black stars are the measured glucose levels using a Yellow Spring Instrument YSI 2300 STAT Plus™ Glucose Analyzer (YSI) device. When modeling each dataset independently (individual models), the green curves apply which show of course the best performance. Depending on the choice of the tuning matrix \(\varPi \), the identified parameters then depend stronger or less strong on each other and an output interval according to (7) can be computed, see the red dashed lines in Fig. 8. Finally, also the mean interval model response is shown, which is computed when using as parameter the average values of all the estimated parameters of the interval model.
Remark 1
Again note the dependency of the results on the tuning matrix \(\varPi \). When choosing \(\varPi =0\) a zero matrix, the corresponding term in the cost function (5) will disappear and the result is an independent estimation of the parameters for each experiment. This also results in the largest possible output interval. In the other extreme, choosing very large values in \(\varPi \) results in very small standard deviations of the model parameters, making them equivalent. This also results in an output interval which degenerates to a single line and is the same result as for a standard multiexperiment identification setup [34].
For an in-depth analysis of the method, it was applied to 10 simulated and 10 clinical datasets. The simulated data was obtained from a time-varying metabolic simulation model [28]. In the following, we will use the term individual model for a model estimated on a single dataset (experiment), the term interval model for a model according to (7), and the term mean interval model when a simulation with the mean value of the interval model parameters is done.
4.2.1 Performance Metrics
Results will be evaluated based on a fit value (8) and on the error in detecting the peak BG. This is visualized in Fig. 9: \(\varDelta BG\) is the difference between the measured and predicted peak BG and \(\varDelta T\) is the difference in time. Both quantities can be positive or negative, where positive means the measured BG peak is higher and later in time than the simulated. The motivation of using those metrics is their importance in diabetes treatment.
4.2.2 Results Using the Simulated Data
First, we will present results from a single simulated patient before presenting statistics of all 10 considered patients. The parameter values and performance metrics reported in Table 2 are from simulated patient number 2. For the interval model, \(\varPi \) (5) was tuned in such a way, that the fit value of the interval model is not significantly less than \(90\,\%\) of the fit of the individual model, and that the ratio between mean value and standard deviation (1 / CV) of each of the four parameters is at least five. In this way, it is ensured that the interval predictions are reasonably tight.
For the presented results in Table 2 and Fig. 10, the elements in the main diagonal of \(\varPi \) are [15000, 125, 20, 0.3] result in \(1/CV=[5.02,-5.90,5.94,6.62]\). The parameter estimation was done on the first 3 days (up to \(t=72\) h). For the individual models, the validation is based on mean parameters of the first 3 days. For the interval models, the performance metrics were calculated based on the exact estimations for days 1–3 and validation was done with the mean interval model.
From Table 2, we see that there is a rather small ratio between mean value and standard deviation of the estimated parameters (especially for \(K_2\))—which would mean that the insulin effect is very different—when the experiments are identified independently of each other. This ratio is greatly increased when applying the proposed interval model estimation, without decreasing the performance (on the three training days). Considering validation, the interval model shows better performance compared to the individual model. Note also that in this particular case, the individually estimated model for day 3 is not useful, because \(K_2<0\), i.e., an insulin injection would result in a glucose increase.
Simulated patient #2: simulated glucose profile (black solid) individual model outputs for the breakfasts (green solid) and worst-case interval model outputs for breakfasts (red dashed). The first 3 days (up to \(t=72\) h) were used for estimation of the models, the remaining 2 days show validation results. Bottom panel shows the scaled impulse carbohydrate (black solid) and insulin (red solid) inputs
The results for all 10 patients are summarized in Table 3 where the mean values of the performance metrics over all 5 days are shown. The interval model shows the highest fit values in every case, and the average error (for all 10 patients) of correct estimation of the BG peak is only 3.79 mg/dl compared to 10.80 mg/dl of the individual models. Note that this rather high error is to a large extent caused by the specific glucose responses to meals by the simulator for some patients, where the response does not have a single maximal value. In those cases there are two almost equivalent high peaks, but separated in time. Such a shape cannot be reproduced with the chosen model structure (1), and the approximation typically lies in the middle of the two peaks. It is of interest to note that from the 30 individual models, there were 8 that showed to be incorrect from a physiological point of view (\(K_1<0\) and/or \(K_2>0\)). Considering the interval model, only for patient #6 no physiologically correct models could be found, as this patient’s glucose dynamics have a high time constant, and BG is only rising in the observational period. To correct this, a longer simulation time would be necessary.
