Abstract
Several filamentous fungi grow on the surface or inside different types of cheese, produce secondary metabolites, and contribute to the organoleptic characteristics of mature cheese. Particularly relevant is the contribution of Penicillium roqueforti to the maturation of blue-veined cheeses (Roquefort, Danablu, Cabrales, etc.). P. roqueforti is inoculated into these cheeses as a secondary starter. This fungus is closely related taxonomically to Penicillium carneum and Penicillium paneum, but these two species are not used as starters because they produce the potent toxin patulin. P. roqueforti Thom has the capability to produce about 20 secondary metabolites of at least seven different families, but it seems that only some of them are produced in microaerobic conditions and accumulate inside the cheese (e.g., andrastins). This article focuses on the biosynthetic pathways, gene clusters, and relevance of the known metabolites of P. roqueforti including roquefortines, PR-toxin and eremofortins, andrastins, mycophenolic acid, clavines (agroclavine and festuclavine), citreoisocoumarin, and orsellinic acid. In addition the biosynthesis of patulin (a P. paneum and P. carneum product) is discussed. Penicillium camemberti grows on the surface of Camembert, Brie, and related white rind cheeses, and the penetration of secondary metabolites inside the cheese is relevant. One of the P. camemberti metabolites, cyclopiazonic acid, is important because of its neurotoxicity and its biosynthesis is reviewed. The removal of toxic metabolites gene clusters by precise gene excision while preserving all other characteristics of the improved starter strains, including enzymes involved in cheese ripening and aroma formation, is now open. A possible strain improvement application to the cheese industry is of great interest.
Access provided by CONRICYT-eBooks. Download reference work entry PDF
Similar content being viewed by others
Keywords
- Cheese fungi
- Blue-veined cheeses
- Penicillium roqueforti
- Penicillium camemberti
- Secondary metabolites biosynthesis
- Roquefortines
- PR-toxin
- Eremofortins
- Andrastins
- Mycophenolic acid
- Clavine alkaloids
- Cyclopiazonic acid
1 Introduction
Many Penicillium roqueforti strains are used in different countries in the world as secondary starters for the production of blue-veined cheese. More than one hundred of these strains have been characterized morphologically [1], and all of them are closely related to the original type strain described by Charles Thom [2] as P. roqueforti Thom ATCC10110. Taxonomically, this strain is referred as P. roqueforti subspecies roqueforti. Some strains of the P. roqueforti cluster (initially identified as P. roqueforti) differ in the metabolites that are separated by thin layer chromatography [3] and in the pigmentation of the reverse of the colonies [4]. These authors found a group of P. roqueforti strains that are less pigmented and produce patulin instead of PR-toxin and named this group P. roqueforti subspecies carneum because they are found associated with spoiled meat products.
Later Boysen et al. [5] using rRNA sequences and RAPD (random amplified polymorphic DNA) techniques divided the “P. roqueforti” strains into three species, namely, P. roqueforti sensu stricto, Penicillium carneum, and Penicillium paneum. The last one was associated with molded bread, flour, and cereal grains.
Recently Houbraken et al. [6] discovered a new member of the P. roqueforti series in cold-preserved apples. This strain, which grows and forms sexual cleistothecia at low temperature, has been classified as Penicillium psychrosexualis. It produces patulin as P. carneum and P. paneum at difference of P. roqueforti (Table 1). So far, P. psychrosexualis has not been found in cheeses and appears to be mainly associated with fruits such as apples and pears, in which it may produce pigmented spots. Therefore, the relevance of the secondary metabolites of this species in cheese is lower than that of P. roqueforti, P. carneum, and P. paneum.
1.1 Fungal Secondary Metabolites
Frequently filamentous fungi produce a few dozens of molecules belonging to different classes of secondary metabolites (polyketides, terpenes, nonribosomal peptides, aromatic compounds, heterocyclic metabolites, etc.) [7]. Usually they are produced as mixtures of chemically related molecules (e.g., roquefortines C, D, L, M or andrastins A to D). Each family of these compounds derives from a set of enzymes encoded by a gene cluster. Genetic information, in the form of gene clusters, for about 15 to 30 secondary metabolites have been found in the sequenced genome of ascomycetes [8, 9]. In some fungi, several gene clusters have been characterized by genetic and biochemical analysis (e.g., A. nidulans, A. fumigatus), whereas in others only a small number of secondary metabolite gene clusters has been identified so far (e.g., P. chrysogenum or P. roqueforti) and many other gene clusters remain cryptic, i.e., encoding unknown products [7, 10]. In addition, a number of gene clusters remain fully silent or nearly silent, although in some cases their expression may be activated by specific methods [10, 11]. In this article, we focus on the study of the secondary metabolites produced by the cheese fung us P. roqueforti (Table 1). The metabolites produced by the related fungi P. paneum and P. carneum, which are only rarely found in blue cheeses, and P. psychrosexualis are listed in Table 1, but they are reviewed succinctly at the end of the chapter (e.g., patulin produced by P. paneum and P. carneum) [12]. Also the biosynthesis of the neurotoxin cyclopiazonic acid by Penicillium camemberti is included in this article.
2 Secondary Metabolites Produced by P. roqueforti
In the last decades, increasing evidence has been reported on the ability of P. roqueforti to produce secondary metabolites in different culture media and inside the blue cheeses [13, 14]. The biosynthetic pathway of some of these metabolites and the gene clusters encoding their pathways have been located in the genome of the producer fungi [15–18], although the pathways for some of the rare secondary metabolites remain unknown. The full genome sequence of P. roqueforti FM164 has been made available [19], and this information will contribute to a better understanding of the ability of this fungus to express the genes encoding secondary metabolites under different growth conditions. So far the information available about the expression of the genes encoding enzymes for secondary metabolites biosynthesis inside the blue-veined cheeses is very scarce.
2.1 Roquefortines
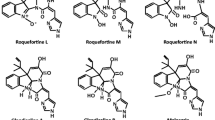
Roquefortines were discovered many decades ago [20, 21] and are among the best known P. roqueforti secondary metabolites [22, 23]. The roquefortine family includes roquefortine C and the related roquefortines D (3, 12-dihydroroquefortine C), 16-hydroxyroquefortine C, roquefortine L, and some other minoritary roquefortines [24]. These compounds are members of the prenylated indole alkaloid class of compounds (Fig. 1) (reviewed in reference [25]). Roquefortine C is produced by P. roqueforti growing in a variety of solid substrates, but its formation in blue cheeses does not occur in significant amounts, and there is a consensus that roquefortines in cheese do not pose a health problem for humans [12, 14, 22, 26].
The compounds of the roquefortine family derive from L-tryptophan, L-histidine, and mevalonate [25]. Roquefortines are produced by several Penicillium species including P. roqueforti, P. chrysogenum [15, 16], and other plant-associated or saprophytic fungi [27, 28]. However, it was unknown if the biosynthetic pathway of roquefortine alkaloids is identical in all these fungi. Recent evidence [18] demonstrated that the roquefortine pathway in P. roqueforti is shorter than that for roquefortine/meleagrin in P. chrysogenum (see below).
2.1.1 Biosynthesis of Roquefortine and Meleagrin
One of the common early intermediates of prenylated indole alkaloids is a molecule of dimethylallyl-tryptophan (DMAT) that is formed by a prenyltransferase that uses L-tryptophan (or a L-tryptophan-containing cyclopiperazine dipeptide) and dimethylallyl diphosphate (DMA-PP) as substrates [15, 16, 25]. In roquefortine alkaloids, precursor condensation of these substrates occurs at C-3 of L-tryptophan with the 3′carbon atom of DMA-PP (named “reverse condensation”) [29].
