Abstract
Microscopy of plant cells has evolved greatly within the past 50 years. Advances in live cell imaging, automation, optics, video microscopy, and the need for high content studies has stimulated the development of computational tools for manipulating, managing, and interpreting quantitative data. These tools automatically and semiautomatically determine, sizes, signal intensities, velocities, classes, and many other features of cells and subcellular structures. Quantitative methods provide data that is a basis for mathematical models and statistical analyses that lead the way to a quantitative systems outlook to cell biology. Four-dimensional video analysis provides vital data concerning the often ignored temporal dynamics within a cell. Here, we will review studies employing technology to detect regions of interest using segmentation, classify data using machine learning and track dynamics in living cells using video analysis. Many of the live cell studies presented would have been impractical without these advanced computational techniques. These examples illustrate the utility and potential for video bioinformatics to augment our knowledge of the dynamics of cells and cellular components in plants.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Video bioinformatics is a relatively new field which can be described as the automated processing, analysis, and mining of biological spatiotemporal information from videos obtained [1]. Advancements in the field of computer vision have given biologists the ability to quantify spatial and temporal dynamics and to do so in a semi automatic and automatic manner. The challenges that arise from bioimage informatics become increasingly more complicated with the addition of the time dimension. Both techniques share very similar applications and challenges including detection of regions of interest (ROIs) via segmentation , registering images, subcellular localization determination, and dealing with large amounts of image data. Here we will discuss the challenges in plant cell biology that can be addressed using automatic quantitative tools such as image and video bioinformatics and the current shortcomings that need to be improved upon as we continue to discover and describe dynamic biological phenomena at the cellular level.
Most of the image data collected to date have been interpreted subjectively, allowing for personal interpretation and a loss of objectivity [2]. In the pursuit of biological discovery, we strive for objectivity and quantitative data that we can manipulate and use to better uncover genuine biological phenomena versus artifacts or biased results. Phenotypes can be continuous and cover a large spectrum, for example, when using chemical genomics to dissect conserved cellular processes [3]. Varying concentrations of bioactive compounds or drugs can illicit proportional phenotypes [4]. Therefore, the need for quantitative image and video data is essential when interpreting data on any time scale.
Ultimately, the quantified data demonstrate the most utility when subjected to statistical analysis. Therefore, it makes sense to quantify enough data to allow for a statistically valuable sample size. This often requires large amounts of data that need. Additionally, high-throughput screens have much to gain from using quantitative metrics to screen for valuable phenotypes [5]. To meet these challenges in a practical manner, quantification needs to be automated. Automation provides decreased analysis time, and allows for reduced inter and intrauser variability. The ability to provide a consistent analysis from sample to sample provides more reliable data. Reliable data are essential to fully understand the nature of any dynamic subcellular process. Dynamic cellular phenomena such as cell division , lipid dynamics , plant defense processes , and cell wall biosynthesis , often require the measurement of various static and dynamic features [6, 7, 8]. The automated detection, tracking, and analysis of these regions of interest summarizes the major goals of video bioinformatics in a cell biological context.
Live cell imaging has become an indispensable tool for discovery throughout the basic and applied sciences. This relatively recent technique has allowed for real-time observation and quantification of dynamic biological processes on the scale of nanometers to meters and milliseconds to days [9]. The advent of green fluorescent protein (GFP) has ignited a live cell imaging revolution and has subsequently enabled the capturing of in vivo spatial and temporal dynamics [10]. Because of their versatility, GFP and its derivatives have become ubiquitous in molecular and cell biology generating large quantities of image and video data. Many of the technical advancements in bioimaging have come from a prolific collaboration between the biological sciences and engineering. The cooperation of these two disciplines has produced indispensable tools to cell biology such as the laser scanning confocal microscope [11], spinning disk confocal microscope [12], mulptiphoton microscope [13], variable-angle epifluorescence microscope (VAEM) [14], and STORM [15] to name a few. All of these imaging modalities produce large quantities of complex multidimensional data. Scientists need to work together with engineers to dissect, manage, manipulate, and ultimately make sense of the image data collected. Practitioners of both disciplines, while still working to improve the acquisition hardware, are also working together to manage and analyze the large amounts of quantifiable image data.