4.2.3 Results Using the Clinical Data
The data used here was collected in [14]. Similar to the previous section, let us first analyze the estimated parameters for a specific patient in Table 4. The ratio between mean value and standard deviation was again greatly increased while the mean performance decrease is only moderate.
A graphical representation of the results obtained for this patient (#0107) is given in Fig. 11. In contrast to the previously simulated cases, there is a considerable difference between CGM and YSI measurements, available only at (infrequent) time intervals. The sampling rate is however small enough such that interpolation with second-order polynomials does not result in excessive overshoots. The figure does not only show that individual and interval model are well suited to represent dynamics of T1DM patients, but also that the estimated parameter intervals are small enough such that min/max simulations (7) result in reasonably small BG prediction intervals. A detailed view of the three breakfast periods is given in Fig. 8 for patient #0115.
Patient #0107: YSI measurements (stars), interpolated with second-order polynomial, CGM readings (black dashed), individual model outputs for the breakfasts (green solid), and worst-case interval model outputs for breakfasts (red dashed). All 3 days were used for estimation of the models. Bottom panel shows the scaled impulse carbohydrate (black solid) and insulin (red solid) inputs
Table 5 complements Table 3 and demonstrates the usefulness of the proposed methodology for real measurements on a group of 28 patients (only 10 are listed for the sake of brevity). Out of this group, only for 2 patients no suitable, i.e., sufficiently small parameter intervals could be found because the variability was too high.
Remark 2
The estimation of the model parameters was done based on YSI measurements, which are probably not available in practice. It should be noted that the proposed model identification procedure is independent of the source of the measurements, the YSI data can be replaced with data from glucose meters which are usually calibrated to show plasma equivalent values. As pointed out in the literature, e.g., in [7], the difference between those two measures can be up to 11 %, with plasma being higher than whole blood values, which directly effects the models. Similarly, a model estimated with YSI data could be used afterwards with a BG meter which introduces an uncertainty in the offset, directly related to the accuracy of the glucose meter.
Remark 3
The proposed model structure approximates BG dynamics based on carbohydrate and insulin inputs. There are several other factors which have an impact, e.g., physical activity, stress, and variation in nutrients in different meals. Activity did not play an important role in the analyzed data, because patients were hospitalized and stress could not be assessed with the available measurements. Complete information of nutrients (carbohydrates, proteins, and lipids) was available, but not utilized. Extended versions of the model structure making use of those data were tested (results not shown), but did not result in improvements. Changing meal compositions cause variations in the individual BG responses and are therefore indirectly captured by the interval model, resulting in enlarged intervals.
5 A Probabilistic Approach
Classical methods use various mathematical techniques to predict a single value in the future (e.g., the expected value in 30 min.). Contrary to that, in the preceding section, interval models were proposed which estimate a predicted range of the future BG. Probabilistic models, the topic of this section, go one step further and are solely concerned with the probability that the future BG will be inside a given region defined on the basis of physiological criteria. This corresponds more to the interest of the clinician, who aims to keep BG in a good range and not to some specific value. This allows also redefining the control task in terms of minimizing a given transition probability.
To estimate the mentioned probabilities for certain BG regions, we propose Gaussian models, in particular Gaussian mixture models (GMM), which are commonly used for modeling of biometric and biological systems with continuous measurements (see for example [15, 16] for an in-depth discussion). According to [39], a GMM is defined as a parametric probability density function represented as a weighted sum of Gaussian component densities.