Following initial precursor studies in P. roqueforti [30, 31], it was established that the tryptophan-histidine cyclopiperazine nucleus of roquefortine C and the related compounds glandicolines A and B, meleagrin, and neoxaline derive from the precursor compounds L-tryptophan and L-histidine. These two amino acids are condensed by a dimodular nonribosomal peptide synthetase, named RDS (roquefortine dipeptide synthetase) consisting of two similar modules with the domain sequence ATCATC, where A indicates adenylation domain (amino acid activation), T thiolation (peptidyl carrier) domain, and C condensation domain. The amino acid specificity of each domain has been elucidated [15, 25]. The cyclodipeptide (cyclo-trp-his) is then prenylated by the roquefortine prenyltransferase (RPT) that introduces an isopentenyl group at C-3 of tryptophan (Fig. 2b). The resulting prenylated compound is roquefortine D (3, 12-dihydroroquefortine C). In the last step of the roquefortine pathway, roquefortine D is oxidized by the roquefortine D dehydrogenase (RDH), losing two H atoms with the formation of a double bond between carbons 3 and 12 resulting in roquefortine C (Fig. 2b). The order of the second and third biosynthetic reactions (prenylation and roquefortine dehydrogenation) is indifferent [16], and therefore, a metabolic grid occurs in these early steps of the pathway [25].
(a) Gene cluster for roquefortine/meleagrin in P. chrysogenum compared to the roquefortine cluster in P. roqueforti. (b) Biosynthetic pathway of roquefortine C (boxed area), meleagrin, neoxaline, and oxaline in P. chrysogenum (Modified from Ref. [28])
In some fungi (e.g., P. chrysogenum and P. oxalicum), roquefortine C is later converted to roquefortine L (or in other fungi to glandicoline A) and then to glandicoline B and meleagrin (see structure in Fig. 1) [31]. The conversion of roquefortine C (or roquefortine L in P. chrysogenum) to the late pathway products involves (i) a carbon scaffold reorganizing oxygenase (SRO), similar to the FtmG oxygenase of Aspergillus fumigatus involved in fumitremorgin biosynthesis, and (ii) a N1 hydroxylase (NOX) and a N-OH methyltransferase (Fig. 2b). The role of each of these enzymes in the roquefortine and meleagrin pathways has been reviewed elsewhere [25, 28].
2.1.2 The Roquefortine/Meleagrin Gene Cluster
Molecular genetic studies on the roquefortine/meleagrin gene cluster (roq/mel) were performed first in P. chrysogenum Wis54-1255 leading to the characterization of the gene cluster and the proposal of a roquefortine/meleagrin biosynthetic pathway [15]. The roquefortine/meleagrin gene cluster was later confirmed by Ali et al. [16] and Ries et al. [24], who reported the formation in P. chrysogenum of roquefortine L (instead of glandicoline A) as an intermediate in the pathway, in addition to other minoritary roquefortines derived from late branches of the pathway. The entire pathway in P. chrysogenum is encoded by a seven-gene cluster (Pc21g15420 to Pc21g15480) (Fig. 2a).
An important question is if the many natural isolates (strains) of P. roqueforti obtained from different geographical areas [1] have genetic differences in their capability to synthesize roquefortine and the related indole alkaloids [14, 26] and whether these differences are due to changes in the roquefortine gene cluster.
The roquefortine gene cluster of P. roqueforti has recently been investigated [18]. The initial steps of roquefortine biosynthesis in P. roqueforti are identical to those of P. chrysogenum, but we found that P. roqueforti lacks the genes that encode the enzymes for the “late” conversion of roquefortine C to roquefortine L, glandicoline B, and meleagrin [18]. A natural short pathway was found in P. roqueforti that is dedicated to the production of roquefortine C but is unable to form derivatives containing the meleagrin scaffold [18, 28].
A comparative analysis of the roq cluster of P. roqueforti and the roq/mel cluster of P. chrysogenum revealed that two key genes located in the central region of the roq/mel cluster in P. chrysogenum (sro and nox) have been lost in P. roqueforti during evolution and the order of two of the conserved genes has changed during gene reorganization. Furthermore, the roqT gene, encoding a transmembrane transport protein in P. chrysogenum, has been rearranged into a pseudogene (Fig. 2a) that encodes only residual peptides [18, 28]. As a result of the roq/mel cluster reorganization, P. roqueforti is unable to convert the roquefortine-type carbon skeleton into a meleagrin-type scaffold and is incapable to produce glandicolines. The cluster reorganization is not a recent event derived from “industrial” strain selection. Rather, it seems to be an ancient phenomenon that occurred probably millions of years ago during adaptation of a progenitor Penicillium to cheese environments [28].
PR-Toxin and Eremofortins
PR-toxin and eremofortins are isoprenoid secondary metabolites. PR-toxin is probably the most potent mycotoxin produced by P. roqueforti [12, 23]. This isoprenoid mycotoxin is clearly toxic for mice, rats, hamsters, and some domestic animals in vivo. Furthermore, PR-toxin has mutagenic action in vitro, as shown in studies using the Ames test. Actually, PR-toxin is considered to be the causative agent of cow toxicosis produced by poorly conserved moldy silages [12]. Fortunately, PR-toxin is modified to less toxic derivatives by P. roqueforti cells, and its toxic form does not seem to be accumulated in large amounts in blue cheese [22].
Recently, we have studied the biosynthesis of PR-toxin and its intermediates the eremofortins [17]. PR-toxin derives from the sesquiterpene (15 carbon atoms) aristolochene ; this first intermediate is formed by aristolochene synthase (encoded by the gene ari1). Hidalgo et al. [17] cloned and sequenced a partial PR-toxin cluster containing four genes that include the ari1 (prx2) gene reported previously in P. roqueforti (Fig. 3a). Gene silencing of each of the four genes, named prx1 to prx4 (prx, abbreviation for PR-toxin), caused a reduction of 65–75 % in the production of PR-toxin indicating that these four genes encode enzymes involved in PR-toxin biosynthesis. An eleven gene cluster (Pc12g06260 to Pc12g06370) that includes the above‐mentioned four prx genes and a 14-TMS (transmembrane spanner domain) drug/H+ antiporter of the MFS family was found in the genome of P. chrysogenum (Fig. 3a). A detailed analysis of the published genome sequence of P. roqueforti FM164 [19] revealed that this strain contains in two subclusters 10 of the 11 prx genes described in P. chrysogenum. The exception is prx10 that was reported as encoding a protein of unknown function [17]. As shown in Fig. 3a, seven of the 10 prx genes (prx 1 to 4, prx 8, 9, and 11) are clustered together in contig Proq02g of P. roqueforti, whereas the three remaining genes (namely, prx5, 6, and 7) are located elsewhere (contig Proq06g) in the genome.
(a) Gene clusters of PR-toxin in P. chrysogenum Wis 54–1255 and P. roqueforti FM164. The location of a pseudogene in the P. chrysogenum cluster (shadowed in black) is shown. In P. roqueforti, three of the prx genes (prx5 to 7, proteins CDM35712 to 35710) are located elsewhere (Contig Proq06g) in the genome in relation to the prx1 to prx11 gene cluster (contig Proq02g, proteins CDM31314 to 31322). (b) Proposed PR-toxin biosynthetic pathway (see text for details)
PR-toxin biosynthesis pathway from farnesyl diphosphate was proposed based on all available evidence [17]. It proceeds to PR-toxin through aristolochene and the eremofortins (Fig. 3b). The PR-toxin pathway is divided in two parts. The first part corresponds to the conversion by oxidative enzymes of the 15-carbon atom aristolochene to 3-hydroxy, 8-oxo, 12-dehydroaristolochene, eremofortin B, and deacetyl-eremofortin A (DAC-EreA), all containing 15 carbon atoms. In the second half, DAC-eremofortin A is acetylated to the 17-carbon eremofortin A by an acetyltransferase encoded by prx11, and then eremofortin A is converted to eremofortin C and finally to PR-toxin.