The traditional method of quantifying image data is to manually draw regions of interest containing the biologically relevant information. This manual measurement is the most popular method of image quantification. Software tools including ImageJ and spin-offs of ImageJ such as Fiji are free [16]. Subcellular phenotyping is time consuming and impractical when performing high-throughput screens, which are necessary for most cell biologists. This data load is only increased when analyzing videos. A recent push toward automation has favored the use of automated microscopes, and robots that perform automated high-throughput sample preparation [17]. This has lead to the development and implementation of and automated semiautomated tools that require modest to little user input [18, 19]. Automated methods can be more consistent and faster since the user does not have to provide information. However, this lack of user input can also lead to reduced flexibility and robustness. On the other hand, semiautomated methods are flexible and possibly more robust due to user input, but can often be slower because the user has to provide prior information to the software. As this analysis becomes more user friendly and practical, the ability to apply a single software tool to multiple biological problems including multidimensional data, will be favored by the biologist thereby most likely favoring the semiautomated methods.
Bioimage informatics has experienced a recent surge in popularity due to the advent of automated microscopes and the subsequent burst of image data. Engineers had to develop methods to manage and interpret large amounts of image data generated by these automated systems. Bioimage informatics relies on several engineering disciplines including computer vision, machine learning , image-processing image analysis and pattern recognition [20]. The application of theses methods aids biologists is rapid detection, quantification, and classification of biological phenomena. Bioimage informatics is generally concerned with two-dimensional data, in the x and y planes, though it is possible to deal with three-dimensional, X, Y, and Z, and four-dimensional data X, Y, Z, and frequency domain [21]. Using these dimensions, data can be accurately extracted when computational techniques are properly applied.
Here we discuss the application of three fields of computer vision as they pertain to plant cell biology including, segmentation , machine learning, and video analysis, while highlighting the recent advances that were possible due to the collaboration of biologists and engineers.
2 Segmentation : Detecting Regions of Interest
Ultimately, biologists want to be able to extract data from acquired multidimensional images. However, the biologist needs to be able to identify those subregions within the image that hold the most data and that are therefore more important. As expert biologists, we can accurately identify the interesting regions of an image intuitively. Segmentation is the process of partitioning the regions of an image into segments [22]. Before we can extract data, we must first detect the objects or regions that are biologically meaningful. Biological images are acquired with various modalities and therefore one segmentation method is not going to be effective for all cases Specialized methods must be applied to each case. Much progress has been made in the domain of confocal microscopy . Bright fluorophores allow for high-contrast images that facilitate robust segmentation. In the realm of plant cell biology, many organelles and protein localization sites resemble bright spots or blobs. This is due to the light diffraction limit which limits the resolution of light microscopy at 250 nm, making small objects appear as fuzzy blobs [23]. Quantifying the number or size of these bright blobs is often done manually and can take several days. Simple segmentation can greatly improve this process which can then lead to feature extraction in both static and dynamic datasets.
Static 2D images are by far the most popular type of microscopy data to analyze because of their relatively short acquisition and analysis time. The majority of subcellular imaging is focused on the localization of proteins of interest. Using fluorescent markers fused to proteins of interest and dyes, cell biologists can understand the proteins that are involved with biological processes by monitoring the abundance, size shape, and localization within organelles. Organelles are of interest to cell biologist because of their diverse and extremely important roles in plant development, homeostasis, and stress responses. Automatic tools are being developed and used to quantify protein localization and spatial features of discrete compartments [24]. Organelles often manifest as punctate dots when imaged using fluorescent confocal laser scanning microscopy . These dots are then quantified per cell area and features extracted such as area, intensity, and number of compartments [19]. Salomon et al. used such a tool to quantify the response of various endomembrane compartment to bacterial infection, cold stress, and dark treatment [25]. Crucial information can also be garnered from the cells themselves. Cell borders can be detected when labeled and size as well as shape information analyzed automatically [25]. This information can then be used to track cell growth and development . Segmentation is the first crucial step to extracting quantitative information from live cell imaging data .
Cells exist in four dimensions, X, Y, Z, and time. If cell biologists want the full complement of information from imaging data, we have to consider all four of these dimensions. Collecting and processing 3D data is computationally more expensive and more difficult to manage but can yield a greater understanding of spatial information. Most confocal microscopes can easily collect data in the Z direction and 3D reconstructions are relatively easy to do now with the capable software. Most of the images captured of dividing plant cell are two-dimensional leaving out the critical third dimension. Miart et al. used 3D constructions of a growing cell plate to understand the role of cellulose synthase complexes in cell plate formation by analyzing time lapse video data of fluorescently labeled cellulose synthase complexes [26]. Although these analyses did not take advantage of automated quantification, the visualization of the 3D cell plate greatly contributed to the understanding of how cellulose synthase complexes (CESAs) are involved in cell plate formation [26]. Quantifying temporal dynamics in a study such as this would lend insight into how fast this process happens and perhaps how the population of CESA complexes shifts from a homeostatic role to an actively dividing role.