5.1 Gaussian and Generalized Gaussian Mixture Models
Let us assume that a given (biological) system has n measured inputs. If we assume that each input is continuously measured with a sampling frequency f, a data vector recorded over given time t will have a total of \(m~=~f\cdot {t}\) samples. Combined together, the individual measurement vectors of the system variables will form a measurement matrix x of dimensions \(n\times m\). A Gaussian Mixture Model of the system is given as
where \(w_i\), \(i = 1,\ldots n\) are the mixture weights and the component Gaussian densities are determined as
with mean vector \(\mu _i\) and covariance matrix \(\varSigma _i, i = 1,\ldots n\). The mixture weights satisfy the constraint that
The complete GMM is represented by the mean vectors, covariance matrices, and mixture weight from all variable densities:
There are several well-established algorithms for estimation of the parameters of GMM from measurement data. Two of them, namely the iterative expectation-maximization (EM) algorithm and the maximum a-posteriori (MAP) from a well-trained prior model are most frequently used. More details about these two algorithms can be found, for example, in [39] or [40].
However, the statistical analysis of the BG data used in this study showed that, in the general case, they cannot be represented by the Gaussian distribution. In principle, this problem can be solved by the approximation of a non-Gaussian distribution. The near-normal distributions, i.e., the distributions which moderately deviate from the Gaussian distribution, can be approximated using disjunction into Gaussian components. The details about this well-known approach can be found, for example, in [27]. Now, modeling of the data with non-Gaussian distributions would be possible using its Gaussian components. This method is known as the independent component analysis (ICA). The fundamental restriction in ICA is that the independent components must be non-Gaussian for ICA to be possible [27]. Under the assumption that the data is distributed according to multivariate Gaussian distribution, i.e., where the components are also distributed according to Gaussian distribution, the ICA method is equivalent to the principal component analysis (PCA). The generalized mixture model using ICA is discussed in [32]. This model is an extension of the GMM where the model components have non-Gaussian distributions. The deviation of Gaussian distribution is modeled using the generalized Gaussian density. It is assumed that function \(p(x|\mu )\) is a differentiable function \(\mu \), so that the log-likelihood is given as:
Thus, the class probabilities for the data matrix x are given as
The detailed discussion about the GMM using ICA, especially about generation of model parameters, can be found, e.g., in [31] or [32]. The cited references also give examples of applications of these models.
5.2 Modeling Method and Model Structure
The main goal of this study was to generate probabilistic mixture models of the blood glucose levels and to use the generated models for short-term predictions, up to 30 min in advance. Those predictions are being calculated on the basis of the current and previous values of BG.
As already mentioned, the models were not designed to predict the exact future BG concentrations in patients. Instead, they predict in which of five predefined regions (which could be easily changed and adapted) the BG would most probably lie after the prediction interval:
-
\(<\)70 [mg/dl] (hypoglycemia)
-
70–120 [mg/dl] (normglycemia)
-
120–180 [mg/dl] (elevated normglycemia)
-
180–300 [mg/dl] (hyperglycemia)
-
\(>\)300 [mg/dl] (acute hyperglycemia)
Data from various patients are, in general case, not normally distributed. Under such circumstances, the blood glucose levels of such patients must be modeled using generalized GMM, as previously discussed in Sect. 5.1.
Several sources have already reported about periodic properties of the BG concentration in the human body. A reader can find in-depth analyses, e.g., in [41] or [22]. Starting from the assumption that the BG exhibits periodic properties, the fast Fourier transformation (FFT) analysis has been conducted in order to determine the dominant periodic components in the measured data. Figure 12 shows the magnitude spectrum of one representative patient (Patient #6 from clinical study [18]). The results are obtained by analyzing a 7-days blood glucose time series. Figure 12 shows the frequencies of the recurring patterns and the estimation of the importance of each cycle in the glucose time series. As it can be seen in the figure, a 12- h pattern repetition appears to be the most significant. Besides the 12- h pattern, the 24- h pattern has also been identified as important due to the circadian nature of glucose control [6]. The confirmation of these results can also be found in the extensive study of frequency characterization of blood glucose dynamics [22].