Both eremofortins and PR-toxin are probably secreted by the MFS transporter encoded by the prx5 gene in the prx cluster, as proposed for several antibiotics and other secondary metabolites [32].
The PR-toxin is converted in vitro and probably also in vivo to PR-amide and PR-imine by reaction of the PR-toxin carboxylic group with ammonium ions or primary amines in the culture medium or in the cells [33, 34], and these derivatives appear to be less toxic than the PR-toxin itself.
2.2 Andrastins
Another important family of P. roqueforti secondary metabolites is the andrastins that belong to the polyketide-isoprenoid class. They are inhibitors of the farnesyltransferase of the ras-encoded oncogenic protein [35, 36]. Prenylation (farnesylation) of the human Ras protein is essential for its biological activity that may cause tumor formation. Therefore, inhibitors of the Ras prenyltransferase activity are interesting for their use as potential antitumor agents [37].
The andrastins belong to the meroterpenoid class of secondary metabolites that include compounds with interesting pharmacological activities [38]. Andrastins A, B, C, and D were discovered by S. Omura (Nobel Prize 2015) and coworkers at the Kitasato Institute in Japan in a screening of antitumoral agents. These compounds were first identified in the culture broth of Penicillium sp. FO4259 [36, 39, 40], and they are produced by several other Penicillium species [41].
Nielsen et al. [13] and Fernández-Bodega et al. [14] found that P. roqueforti produces andrastins and that andrastin A (the final product of the biosynthetic pathway) is accumulated inside blue cheeses inoculated with P. roqueforti as a secondary starter. Andrastin A concentrations in different blue cheeses such as Roquefort, Danablu, Cabrales, Bejes-Tresviso, and Valdeón vary depending on the particular P. roqueforti strain used as starter and ripening conditions [14]. Andrastins are considered to be beneficial for human health because of their ras prenyltransferase inhibitory activity, but there are no studies that support its lack of toxicity when accumulated in high concentrations in cheese.
2.2.1 Biosynthesis of Andrastins
Initial precursor incorporation studies showed that the andrastins derive from 3, 5-dimethylorsellinic acid (DMOA) and the terpene precursor farnesyl diphosphate (FPP) [39]. In fungi, orsellinic acid is formed by the condensation of one unit of acetyl-CoA (starter unit) and three units of malonyl-CoA (elongation unit) followed by cyclization of the tetraketide to form the aromatic ring of orsellinic acid . These reactions are catalyzed by a specific nonreducing polyketide synthase (nr-PKS). The precursor incorporation studies suggested that the two methyl groups of DMOA derive from methionine [39] although it is not entirely clear if the incorporation of the methyl groups occurs during polyketide elongation or after orsellinic acid is formed. Based on the information available on the molecular genetics of the biosynthesis of other farnesylated-DMOA-derived fungal metabolites (e.g., austinol or terretonin), Matsuda et al. [42] identified a gene cluster encoding enzymes for andrastin biosynthesis in P. chrysogenum. The biosynthetic pathway of farnesyl-DMOA containing meroterpenoids [38] indicates that DMOA is converted to farnesyl-DMOA by a specific farnesyltransferase and then the farnesyl-DMOA is converted into farnesyl-DMOA methyl ester by the action of methyltransferase. A FAD-dependent monooxygenase converts the terminal double bond of farnesyl-DMOA methyl ester into its epoxy derivative (Fig. 4b).
(a) Gene cluster for andrastin in P. chrysogenum (see text for details). (b) Biosynthetic pathway of andrastins (Modified from Ref. [42])
The epoxy farnesyl-DMOA methylester is later cyclized to a polycyclic meroterpenoid by a characteristic terpene cyclase. The cyclases of each meroterpenoid gene cluster may yield a (slightly) different cyclic structure [43] that are converted to different final meroterpenoid molecules by “late” modification enzymes (so-called tailoring enzymes) (Fig. 4b).
2.2.2 Andrastins Gene Cluster
The andrastin A gene cluster of P. chrysogenum (Fig. 4b) comprises eleven genes (Pc22g22820 to Pc22g22920) of which nine correspond to enzymes that are directly involved in andrastin A (the most modified final product) biosynthesis. These nine enzymes include (i) an iterative type I, nonreductive polyketide synthase that forms DMOA (named AdrD); (ii) a prenyltransferase that attaches the farnesyl group of FPP to the DMOA moiety (AdrG); (iii) a methyltransferase that methylates the carboxyl group of farnesyl-DMOA forming farnesyl-DMOA methyl ester (AdrK); (iv) a FAD-dependent monooxygenase that converts farnesyl-DMOA methyl ester to epoxy-farnesyl-DMOA methyl ester (AdrH); (v) a terpene cyclase (AdrI) that cyclizes the epoxyfarnesyl-DMOA methyl ester intermediate to form andrastin E, the first member of the andrastin family; and (vi) four additional tailoring enzymes that convert andrastin E to andrastins D, F, C, B, and A (final product) (Fig. 4b). These tailoring enzymes include a short-chain dehydrogenase (AdrF), a ketoreductase (AdrE), an acetyltransferase (AdrJ) forming andrastin C, and finally a P450 monooxygenase (AdrA) involved in the consecutive oxidations of the C-23 methyl group of andrastin C to form andrastin B and then andrastin A that contain an alcohol and an aldehyde group at the C-23 position, respectively (Fig. 4b). The involvement of these 9 genes in andrastin A biosynthesis was confirmed by heterologous expression of a reconstructed gene cluster in Aspergillus oryzae that resulted in the production of andrastin A [42]. The andrastin gene cluster of P. roqueforti has not been characterized so far, although is likely to be similar to that of P. chrysogenum.
2.3 Mycophenolic Acid
Another important secondary metabolite of P. roqueforti is mycophenolic acid (MPA). This compound was already known at the beginning of the twentieth century, before the discovery of penicillin, as an antibiotic active against Bacillus anthracis, produced by a Penicillium sp. strain. Production of MPA in liquid cultures has been studied in Penicillium brevicompactum [44], the fungus which is used for MPA industrial production and in P. roqueforti [17]. Mycophenolic acid, discovered initially as antibacterial agent, was later found to have other important biological activities [45]. Particularly relevant is its activity as immunosuppressant used successfully to prevent organ rejection in transplants [46]. In addition, MPA has antitumor, antiviral, and antifungal activities and is used in the treatment of psoriasis [47–51].
2.3.1 Mycophenolic Acid Biosynthesis and Resistance Genes
Initial precursor incorporation studies [52] suggested that MPA is a compound synthesized through the hybrid polyketide-terpene pathway. Recently, the mpa gene cluster was cloned from a P. brevicompactum strain [53] and later confirmed in the sequenced genome of a different P. brevicompactum strain [54]. In both strains, the mpa cluster comprises seven genes (Fig. 5), namely, mpaA (encoding a prenyltransferase), mpaB (encoding a protein of unknown function), mpaC (encoding a polyketide synthase), mpaDE (encoding a bifunctional fused protein with two domains corresponding to a P450 monooxygenase and a hydrolase), mpaF (encoding an inosine-5′-phosphate dehydrogenase), mpaG (encoding an O-methyltransferase), and mpaH (encoding an oxidative cleavage enzyme).