Four-dimensional data including 3D movies of cellular phenomena, will become more popular as the tools to analyze this data become more sophisticated and more user friendly. Automated 4D analysis tools are already being used by cell biologists to analyze trichome development [9]. This system extracts the leaf surface, segmenting the mid-plane of the young leaf and detects the developing trichomes using a Hough transform which can detect circles [27]. One 3D image is registered to the next 3D image in the time series to maintain consistency and to track and compare its growth over time [28]. These tools will need to be adapted from analyzing gross morphology to tracking moving cellular structures over time.
3 Machine Learning Allows Automatic Classification of Cellular Components
Machine learning is a subdiscipline of artificial intelligence that is focused on the development of algorithms that can learn from given data [29]. These methods often require the use of training data and user input to learn how the data should be classified. Training the algorithm allows it to correctly identify the class to which each sample belongs. A simple example is the spam filter on most email accounts that can discern between those messages that are spam and those that are important.
A logical application of machine learning in cell biology was determining the subcellular localization of fluorescent markers based on extracted features. Traditionally, cell biologists have to colocalize their protein of interest and markers of known localization to determine where the protein is located. Biologists could simply analyze a confocal micrograph with a machine learning program and receive the location of their protein of interest. An additional advantage to the machine learning methods over traditional cellular methods, other than reduced time is that these methods provide statistics as to how likely the determined localization is to be true [30]. Though there seems to be reasonable progress in determining subcellular localization using machine learning, the biological community has yet to adopt the methodology. Prediction of subcellular localization will streamline experimental design and support traditional colocalization assays.
Machine learning is a powerful tool for gene discovery and organelle dynamics. It can help uncover relationships that we otherwise could not. Because organelle dynamics can be complex and variable, it is valuable to simplify dynamics. Using Baysian networks, Collinet et al. found that endosome number, size, concentration of cargo, and position are mediated by genetic regulation and not random [31]. Furthermore, they used this method to discover novel components regulating endocytosis by clustering endocytic phenotypes caused by screening siRNA libraries [31]. Statistical analysis was similarly used to summarize and classify organelle movement in Arabidopsis stomata (Higaki 2012). The result is an atlas of organelle movement in stomata that can be compared to various conditions. Organelle movement patterns were compared between open and closed stomata revealing differences in ER position in response to stomatal opening . These new findings emphasize the need for statistical methods to manage complex data and present this data in forms we can easily understand and manipulate.
Though we are interested in cell autonomous processes, cells do not exist in a vacuum. We are also interested in how a cell influences the development and function of its neighboring cells. To address this challenge, segmentation coupled with machine learning was used to jointly detect and classify cell types in whole tissues. three-dimensional images of propidium iodide stained roots were used to automatically find cell files in longitudinal and transverse sections using watershed segmentation and a support vector machine to classify cell types [32]. An alternative approach used histological sections of Arabidopsis hypocotyls to differentiate tissue layers and predict the location of phloem bundle cells [33]. The true utility of these tools will be realized when they are used to compare wild-type cell profiles with mutants, possibly being used in large content screening.
4 Quantifying Temporal Dynamics Adds a New Dimension of Data
Once an object has been detected and classified, it is often very important to follow its movement through time and space. This extremely important problem of tracking has been tackled by many engineers developing the field of computer vision. A multitude of tools are available for tracking cells and organelles, most of these being manual and semi automated [19]. Tracking organelles are difficult because rarely do they have a straight forward movement model. It is because of the diversity and variability of tracking problems that semiautomated methods are the most widely used. Common problems include object moving out of the plane of focus when using 2D images. It is because of this issue that 3D movies are such valuable data sets [34]. Therefore, automatically tracking object in a 3D image set is an invaluable tool [35]. Other challenges include maintaining identity when two objects fuse or break off from one another, and maintaining multiple tracks at the same time. A perfect tracking algorithm would overcome all of these problems, while maintaining minimal user input and accurate segmentation .