Thus, based on the previous considerations, a model structure shown in Fig. 13 has been selected. It includes the previous levels of blood glucose as inputs, specifically the ones measured 24 h before (\(BG_{t-24h}\)), 12 h before (\(BG_{t-12h}\)) and the last available measurement (\(BG_{t-1}\)). The model structure also includes the intake of insulin (I) and carbohydrates (C). It is important to note that, differently from the continuous 24-h blood glucose level measurements, the intake of insulin and carbohydrates were discrete events typically occurring 3–6 times a day during the clinical stay of the patients. It is also important to mention that the carbohydrate values used are not measured but estimated on the basis of the actual content of patients meals.
5.3 Modeling Results
The data used here were collected in the clinical trial [18] conducted at the Institute for Diabetes-Technology GmbH in Ulm, Germany between October and December 2011. In this study, a total of 12 patients with type-1 diabetes spent 7 days hospitalized and were equipped with six CGM sensors in parallel. For the purpose of the present study, the data from one of the sensors (a DexCom™Seven® Plus sensor by DexCom, San Diego, CA) was used which provides continuous information on subcutaneous glucose for 7 days with a sampling time of 5 min. Additionally, we used recorded information about meal intakes and bolus insulin injections.
The measurement data from clinical patients used in this study were divided into two parts. The larger part of the data which corresponded to the first 6 clinical days, was used to train the GMM. The smaller part of the data corresponded to the final (seventh) clinical day and was used for prediction and testing of the trained models. The models were used first to predict the blood glucose levels during the seventh clinical day and then to compare the prediction with the actual measurements in order to assess the correctness of the model.
Figure 14 shows the results of prediction of the BG levels (concentrations) for a clinical patient (designated for the purpose of this study as Patient #6), in particular the predictions of the final clinical day (24 h). The measured blood glucose levels from the previous 6 clinical days are used to generate the prediction model. The measurements and predictions in Fig. 14 are shown in 10 min equidistance (or time slices). To prevent possible misunderstanding, it is important to underline that, although the results are shown in 10 min time slices, the figures show future trends 10 min from the observation point at any particular time.
Figure 14 is divided into two parts. The upper part shows the actual (measured) blood glucose levels (showed using black dots) and the predicted blood glucose intervals (showed as vertical green lines limited at their ends by dots). The lower portion of figures indicates if the prediction above was correct or incorrect. A particular prediction was considered as a correct one, in a strict sense, if the actual measurement fell within its limits.
However, some of the predictions which are classified as incorrect can be found at or very close to the limits of the prediction segments and they are actually predicting the general trend of the blood glucose signal well. Examples of that can be seen in Fig. 14 (e.g., samples #70 or #132). Therefore, some of those slightly incorrect predictions can be considered to be borderline cases. This was one of the reasons to define an evaluation algorithm which could cope with different kinds of borderline behavior of the BG models. The evaluation algorithm, therefore, includes the following rules:
-
Segment overlapping: each two neighboring segments are overlapped for 5 [mg/dl]; therefore, at some prediction instances the modeling algorithm can extend the width of the prediction segments.
-
Persistence principle: in order to eliminate the prediction outliers, all single prediction segments which are in opposition to their neighboring prediction segments (from left and right) are changed to correspond to their neighboring predictions; this rule is not applied in cases when there is a persistent transition from one to another prediction segment.
The results shown in Fig. 14 are obtained by the application of so defined evaluation algorithm. The model has 96.53 % correct prediction rate, having only 3.47 % incorrect predictions during the final 24-h period.