(a) Gene cluster of mycophenolic acid in P. brevicompactum. (b) Proposed biosynthetic pathway for mycophenolic acid (Modified from Ref. [53])
A key enzyme in MPA biosynthesis is the non-reductive iterative PKS encoded by mpaC. This protein contains the following domains: a starter unit acyltransferase (SAT), a ketosynthase (KS), an acyl-carrier protein (ACP), a methyltransferase (MT), and a standard acyltransferase (AT). These activities are required for the synthesis of the MPA intermediate 5-methylorsellinic acid from one starter acetyl-CoA, three malonyl-CoA extender units, and a methyl group.
Involvement of the mpaC gene encoding the non-reductive PKS and mpaDE encoding the bifunctional P450 monooxygenase-hydrolase in MPA biosynthesis has been confirmed by disruption of these genes in P. brevicompactum and by their expression in the heterologous host Aspergillus nidulans, a nonproducer of MPA that lacks the orthologous genes [53, 55]. More recently Zhang et al. [54] proved that mpaG encodes a S-adenosylmethionine (SAM)-dependent O-methyltransferase that converts in vitro demethylmycophenolic acid to MPA, the last step in the pathway (Fig. 5). This methyltransferase was purified after expression of the mpaG gene in E. coli. The enzyme showed similar substrate kinetics to O-methyltransferase obtained from P. stoloniferum (gene not yet cloned), another mycophenolic acid-producing fungus.
The MPA-producing fungi have to protect themselves against the antifungal activity of MPA. This resistance to MPA appears to be exerted by an IMP-dehydrogenase encoded by mpaF [56], although other mechanisms such as active MPA secretion and lack of uptake of the secreted extracellular MPA may also contribute to the resistance as occurs with other secreted metabolites [32].
Mycophenolic acid is active against fungi and human lymphocytes (involved in immune response) because it exerts a strong inhibition of the inosine-5′-phosphate (IMP) dehydrogenase, a key enzyme in de novo purine biosynthesis in those cells that lack the purine recycling pathway (as it is the case in lymphocytes). Overexpression of the mpaF gene in A. nidulans drastically increases the resistance to MPA in this fungus [55, 56]. Indeed, these authors reported that six different fungi, including those that produce MPA and also some putative nonproducers, contain two IMP dehydrogenase genes, one of them presumably located within the mpa gene cluster [56].
While this manuscript was in the proof stage, a recent report described the mycophenolic acid gene cluster in P. roqueforti (see note added in proof).
2.4 Agroclavine and Festuclavine
There are two subgroups of alkaloids produced by fungi: (i) the clavine alkaloids represented by fumigaclavine, synthesized by A. fumigatus [57], and agroclavine and festuclavine produced by P. roqueforti, and (ii) the lysergic acid-containing ergot peptide alkaloids produced by species of Claviceps [58]. Several P. roqueforti strains of different origins produce the clavine-type alkaloids agroclavine and festuclavine (Fig. 1). Festuclavine is also produced by P. carneum [12].
The clavine alkaloids have a tricyclic or tetracyclic structure with small structural differences between them. There are no detailed studies on the biosynthesis of agroclavine and festuclavine in P. roqueforti, but the biosynthesis of fumigaclavines in A. fumigatus [57] and ergot alkaloids in Claviceps purpurea [58] has been extensively studied.
2.4.1 Biosynthesis of Agroclavine and Festuclavine
All these compounds derive from the precursors L-tryptophan, dimethylallyl diphosphate, and the methyl group of methionine. The first step in the biosynthesis of these clavine alkaloids is prenylation of L-tryptophan at C-4 to form 4-dimethylallyltryptophan (DMAT) by the enzyme DMAT synthase, a prenyltranferase encoded by the gene named fgaPT2 in A. fumigatus (or dmaW in Claviceps). In the second step, the primary amino group of DMAT is methylated using S-adenosylmethionine as methyl donor. The N-methyltransferase is encoded by the fgaMT gene located in an 11-gene cluster that comprises all genes involved in the pathway [59, 60]. The N-methyl-DMAT is then cyclized and oxidized to chanoclavin-1 by a FAD-containing oxidoreductase named chanoclavine synthase, encoded by the casA gene (also named case by other authors) [61]. This conversion also requires the product of a cluster-located gene, casC, encoding a catalase-like protein that is involved in oxidation of the 3′-methyl group of the dimethylallyl moiety (derived from DMA-PP) to a CH2OH [62]. Disruption of this catalase-like gene results in the interruption of the clavine or ergot alkaloids biosynthetic pathways in the producer organisms with accumulation of the N-methyl-DMAT intermediate. The exocyclic alcohol group of chanoclavine-I is then oxidized to form chanoclavine-1-aldehyde by the enzyme chanoclavine-1 dehydrogenase encoded by the fgaDH gen (also named casD) [58, 63]. The chanoclavine-1-aldehyde is the branching point intermediate in the biosynthesis of the different clavines and lysergic acid-containing alkaloids in different fungi. P. roqueforti produces the tetracyclic compound festuclavine that in A. fumigatus (but apparently not in most P. roqueforti strains) is later converted to fumigaclavine. The conversion of the tricyclic intermediate chanoclavine-1-aldehyde to festuclavine requires two enzymes encoded by the genes fgaFS and fgaOx3 [64]. The exact mechanism by which these two enzymes convert chanoclavine-1-aldehyde to festuclavine is still a matter of debate [64, 65]
The second tetracyclic clavine alkaloid produced by P. roqueforti is agroclavine. Agroclavine differs from festuclavine in that the former has a double bond between carbons 8 and 9 that is already present in the previous intermediate chanoclavine-1-aldehyde but is saturated (reduced) in festuclavine (Fig. 1), suggesting that there is an enzyme activity involved in the reduction of the double bond. At difference of P. roqueforti, A. fumigatus does not seem to accumulate agroclavine, probably because the pathway continues to fumigaclavine.
Formation of agroclavine from chanoclavine -1-aldehyde (the branching point intermediate) has been reported in Claviceps [66]. The agroclavine synthase EasG of Claviceps is a homologous enzyme to festuclavine synthase of A. fumigatus. Indeed, Cheng et al. [65] reported that the festuclavine synthase of A. fumigatus (about 65 % similarity to the agroclavine synthase of Claviceps) is able to produce agroclavine when incubated with the substrate chanoclavine-1-aldehyde in the presence of a FgaOx3 enzyme from Neotyphodium lolii. The difference between both homologous enzymes may explain the lack of the hydrogenase (reductase) activity characteristic of A. fumigatus (a festuclavine producer) in the agroclavine producers, such as Claviceps purpurea. Interestingly, P. roqueforti produces both agroclavine and festuclavine [12]. In summary, it seems likely that P. roqueforti synthesizes agroclavine by the action of a FgaFS-homologous enzyme as a less reduced product of the pathway.
Other P. roqueforti Metabolites
Several strains of P. roqueforti are also known to produce citreoisocoumarin and small amounts of orsellinic acid. Very little is known about the biosynthesis of these compounds.