The purpose of quantifying movement and movement patterns is to gain useful biological insight such as diffusion rates , types of motion including Brownian motion , non-Brownian motion, confined motion, directed motion, or anomalous diffusion [36]. Ung et al. correlated multiple dynamic features which suggested that when tobacco pollen tubes where treated with specific bioactive compounds the contained Golgi bodies increased in size and this increase in size was correlated to an increase in signal intensity and a decrease in straightness suggesting that these were possibly multiple fused Golgi and that this fusion disrupted movement [19]. Similar correlations were made by collinet et al. when examining endocytosis in mammalian cells [31]. Indeed these data are consistent with the hypothesis proposed by Hamilton et al. (2007) including conservation of surface area, measurement of volume, flux across a membrane, the role of pressure and tension and vesicle fusion . These biological details would not be obtainable without quantitative video analysis.
Although each challenge was presented separately, they are by no means mutually exclusive. The vast majority of image analysis problems require identification of regions of interest before they can be quantified, tracked, or classified.
5 Opportunities for Innovation in Cellular Video Bioinformatics
As video bioinformatics tools become increasingly accurate and biologist friendly, they will be more widely used in biological studies. The future of video analysis is moving toward automatic quantification of cellular dynamics in four dimensions (3D time lapse images ). The amount of data that can be extracted from 3D movies will increase with the availability and ease of use of software. Biologists will be able to quantify difference in movement possibly identifying underlying principals of movement and other components essential to cellular dynamics. As these video analysis tools become more fully automated, it will be more practical to screen for factors that influence dynamics. In this manner, biologists will be able to directly screen for changes in cellular dynamics.
Creating the tools to quantify cellular dynamics is futile unless biologists use them to produce data. The pipeline from engineer to the biological community needs to be stronger. This could be enhanced by taking advantage of open source repositories of image analysis tools. A small pool of these repositories currently available and will grow in popularity as the need for these programs becomes greater [37]. As we take advantages of quantitative methods we will produce large amounts of data that has the potential to fuel mathematical models or other future studies [38]. Many mathematical models require know numerical parameters to be of use.
As live cell imaging modalities and acquisition methods become more advanced including super resolution methods and as biological systems change, our analysis methods will to have to adapt. In the future, these efforts will be spearheaded by a handful of interdisciplinary scientists that will be trained in biological principals, experimental design, computer programming, and image analysis’s tool design. Future biologists will have to be well-versed in computer programming basics and be able to design tools that are applicable to their specific research topic, while having a basic understanding of the appropriate algorithms all while being able to communicate with engineers. Engineers on the other hand, will have to understand biological limitations, know which features are useful, experimental design, and acquisition methods (Figs. 10.1, 10.2 and 10.3).
Pattern recognition of membrane compartments in leaf epidermal tissue at cellular and subcellular resolution. Merged confocal laser microscopic images show Arabidopsis leaf epidermal cells. Top section GFP-2xFYVE plants were imaged at 403 magnification (scale bar = 20 mm) and images analyzed by the endomembrane script. Bottom section GFP-PEN1 plants were imaged at 203 magnification (scale bar = 50 mm) and images analyzed by the plasma membrane microdomain script. a and b, Merged pseudo images. c and d, Recognition of epidermal cells is shown by colored lines. e, Recognition of GFP-2xFYVE-labeled endosomal compartments is shown by colored circles. f, Recognition of (b). Graminis induced GFP-PEN1 accumulation beneath attempted fungal entry sites (indicated by arrowheads) is shown by colored circles. Color coding is random, different colors indicate individual cells compartments [25]
Cell wall stained by Propidium iodide : a Raw data of a test image. b After anisotropic diffusion . c Watershed segmentation (from local minima) shown in random color. d Final segmentation result. e Classification of the segments. f Manually generated ground-truth for segmentation and classification (modified from Liu et al. [32])