The evaluation algorithm was able to deliver usable predictions of blood glucose trends up to 30 min in the future. The future predictions are generated stepwise, in 10-min steps, where the second (20-min prediction) and the third step (30-min prediction) are using the predictions from their preceding steps in calculations. The 20-min predictions are, therefore, calculated on the basis of the preceding 10-min. predictions, and the 30-min predictions are calculated on the basis of the preceding 20-min predictions. As a consequence, the evaluation algorithm was able to make corrections of the results with 10 min delay.
Figures 15, 16 and 17 show blood glucose prediction results for a clinical patient (Patient #4). The figures show the 10-, 20-, and 30-min predictions of the final (seventh) clinical day of Patient #4. Similarly as in the case of Patient #6, the measured blood glucose levels from the previous six clinical days are used to generate the prediction model. The model performance for the tests shown in Figs. 15, 16 and 17 are summarized in Table 6.
For the purpose of comparison, the blood glucose levels of the same patient (Patient #4) are modeled using linear second-order autoregressive models (AR) for the same prediction horizons (10-, 20-, and 30-min). The linear AR models used the same data as the GMM models. However, due to the fundamental difference between these two model types (the linear AR models predict single values, while the proposed prediction method using GMM models predicts intervals with certain probabilities), in order to make the comparison possible, the single value predictions of the linear AR models are used to predict the blood glucose intervals. Figure 18 shows both the single value predictions as well as the corresponding interval predictions of blood glucose level; the model performance for the tests shown in Fig. 18 is also summarized in Table 6.
In general, based on the results summarized in Table 6, it can be concluded that the GMM models deliver mostly correct predictions. The correct prediction rate for the 20- and 30-min predictions is expectedly lower than for the 10-min prediction because, as explained earlier, the 20- and 30-min predictions use the outputs from their preceding predictions which are already uncertain to the some extent.
The comparison of the GMM models with the linear AR models showed that the linear AR models exhibit slightly better 10-min predictions, while the GMM models exhibit superior results for 20- and especially 30-min predictions. However, the performance of the linear AR models in predicting intervals should be taken with caution. The linear AR models predict single values of blood glucose concentration. Thus, a probability could not be assigned to the BG intervals detected using those single values predictions of the linear AR models, making them not entirely comparable to the GMM models. Instead, if a fit value is used to assess the performance of the linear AR models, they showed much worse results: 84.76 %, 68.82 %, and 54.01 % fit for 10-, 20-, and 30-min predictions, respectively. Thus it can be concluded that the proposed method using GMM models showed superior performance especially as the prediction window increased. It is important to stress that in no case a wrong prediction of the proposed method using GMM models could have led to a dangerous patient decision (as in the case of a hyperglycemia predicted when the patient was in fact in hypoglycemia).
6 Conclusion and Outlook
The increasing availability of new sensors, smart insulin pumps, and more in general of computing power opens new chances for personalized diabetes care, in particular for closed-loop control, as in the artificial pancreas. First results in this direction are promising.
Nevertheless, variability, in particular intrapatient variability, remains a challenge. It also means that some methods may work well for some patient in most cases, but not for all patients and all cases. Of course, having a perfect model which can predict the future BG value with a high precision would be the best. Clinical experience, however, shows that some conditions, like anger, can affect enormously the BG development without being predictable.
This chapter intended to show that it might be wiser to accept this limitation from the very beginning and to look for “range” models, either in the form of moving or of prefixed ranges. While they will almost never provide an exact prediction of the next BG values, their error can be limited in such a way to be clinically easier to cope with than classical models.
Of course, the validity of the proposed approach needs testing in a clinical study and there is still much work to be done to tailor the methods to the specific problem.