Orsellinic Acid. The biosynthesis of orsellinic acid is related to that of methylorsellinic acid and dimethylorsellinic acid described above. In the absence of experimental information in P. roqueforti, it is unclear if there is a separate polyketide synthase without the methylation domain, specific for orsellinic acid biosynthesis or whether this compound is formed by the 5-methylorsellinic acid PKS of the mycophenolic acid pathway (encoded by mpaC) when the methyltransferase domain is bypassed by the “domain skipping” mechanism occurring in some of these synthases. In support of this last possibility is the fact that orsellinic acid is produced in very small amounts in the tested strains of P. roqueforti [12]. A similar nr-PKS is the DMOA synthase involved in andrastins biosynthesis (see above).
Citreoisocoumarin. Another metabolite produced by R. roqueforti is citreoisocoumarin. There is no information on the biosynthesis of this compound in P. roqueforti, but in Fusarium species, citreoisocoumarin is known to be a byproduct of the biosynthesis of aurofusarin. Both compounds derive from a precursor polyketide that may be cyclized by (i) a carbon-to-carbon (C-C) Claysen-type condensation giving aurofusarin or (ii) by formation of an internal lactone resulting in citreoisocoumarin [67]. This last type of cyclization (lactone formation) appears to predominate in P. roqueforti, but the enzyme(s) and molecular basis underlying citreoisocoumarin biosynthesis are still unknown.
3 Metabolites of Penicillium carneum and Penicillium paneum
P. carneum and P. paneum are closely related to P. roqueforti Thom, although as indicated in the Introduction section, they are classified as separate species [5, 12]. Indeed, P. carneum and P. paneum differ from P. roqueforti and among themselves in their ability to produce some secondary metabolites (Table 1). P. carneum predominates in some spoiled meat products, whereas P. paneum is associated with molded bread or grains and grass silages. Both fungi may occur in the surface of some cheeses, but they are not used as secondary starters in blue-veined cheese [1, 23, 26], although they may be present in homemade artisanal blue cheeses, particularly in those produced in some developing countries. P. carneum and P. paneum may be included in the group of cheese contaminant fungi [22], and their absence in most blue cheeses makes their secondary metabolites less relevant for human health.
The main mycotoxins produced by these fungi are patulin, marcfortins, penitrem, and botryodiplodin (Table 1). P. paneum is more different from P. roqueforti; it lacks the ability to produce PR-toxin but synthesizes patulin that is not found in P. roqueforti. P. carneum produces most of the described metabolites of P. roqueforti and also patulin. Penitrem A is produced by P. carneum but not by P. roqueforti or P. paneum. The marcfortins A, B, C are produced only by P. paneum. On the other hand, PR-toxin and the intermediates eremofortins A, B, and C are produced exclusively by P. roqueforti [12]. In summary, P. paneum is different from the other two related species in its set of secondary metabolites, whereas P. carneum is more similar to P. roqueforti. The biosynthesis of these metabolites is poorly known with exception of that of patulin.
3.1 Patulin
Patulin is a potent mycotoxin that causes neurological and immunological disorders and gastrointestinal alterations in humans [68]. Patulin is a common mycotoxin produced by many species of Penicillium (including P. paneum and P. carneum), Aspergillus, Paecilomyces, and Byssochlamys nivea [69, 70]. Patulin is frequent in fungi-spoiled apples, and the levels of patulin allowed in apple-derived products, such as cider, apple jellies, or apple-derived infant foods, are strictly limited by the food safety agencies of many western countries. Among the producer fungi, Penicillium expansum is known to cause the soft rot of apples and pears and appears to be the major producer of patulin in fruits [71].
Fragmented evidence reported over the last three decades has shown that the first intermediate, 6-methylsalicylic acid (6-MSA), is converted to patulin through the intermediates m-cresol, m-hydroxybenzyl alcohol, and isoepoxydon. Biochemical and genetic studies in different producer fungi, including P. paneum, identified two enzymes, namely, 6-methyl salicylic acid synthase (6-MSAS) and isoepoxydon dehydrogenase (IDH) involved in patulin biosynthesis [72–74]. More recently two P450 monooxygenases have been found to be involved in the conversion of 6-MSA to m-cresol and m-hydroxybenzyl alcohol [75]. When the genome of the patulin producer Aspergillus clavatus was sequenced (TIGR http://www.aspergillus.org.uk/indexhome.htm?secure/sequence_info/index.php ~ main), a 15-gene cluster putatively encoding the entire patulin pathway was identified. This cluster includes the msas gene (encoding 6-MSA synthase), the idh (encoding the IDH), and the two P450 monooxygenase encoding genes. The patulin gene cluster of P. paneum or P. carneum has not been reported yet but is likely to be similar to that of A. clavatus.
4 Penicillium camemberti: Cyclopiazonic Acid
Several filamentous fungi may grow on the surface of cheeses during the ripening process. Most of them are strictly external, but some of them, e.g., P. camemberti and P. nalgiovense, may contribute to the organoleptic characteristics of mature cheeses.
P. camemberti is associated with ripening of white rind soft cheeses such as Camembert and Brie cheeses [76]. Selected strains are routinely used in production of soft cheeses. Although many of the P. camemberti secondary metabolites remain unexplored, one of them, cyclopiazonic acid, acquired relevance because of its well-known neurotoxicity. Cyclopiazonic acid (CPA) is a highly active inhibitor of Ca2+-dependent ATPases of animal cells and is a potent neurotoxin for humans and other mammals [77].
Cyclopiazonic acid is a prenylated indole alkaloid containing a tetramic acid ring produced by several Aspergillus and Penicillium species, including P. camemberti. The biosynthesis of CPA has been studied in Aspergillus flavus and A. oryzae but not in detail in P. camemberti, although the biosynthetic pathway is likely to be conserved in all fungi.
CPA derives from L-tryptophan, a four-carbon unit, and a dimethylallyl diphosphate units [77]. The four-carbon unit is formed by condensation of an acetyl-CoA and a malonyl-CoA. The first step in the CPA biosynthetic pathway is catalyzed by a hybrid polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) that activates L-tryptophan and condenses this amino acid with acetyl-CoA and malonyl-CoA units forming the acetoacetyl-L-tryptophan (AA-L-trp) intermediate. This key enzyme is encoded by the cpaA (also named cpaS) gene. The NRPS component of the hybrid PKS-NRPS has four domains C-A-T-R* with activities for condensation, L-tryptophan activation and thiolation (peptidyl/acyl carrier), and peptide product release. The R* domain is a reductase that lacks the catalytic triad of NADH-dependent reductases and is proposed to release the N-acetoacetyl-tryptophan intermediate by cyclization via a Dieckmann condensation to form the cyclo-acetoacetyl-L-tryptophan intermediate that is released from the phosphopantetheinyl arm of the T domain of the enzyme [78].
In the second step of the CPA biosynthetic pathway, the cyclo-acetoacetyl-L-tryptophan intermediate is prenylated at C-4 of the indole nucleus by the enzyme cyclo-AA-trp prenyltransferase that introduces an isopentenyl group from the DMA-PP donor. The enzyme encoded by the cpaD gene is a member of the well-known family of prenyltransferases [79]. In A. clavatus and A. oryzae, there is a third step of the pathway catalyzed by a FADH-dependent oxidoreductase cpaO that converts β-cyclo-acetoacetyl-tryptophan into the final product, CPA [77, 80, 81]. The cpa gene cluster of A. flavus includes in addition to cpaA-epaD-cpaO structural genes two additional genes, epaM that encodes a transmembrane protein of the MFS family and cpaR that encodes a regulatory protein of the C6 Zn2+ finger type [82]. The CpaM transporter presumably is involved in CPA secretion [32].
CPA is the final product of the biosynthetic pathway in A. flavus and probably also in P. camemberti since CPA is secreted and accumulated in these two fungi. However, A. oryzae that is considered to be a “domesticated” variant of A. flavus widely used in Japanese food industries contains an additional gene in the cluster, cpaH, that encodes a P450 monooxygenase which converts CPA to 2-oxo-CPA. This compound is much less toxic than CPA [83], and the authors proposed that this P450 monooxygenase is a toxicity-reducing “safeguard” enzyme evolved in A. oryzae during its adaptation to grow in fermented foods [83]. The mechanism of adaptive safeguard appears to be more complex since some A. oryzae strains have lost completely the ability to produce CPA and 2-oxo-CPA due to a mutation in the N-terminal region of the CpaA hybrid PKS-NRPS. It will be interesting to confirm that P. camemberti lacks the “safeguard” cpaH gene and therefore maintains the high toxicity characteristic of CPA.
Other secondary metabolites have been reported to be produced by P. camemberti, e.g., asperenone, asperrubrol-like compounds, methyl-isoborneol, and hadacidin [27], but it is unknown if these compounds pose a health problem for humans.
5 Conclusions and Future Perspectives
Blue-veined cheese is a gourmet food consumed all over the world. These cheeses are maturated in different countries with P. roqueforti as a secondary starter that grows inside the cheese in microaerobic conditions. As described in this article, P. roqueforti is able to produce about 20 different secondary metabolites belonging to at least seven different families. Some of these compounds are highly toxic, e.g., PR-toxin, whereas others, e.g., andrastins, are considered to be beneficial for human health. Selection of fungal strains over centuries has favored the use of P. roqueforti Thom that lacks production of patulin, over the closely related P. carneum and P. paneum that produce this mycotoxin. Most P. roqueforti secondary metabolites are produced in rich solid (agar plates) and liquid cultures, but there is very limited information on the production and accumulation of secondary metabolites inside the cheese, under the microaerobic conditions. Andrastin A is known to be formed and accumulated in the blue-veined cheese s [13, 14]. Humans and other animals consume a variety of plants and fungal secondary metabolites; many of them are probably nontoxic, but in other cases, animals reject certain plants or mushrooms because they contain toxic secondary metabolites known as feeding deterrents. It is possible now to construct strains lacking certain secondary metabolite gene clusters. The possible removal of toxic metabolites gene clusters by precise gene excision, while preserving all other characteristics, including enzymes involved in cheese ripening and aroma formation, is now a possible application of the molecular genetics of P. roqueforti to the cheese industry. This “domestication” process would be equivalent to the “natural domestication” process that has evolved in A. oryzae for removal of CPA.
6 Authors’ Note
When this article was in press Del Cid and coworkers reported the mycophenolic acid gene cluster of P. roqueforti (Del-Cid A, Gil-Durán C, Vaca I, Rojas-Aedo JF, García-Rico RO, Levicán G, Chávez R. (2016) Identification and Functional Analysis of the Mycophenolic Acid Gene Cluster of Penicillium roqueforti. PLoS One. 1(1):e0147047.
The P. roqueforti mpa cluster is almost identical to that known for P. brevicompactum (Fig. 5) and the conclusion obtained by these authors are similar for both fungi.
Abbreviations
- ACP:
-
Acyl-carrier protein
- AT:
-
Acyltransferase
- ATCC:
-
American Type Culture Collection
- DMA-PP:
-
Dimethylallyl diphosphate
- DMAT:
-
Dimethylallyltryptophan
- DMOA:
-
3,5-Dimethylorsellinic acid
- FPP:
-
Farnesyl diphosphate
- KS:
-
Ketosynthase
- MFS:
-
Major facilitator superfamily
- MPA:
-
Mycophenolic acid
- 6-MSAS:
-
6-Methyl salicylic acid synthase
- MT:
-
Methyltransferase
- nr-PKS:
-
Non-reductive polyketide synthase
- NOX:
-
N1 hydroxylase
- RAPD:
-
Random amplified polymorphic DNA
- RDH:
-
Roquefortine D dehydrogenase
- RPT:
-
Roquefortine prenyltransferase
- SAR:
-
Starter unit acyltransferase
References
Gillot G, Jany J-L, Coton M, Le Floch G, Debaets S, Ropars J, López-Villavicencio M, Dupont J, Branca A, Giraud T, Coton E (2015) Insights into Penicillium roqueforti morphological and genetic diversity. Plos One 10(6), e0129849
Thom C (1906) Fungi in cheese ripening: Camembert and Roquefort. USDA Bureau of Animal Industrial Bulletin 82:1–39
Engel G, Teuber M (1989) Toxic metabolites from fungal cheese starter cultures (Penicillium camemberti and Penicillium roqueforti). In: van Egmond HP (ed) Mycotoxins in dairy products. Elsevier Applied Science, London
Frisvad JC, Filtenborg O (1989) Terverticillate penicillia: chemotaxonomy and mycotoxin production. Mycologia 81:837–861
Boysen M, Skouboe P, Frisvad J, Rossen L (1996) Reclassification of the Penicillium roqueforti group into three species on the basis of molecular genetic and biochemical profiles. Microbiology 142:541–549
Houbraken J, Frisvad JC, Samson RA (2010) Sex in Penicillium series Roqueforti. IMA Fungus 1:171–180
Zeilinger S, Martín JF, García-Estrada C (2015) Fungal secondary metabolites in the OMICS era. In: Zeilinger S, Martín JF, García-Estrada C (eds) Biosynthesis and molecular, vol II. Springer, New York
Martín JF, García-Estrada C, Zeilinger S (eds) (2014) Biosynthesis and molecular genetics of fungal secondary metabolites. Springer, New York
Zeilinger S, Martín JF, García-Estrada C (eds) (2015) Biosynthesis and molecular genetics of fungal secondary metabolites, vol II. Springer, New York
Martín JF, Liras P (2015) Novel antimicrobial and other bioactive metabolites obtained from silent gene clusters. In: Demain AL, Sánchez S (eds) Antibiotics: current innovations and future trends. Horizon Scientific Press and Caister Academic Press, Norfolk
Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C (2007) Genomic-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol 3:213–217
Nielsen KF, Sumarah MW, Frisvad JC, Miller JD (2006) Production of metabolites from Penicillium roqueforti complex. J Agric Food Chem 54:3756–3763
Nielsen KF, Dalsgaard PW, Smedsgaard J, Larsen TO (2005) Andrastins A-D, Penicillium roqueforti metabolites consistently produced in blue-mold ripened cheese. J Agric Food Chem 53:2908–2913
Fernández-Bodega MA, Mauriz E, Gómez Martín JF (2009) Proteolytic activity, mycotoxins and andrastin A in Penicillium roqueforti strains isolated from Cabrales, Valdeón and Bejes-Tresviso local varieties of blue-veined cheeses. Int J Food Microbiol 136:18–25