References
Bequette, B.W.: Challenges and recent progress in the development of a closed-loop artificial pancreas. Annu. Rev. Control 36(2), 255–266 (2012). doi:10.1016/j.arcontrol.2012.09.007
Bergman, R.N.: Minimal model: perspective from 2005. Horm. Res. 64(suppl 3), 8–15 (2005)
Bergman, R.N., Ider, Y.Z., Bowden, C.R., Cobelli, C.: Quantitative estimation of insulin sensitivity. Am. J. Physiol. 236(6), E667–677 (1979)
Boiroux, D., Schmidt, S., Frøssing, L., Nørgaard, K., Madsbad, S., Skyggebjerg, O., Duun-Henriksen, A.K., Poulsen, N.K., Madsen, H., Jørgensen, J.B.: Control of blood glucose for people with type 1 diabetes: an in-vivo study. In: Proceedings of the 17th Nordic Process Control Workshop, pp. 133–140 (2012)
Castillo Estrada, G., Del Re, L., Renard, E.: Nonlinear gain in online prediction of blood glucose profile in type 1 diabetic patients. In: Proceedings of the 49th IEEE Conference on Decision and Control (CDC), pp. 1668–1673 (2010)
Castillo Estrada, G.E.: Identification of glucose dynamics in type 1 diabetes patients using multiple daily injection therapy. Ph.D. thesis, Johannes Kepler University Linz, Austria (2011)
Cengiz, E., Tamborlane, W.V.: A tale of two compartments: interstitial versus blood glucose monitoring. Diabetes Tech. Ther. 11(S1), S11–S16 (2009)
Dalla Man, C., Guerra, S., Sparacino, G., Renard, E., Cobelli, C.: A reduced type 1 diabetes model for model predictive control. In: Book of abstracts, 3rd International Conference on Advanced Technologies and Treatments for Diabetes (ATTD), Basel, Switzerland (2010)
Cescon, M., Stemmann, M., Johansson, R.: Impulsive predictive control of T1DM glycemia: an in-silico study. In: 2012 ASME Dynamic Systems and Control Conference. Fort Lauderdale, FL, USA (2012)
Cescon, M., Johansson, R., Renard, E., Maran, A.: Identification of individualised empirical models of carbohydrate and insulin effects on T1DM blood glucose dynamics. Int. J. Control 87(7), 1438–1453 (2014). doi:10.1080/00207179.2014.883171
Dalla Man, C., Micheletto, F., Lv, D., Breton, M., Kovatchev, B., Cobelli, C.: The UVA/PADOVA type 1 diabetes simulator: new features. J. Diabetes Sci. Technol. 8(1), 26–34 (2014)
Dalla Man, C., Rizza, R.A., Cobelli, C.: Meal simulation model of the glucose-insulin system. IEEE Trans. Biomed. Eng. 54(10), 1740–1749 (2007). doi:10.1109/TBME.2007.893506
del Re, L., Grünbacher, E., Pichler, K.: Verfahren und Vorrichtung zum Diagnostizieren des Zustandes eines Maschinenbauteils (2008)
DIAdvisor: European commission IST programme FP7 project DIAdvisor, \(n^{\circ }\) 216592. http://www.diadvisor.org
Duda, R., Hart, P.: Pattern Classification and Scene Analysis. Wiley, New York (1973)
Duda, R.O., Hart, P.E., Stork, D.G.: Pattern Classification, 2 edn. Wiley (2000). ISBN 978-0-471-05669-0
Fang, Q., Yu, L., Li, P.: A new insulin-glucose metabolic model of type 1diabetes mellitus: an in silico study. Comput. Method. Program. Biomed. 120, 16–26 (2015)
Freckmann, G., Pleus, S., Link, M., Zschornack, E., Klötzer, H.M., Haug, C.: Performance evaluation of three continuous glucose monitoring systems: comparison of six sensors per subject in parallel. J. Diabetes Sci. Technol. 7(4), 842–853 (2013)
Garnier, H., Mensler, M., Richard, A.: Continuous-time model identification from sampled data: implementation issues and performance evaluation. Int. J. Control 76, 1337–1357 (2003)