García-Estrada C, Ullán RV, Albillos SM, Fernández-Bodega MÁ, Durek P, von Döhren H, Martín JF (2011) A single cluster of coregulated genes encodes the biosynthesis of the mycotoxins roquefortine C and meleagrin in Penicillium chrysogenum. Chem Biol 18:1499–1512
Ali H, Ries MI, Nijland JG, Lankhorst PP, Hankemeier T, Bovenberg R, Vreeken RJ, Driesen AJM (2013) A branched biosynthetic pathway is involved in production of roquefortine and related compounds in Penicillium roqueforti. PlosOne 8, e65328
Hidalgo PI, Ullán RV, Albillos SM, Montero O, Fernández-Bodega MÁ, García-Estrada C, Fernández-Aguado M, Martín JF (2014) Molecular characterization of the PR-toxin gene cluster in Penicillium roqueforti and Penicillium chrysogenum: cross talk of secondary metabolite pathways. Fungal Gen Biol 62:11–24
Kosalková K, Domínguez-Santos R, Coton M, Coton E, García-Estrada C, Liras P, Martín JF (2015) A natural short pathway synthesizes roquefortine C but not meleagrin in three different Penicillium roqueforti strains. Appl Microbiol Biotechnol 99:7601–7612
Cheeseman K, Ropars J, Renault P, Dupont J, Gouzy J, Branca A, Abraham AL, Ceppi M, Conseiller E, Debuchy R, Malagnac F, Goarin A, Silar P, Lacoste S, Sallet E, Bensimon A, Giraud T, Brygoo Y (2014) Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nat Commun 5:2876
Ohmomo S, Sato T, Utagawa T, Abe M (1975) Production of alkaloids and related substances by fungi. Isolation of festuclavine and three new indole alkaloids, roquefortine A, B, and C from cultures of Penicillium roqueforti. Nippon Nogei Kagaku Kaishi 49:615–623
Scott PM, Merrien MA, Polonsky J (1976) Roquefortine and isofumigaclavine A, metabolites from Penicillium roqueforti. Experientia 32:140–142
Hymery N, Vasseur V, Coton M, Mounier J, Jany J-L, Barbier G, Coton E (2014) Filamentous fungi and mycotoxins in cheese: a review. Compr Rev Food Sci Food Saf 13:437–456
Martín JF, Coton M (2016) Blue cheese: microbiota and fungal metabolites. In: Frias J, Martínez-Villaluenga C, Peñas E (eds) Fermented foods in health and disease prevention. Elsevier, New York
Ries MI, Ali H, Lankhorst PP, Hankemeier T, Bovenberg RA, Driessen AJ, Vreeken RJ (2013) Novel key metabolites reveal further branching of the roquefortine/meleagrin biosynthetic pathway. J Biol Chem 288:37289–32195
Martín JF, Liras P, García-Estrada C (2014) Roquefortine and Prenylated Indole Alkaloids. In: Martín JF, Garcia-Estrada C, Zeilinger S (eds) Biosynthesis and molecular genetics of fungal secondary metabolites. Springer, New York
Fontaine K, Passeró E, Vallones L, Hymery N, Coton M, Jany JL, Mounier J, Coton E (2015) Occurrence of roquefortine C, mycophenolic acid and aflatoxin M1 mycotoxins in blue-veined cheeses. Food Control 47:634–640
Frisvad JC, Smedsgaard J, Larsen TO, Samson RA (2004) Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol 49:201–241
Martín JF, Liras P (2016) Evolutionary formation of gene clusters by reorganization: the meleagrin/roquefortine paradigm in different fungi. Appl Microbiol Biotechnol 100:1579–1587
Li SM (2009) Evolution of aromatic prenyltransferases in the biosynthesis of indole derivatives. Phytochemistry 70:1746–1757
Ohmomo S, Oguma K, Ohashi T, Abe M (1978) Isolation of a new indole alkaloid, roquefortine D from cultures of Penicillium roqueforti. Agric Biol Chem 42:2387–2389
Reshetilova TA, Vinokurova NG, Khmelenina VN, Kozlovsky AG (1995) The role of roquefortine in the synthesis of alkaloids meleagrin, glandicolines A and B, and oxaline in fungi Penicillium glandicola and P. atramentosum. Microbiology 64:27–29
Martín JF, Casqueiro J, Liras P (2005) Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr Opin Microbiol 8:282–293
Moreau S, Gaudemer A, Lablache-Combier A, Biguet J (1976) Metabolites de Penicillium roqueforti: PR-toxine et metabolites associes. Tetrahedron Lett 11:833–834
Chang SC, Lu KL, Yeh SF (1993) Secondary metabolites resulting from degradation of PR-toxin by Penicillium roqueforti. Appl Environ Microbiol 59:981–986
Overy DP, Nielsen KF, Smedsgaard J (2005) Roquefortine/oxaline biosynthesis pathways metabolites in Penicillium ser Corymbifera: in planta production and implications for competitive fitness. J Chem Ecol 31:2373–2390
Omura S, Inokoshi J, Uchida R, Shiomi K, Masuma R, Kawakubo R, Tanaka H, Iwai Y, Kosemura S, Yamamura S (1996) Andrastins A-C, new protein farnesyltransferase inhibitors produced by Penicillium sp. FO-3929. I. Producing strain, fermentation, isolation, and biological. Tetrahedron Lett 37:1265–1268
Vilella D, Sánchez M, Platas G, Salazar O, Genilloud O, Royo I, Cascales C, Martín I, Díez T, Silverman KC, Lingham RB, Singh SB, Jayasuriya H, Peláez F (2000) Inhibitors of farnesylation of Ras from a microbial natural products screening program. J Ind Microbiol Biotechnol 25:315–327
Matsuda Y, Abe I (2014) Meroterpenoids. In: Martín JF, García-Estrada C, Zeilinger S (eds) Biosynthesis and molecular genetics of fungal secondary metabolites. Springer, New York
Uchida R, Shiomi K, Inokoshi J, Sunazuka T, Tanaka H, Iwai Y, Takayanagi H, Omura S (1996) Andrastins A-C, new protein farnesyltransferase inhibitors produced by Penicillium sp. FO-3929. II. Structure elucidation and biosynthesis. J Antibiot (Tokyo) 49:418–424
Uchida R, Shiomi K, Inokoshi J, Tanakaf H, Iwai Y, Omura S (1996) Andrastin D, Novel protein farnesyltransferase inhibitor produced by Penicillium sp. FO-3929. J Antibiot (Tokyo) 49:1278–1280
Nicoletti R, Ciavatta L, Buommino E, Tufano MA (2008) Antitumor extrolites produced by Penicillium species. Int J Biomed Pharm Sci 2:1
Matsuda Y, Awakawa T, Abe I (2013) Reconstituted biosynthesis of fungal meroterpenoid andrastin A. Tetrahedron 69:8199e8204
Matsuda Y, Awakawa T, Itoh T, Wakimoto T, Kushiro T, Fujii I, Ebizuka Y, Abe I (2012) Terretonin biosynthesis requires methylation as essential step for cyclization. Chembiochem 13:1738–17341
Artigot MP, Loiseau N, Laffitte J, Mas-Reguieg L, Bartman CD, Doerfler DL, Bird BA, Remaley AT, Peace JN, Campbell IM (1981) Mycophenolic acid production by Penicillium brevicompactum on solid media. Appl Environ Microbiol 41:729–736
Bentley R (2000) Mycophenolic acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem Rev 100:3801–3826
Meier-Kriesche HU, Li S, Gruessner RW, Fung JJ, Bustami RT, Barr ML, Leichtman AB (2006) Immunosuppression: evolution in practice and trends, 1994–2004. Am J Transplant 6:1111–1131
Borroto-Esoda K, Myrick F, Feng J, Jeffrey J, Furman P (2004) In vitro combination of amdoxovir and the inosine monophosphate dehydrogenase inhibitors mycophenolic acid and ribavirin demonstrates potent activity against wild-type and drug-resistant variants of human immunodeficiency virus type 1. Antimicrob Ag Chemother 48:4387–4394
Diamond MS, Zachariah M, Harris E (2002) Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304:211–221