Garnier, H., Young, P.: What does continuous-time model identification have to offer? In: Proceedings of the 16th IFAC Symposium on System Identification, vol. 16 (2012)
Goodyear, L.J., Kahn, B.B.: Exercise, glucose transport, and insulin sensitivity. Annu. Rev. Med. 49, 235–261 (1998)
Gough, D.A., Kreutz-Delgado, K., Bremer, T.M.: Frequency characterization of blood glucose dynamics. Ann. Biomed. Eng. 31(3), 91–97 (2003)
Guyton, A.C.: Textbook of Medical Physiology, 11th edn. Elsevier Inc., Philadelphia (2006)
Hinshaw, L., Dalla Man, C., Nandy, D.K., Saad, A., Bharucha, A.E., Levine, J.A., Rizza, R.A., Basu, R., Carter, R.E., Cobelli, C., Kudva, Y.C., Basu, A.: Diurnal pattern of insulin action in type 1 diabetes, implications for a closed-loop system. Diabetes 62(7), 2223–2229 (2013)
Hovorka, R., Canonico, V., Chassin, L.J., Haueter, U., Massi-Benedetti, M., Federici, M.O., Pieber, T.R., Schaller, H.C., Schaupp, L., Vering, T., Wilinska, M.E.: Nonlinear model predictive control of glucose concentration in subjects with type 1 diabetes. Physiol. Meas. 25(4), 905–920 (2004)
Hovorka, R., Elleri, D., Thabit, H., Allen, J.M., Leelarathna, L., El-Khairi, R., Kumareswaran, K., Caldwell, K., Calhoun, P., Kollman, C., Murphy, H.R., Acerini, C.L., Wilinska, M.E., Nodale, M., Dunger, D.B.: Overnight closed-loop insulin delivery in young people with type 1 diabetes: a free-living, randomized clinical trial. Diabetes Care 37(5), 1204–1211 (2014). doi:10.2337/dc13-2644
Hyvärinen, A., Karhunen, J., Oja, E.: Independent Component Analysis. Wiley, New Jersey (2001)
Kirchsteiger, H., del Re, L.: Robust tube-based predictive control of blood glucose concentration in type 1 diabetes. In: IEEE 52nd Annual Conference on Decision and Control (CDC), 2013, pp. 2084–2089 (2013). doi:10.1109/CDC.2013.6760189
Kirchsteiger, H., Castillo Estrada, G., Pölzer, S., Renard, E., Del Re, L.: Estimating interval process models for type 1 diabetes for robust control design. In: Proceedings of the 18th IFAC World Congress, pp. 11,761–11,766. Milano, Italy (2011)
Kirchsteiger, H., Johansson, R., Renard, E., Re, L.D.: Continuous-time interval model identification of blood glucose dynamics for type 1 diabetes. Int. J. Control 87(7), 1454–1466 (2014). doi:10.1080/00207179.2014.897004
Lee, T.W., Lewicki, M.S.: The generalized gaussian mixture model using ica. In: Proceedings of International Workshop ICA, pp. 239–244 (2000)
Lee, T.W., Lewicki, M., Sejnowski, T.: Unsupervised classification with non-gaussian mixture models using ICA. Adv. Neural Inf. Process. Syst. 11, 508–514 (1999)
Leelarathna, L., Dellweg, S., Mader, J.K., Allen, J.M., Benesch, C., Doll, W., Ellmerer, M., Hartnell, S., Heinemann, L., Kojzar, H., Michalewski, L., Nodale, M., Thabit, H., Wilinska Malgorzata, E., Pieber, T.R., Arnolds, S., Evans, M.L., Hovorka, R.: On behalf of the AP@home Consortium: day and night home closed-loop insulin delivery in adults with type 1 diabetes: three-center randomized crossover study. Diabetes 37, 1931–1937 (2014)
Ljung, L.: System Identification, 2nd edn. PTR Prentice Hall (1999)
Milanese, M., Norton, J., Piet-Lahanier, H., Walter, E. (eds.): Bounding Approaches to System Identification. Kluwer Academic/Plenum Publishing Corporation, London (1996)
Percival, M.W., Bevier, W.C., Wang, Y., Dassau, E., Zisser, H.C., Jovanovic, L., Doyle, F.J.: Modeling the effects of subcutaneous insulin administration and carbohydrate consumption on blood glucose. J. Diabetes Sci. Technol. 4(5), 1214–1228 (2010)
Reiterer, F., Kirchsteiger, H., Assalone, A., Freckmann, G., del Re, L.: Performance assessment of estimation methods for CIR/ISF in bolus calculators. In: Proceedings of the 9th IFAC Symposium on Biological and Medical Systems, pp. 231–236 (2015)
Reiterer, F., Kirchsteiger, H., Freckmann, G., del Re, L.: Identification of diurnal patterns in insulin action from measured CGM data for patients with T1DM. In: Proceedings of the 2015 European Control Conference, pp. 1–6 (2015)
Reynolds, D.: Encyclopedia of Biometric Recognition, chap. Gaussian mixture models, p. 1217. MIT Lincoln Laboratory, Cambridge, MA (2008)
Reynolds, D., Rose, R.: Robust text-independent speaker identification using gaussian mixture speaker models. IEEE Trans. Speech Audio Process. 3, 72–83 (1995)
Riva, A., Bellazzi, R.: Learning temporal probabilistic causal models from longitudinal data. Artif. Intell. Med. 8, 217234 (1996)
Saad, A., Dalla Man, C., Nandy, D.K., Levine, J.A., Bharucha, A.E., Rizza, R.A., Basu, R., Carter Rickey, E., Cobelli, C., Kudva, Y.C., Basu, A.: Diurnal pattern to insulin secretion and insulin action in healthy individuals. Diabetes 61(11), 2691–2700 (2012)
Söderström, T., Stoica, P.: System Identification. Prentice Hall International, Hemel Hempstead (1989)
Sorensen, J.T.: A physiologic model of glucose metabolism in man and its use to design and assess improved insulin therapies for diabetes. Ph.D. thesis, Massachusets Institute of Technology (1978)
Trogmann, H., Kirchsteiger, H., Castillo Estrada, G., del Re, L.: Fast estimation of meal/insulin bolus effects in t1dm for in silico testing using hybrid approximation of physiological meal/insulin model. 70th Scientific Sessions of the American Diabetes Association (2010). Abstract 504-P
Van Cauter, E., Polonsky, K.S., Scheen, A.J.: Role of circadian rythmicity and sleep in human glucose regulation. Endocr. Rev. 18(5), 716–738 (1997)
Visentin, R., Dalla Man, C., Kovatchev, B., Cobelli, C.: The university of virginia/padova type 1 diabetes simulator matches the glucose traces of a clinical trial. Diabetes Technol. Ther. 16(7), 428–434 (2014)
Acknowledgments
This chapter is derived in part from an article published in the International Journal of Control, published online 25 Mar 2014, copyright by Informa UK Limited trading as Taylor & Francis Group, available online: http://www.tandfonline.com/doi/full/10.1080/00207179.2014.897004, as well as in part from an article published in the Proceedings of the 2014 22nd Mediterranean Conference of Control and Automation (MED), copyright by The Institute of Electrical and Electronics Engineers, Incorporated (IEEE), available online: http://dx.doi.org/10.1109/MED.2014.6961587 .
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Kirchsteiger, H., Efendic, H., Reiterer, F., del Re, L. (2016). Alternative Frameworks for Personalized Insulin–Glucose Models. In: Kirchsteiger, H., Jørgensen, J., Renard, E., del Re, L. (eds) Prediction Methods for Blood Glucose Concentration. Lecture Notes in Bioengineering. Springer, Cham. https://doi.org/10.1007/978-3-319-25913-0_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-25913-0_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-25911-6
Online ISBN: 978-3-319-25913-0
eBook Packages: EngineeringEngineering (R0)