Nicoletti R, De Stefano M, De Stefano S, Trincone A, Marziano F (2004) Identification of fungitoxic metabolites produced by some Penicillium isolates antagonistic to Rhizoctonia solani. Mycopathologia 158:465–474
Tressler RJ, Garvin LJ, Slate DL (1994) Anti‐tumor activity of mycophenolate mofetil against human and mouse tumors in vivo. Int J Cancer 57:568–573
Epinette WW, Parker CM, Jones EL, Greist MC (1987) Mycophenolic acid for psoriasis. A review of pharmacology, long-term efficacy, and safety. J Am Acad Dermatol 17:962–971
Nulton CP, Naworal JD, Campbell IM, Grotzinger EW (1976) A combined radiogas chromatograph/mass spectrometer detects intermediates in mycophenolic acid biosynthesis. Anal Biochem 75:219–233
Regueira TB, Kildgaard KR, Hansen BG, Mortensen UH, Hertweck C, Nielsen J (2011) Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl Environ Microbiol 77:3035–3043
Zhang W, Cao S, Qiu L, Qi F, Li Z, Yang Y, Huang S, Bai F, Liu C, Wan X, Li S (2015) Functional characterization of MpaG′, the O-methyltransferase involved in the biosynthesis of mycophenolic acid. Chembiochem 16:565–569
Hansen BG, Salomonsen B, Nielsen MT, Nielsen JB, Hansen NB, Nielsen K, Regueira TB, Nielsen J, Patil KR, Mortensen UH (2011) Versatile enzyme expression and characterization system for Aspergillus nidulans with the Penicillium brevicompactum polyketide synthetase gene from the mycophenolic acid gene cluster as a test case. Appl Environ Microbiol 77:3044–3051
Hansen BG, Genee HJ, Kaas CS, Nielsen JB, Regueira TB, Mortensen UH, Frisvad JC, Patil KR (2011) A new class of IMP dehydrogenase with a role in self-resistance of mycophenolic acid producing fungi. BMC Microbiol 11:202
Rigbers O, Lin S-M (2008) Ergot alkaloid biosynthesis in Aspergillus fumigatus overproduction and biochemical characterization of a 4-dimethylallyltryptophan N-methyltransferase. J Biol Chem 283:26859–26868
Tudzynski P, Neubauer L (2014) Ergot Alkaloids. In: Martín JF, García-Estrada C, Zeilinger S (eds) Biosynthesis and molecular genetics of fungal secondary metabolites. Springer, New York
Lorenz N, Haarmann T, Paqoutová S, Jung M, Tudzynski P (2009) The ergot alkaloid gene cluster: functional analyses and evolutionary aspects. Phytochemistry 70:1822–1832
Panaccione DG (2010) Ergot alkaloids. In: Hofrichter M (ed) The mycota. Springer, Berlin
Lorenz N, Olnovská J, Kulc M, Tudzynski P (2010) Alkaloid cluster gene ccsA of the ergot fungus Claviceps purpurea encodes chanoclavine I synthase, a Xavin adenine dinucleotide-containing oxidoreductase mediating the transformation of N-methyl-dimethylallyltryptophan to chanoclavine I. Appl Environ Microbiol 76:1822–1830
Goetz KE, Coyle CM, Cheng JZ, O’Connor SE, Panaccione DG (2011) Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Curr Genet 57:201–211
Wallwey C, Matuschek M, Li S (2010) Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Arch Microbiol 192:127–134
Wallwey C, Matuschek M, Xie X, Li S (2010) Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I aldehyde to festuclavine by the festuclavine synthase FgaFS in the presence of the old yellow enzyme FgaOx3. Org Biomol Chem 8:3500–3508
Cheng JZ, Coyle CM, Panaccione DG, O’Connor SE (2010) Controlling a structural branch point in ergot alkaloid biosynthesis. J Am Chem Soc 132:12835–12837
Matuschek M, Wallwey C, Xie X, Li SM (2011) New insights into ergot alkaloid biosynthesis in Claviceps purpurea: an agroclavine synthase EasG catalyses, via a non-enzymatic adduct with reduced glutathione, the conversion of chanoclavine-I aldehyde to agroclavine. Org Biomol Chem 9:4328–4335
Sorensen JL, Nielsen KF, Sondergaard TE (2012) Redirection of pigment biosynthesis to isocoumarins in Fusarium. Fungal Genet Biol 49:613–618
Puel O, Galtier P, Oswald IP (2010) Biosynthesis and toxicological effects of patulin. Toxins 2:613–631
Houbraken J, Samson RA, Frisvad JC (2006) Byssochlamys: significance of heat resistance and mycotoxin production. Adv Exp Med Biol 571:211–224
Varga J, Due M, Frisvad J, Samson RA (2007) Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Stud Mycol 59:89–106
McKinley ER, Carlton WW (1991) Patulin. In: Salunkhe DK, Sharma RP (eds) Mycotoxins and phytoalexins. CRC Press, Boca Raton
Beck J, Ripka S, Siegner A, Schiltz E, Schweizer E (1990) The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur J Biochem 192:487–498
Wang IK, Reeves C, Gaucher GM (1991) Isolation and sequencing of a genomic DNA clone containing the 3′terminus of the 6-methylsalicylic acid polyketide synthetase gene of Penicillium urticae. Can J Microbiol 37:86–95
Dombrink-Kurtzman MA (2007) The sequence of the isoepoxydon dehydrogenase gene of the patulin biosynthetic pathway in Penicillium species. Antonie Van Leeuwenhoek 91:179–189
Artigot MP, Loiseau N, Laffitte J, Mas-Reguieg L, Tadrist S, Oswald IP, Puel O (2009) Molecular cloning and functional characterization of two CYP619 cytochrome P450s involved in biosynthesis of patulin in Aspergillus clavatus. Microbiology 155:1738–1747
Abbas A, Dobson ADW (2011) Yeasts and molds: Penicillium camemberti. In: Funquay JW (ed) Encyclopedia of dairy sciences. Academic, San Diego
Chang PK, Horn BW, Dorner JW (2009) Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet Biol 46:176–182
Liu X, Walsh CT (2009) Cyclopiazonic acid biosynthesis in Aspergillus sp.: characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-L-tryptophan. Biochemistry 48:8746–8757
Liu X, Walsh CT (2009) Characterization of cyclo-acetoacetyl-L-tryptophan dimethylallyltransferase in cyclopiazonic acid biosynthesis: substrate promiscuity and site directed mutagenesis studies. Biochemistry 48:11032–11044
Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y (2008) Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet Biol 45:1608–1615
Holzapfel CW, Wilkins DC (1971) On the biosynthesis of cyclopiazonic acid. Phitochem 10:351–358
Shinohara Y, Tokuoka M, Koyama Y (2011) Functional analysis of the cyclopiazonic acid biosynthesis gene cluster in Aspergillus oryzae RIB 40. Biosci Biotechnol Biochem 75:2249–2252
Kato N, Tokuoka M, Shinohara Y, Kawatani M, Uramoto M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Takahashi S, Koyama Y, Osada H (2011) Genetic safeguard against mycotoxin cyclopiazonic acid production in Aspergillus oryzae. Chembiochem 12:1376–1382
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this entry
Cite this entry
Martín, J.F., Liras, P. (2017). Secondary Metabolites in Cheese Fungi. In: Mérillon, JM., Ramawat, K. (eds) Fungal Metabolites. Reference Series in Phytochemistry. Springer, Cham. https://doi.org/10.1007/978-3-319-25001-4_37
Download citation
DOI: https://doi.org/10.1007/978-3-319-25001-4_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-25000-7
Online ISBN: 978-3-319-25001-4
eBook Packages: Chemistry and Materials ScienceReference Module Physical and Materials ScienceReference Module Chemistry, Materials and Physics