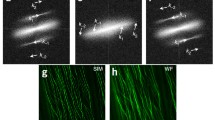
Representation obtained by automatic tracking method of the same tracking result from a cell tagged with GFP-Rab6 . a 3D trajectories of the detected objects. The color indicates the time window when an object was detected. The z projection of the first 3D image of the time series is shown below the trajectories to visualize the cell morphology (modified from Racine et al. [35])
References
Bhanu B (2009) IGERT Program. In: The UC Riverside integrated graduate education research and training program in video bioinformatics. Univeristy of California, Riverside, 26 Oct 2009. Web. 24 Mar 2014
Peterson RC, Wolffsohn JS (2007) Sensitivity and reliability of objective image analysis compared to subjective grading of bulbar hyperaemia. Br J Ophthalmol 91(11):1464–1466
Drakakaki G, Robert S, Szatmari AM, Brown MQ, Nagawa S, Van Damme D, Leonard M, Yang Z, Girke T, Schmid SL, Russinova E, Friml J, Raikhel NV, Hicks GR (2011) Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc Natl Acad Sci USA 108(43):17850–17855
Robert S, Chary SN, Drakakaki G, Li S, Yang Z, Raikhel NV, Hicks GR (2008) Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc Natl Acad Sci USA 105(24):8464–8469
Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S (2012) Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24(10):4205–4219
Tataw OM, Liu M, Roy-Chowdhurry A, Yadav RK, Reddy GV (2010) Pattern analysis of stem cell growth dynamics in the shoot apex of arabidopsis. In: 17th IEEE international conference on image processing (ICIP), pp 3617–3620
Liu J, Elmore JM, Lin ZJ, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9(2):137–146
Sampathkumar A, Gutierrez R, McFarlane HE, Bringmann M, Lindeboom J, Emons AM, Samuels L, Ketelaar T, Ehrhardt DW, Persson S (2013) Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol 162(2):675–688
Domozych DS (2012) The quest for four-dimensional imaging in plant cell biology: it’s just a matter of time. Ann Bot 110(2):461–474
Brandizzi F, Fricker M, Hawes C (2002) A greener world: the revolution in plant bioimaging. Nat Rev Mol Cell Biol 3(7):520–530
Rajadhyaksha M, Anderson R, Webb RH (1999) Video-rate confocal scanning laser microscope for imaging human tissues; In Vivo. Appl Opt 38(10):2105–2115
Nakano A (2002) Spinning-disk confocal microscopy–a cutting-edge tool for imaging of membrane traffic. Cell Struct Funct 27(5):349–355
Meyer AJ, Fricker MD (2000) Direct measurement of glutathione in epidermal cells of intact Arabidopsis roots by two-photon laser scanning microscopy. J Microsc 198(3):174–181
Konopka CA, Bednarek SY (2008) Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J 53(1):186–196
Rust MJ, Bates M, Zhuang X (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3(10):793–796
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682
Hicks GR, Raikhel NV (2009) Opportunities and challenges in plant chemical biology. Nat Chem Biol 5(5):268–272
Kuehn M, Hausner M, Bungartz HJ, Wagner M, Wilderer PA, Wuertz S (1998) Automated confocal laser scanning microscopy and semiautomated image processing for analysis of biofilms. Appl Environ Microbiol 64(11):4115–4127
Ung N, Brown MQ, Hicks GR, Raikhel NV (2012) An approach to quantify endomembrane dynamics in pollen utilizing bioactive chemicals. Mol Plant
Eils R, Athale C (2003) Computational imaging in cell biology. J Cell Biol 161(3):477–481
Cheung G, Cousin MA (2011) Quantitative analysis of synaptic vesicle pool replenishment in cultured cerebellar granule neurons using FM dyes. J Vis Exp (57)
Shapiro LG, Stockman GC (2001) Computer Vision. New Jersey, Prentice-Hall, pp 279–325. ISBN 0-13-030796-3
de Lange F, Cambi A, Huijbens R, de Bakker B, Rensen W, Garcia-Parajo M, van Hulst N, Figdor CG (2001) Cell biology beyond the diffraction limit: near-field scanning optical microscopy. J Cell Sci 114(23):4153–4160
Sethuraman V, Taylor S, Pridmore T, French A, Wells D (2009) Segmentation and tracking of confocal images of Arabidopsis thaliana root cells using automatically-initialized Network Snakes. In: 3rd international conference on bioinformatics and biomedical engineering, 2009. ICBBE 2009
Salomon S, Grunewald D, Stüber K, Schaaf S, MacLean D, Schulze-Lefert P, Robatzek S (2010) High-throughput confocal imaging of intact live tissue enables quantification of membrane trafficking in arabidopsis. Plant physiol 154(3):1096
Miart F, Desprez T, Biot E, Morin H, Belcram K, Höfte H, Gonneau M, Vernhettes S (2013) Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J
Illingworth J, Kittler J (1987) The adaptive Hough transform. IEEE Trans Pattern Anal Mach Intell 5:690–698
Bensch R, Ronneberger O, Greese B, Fleck C, Wester K, Hulskamp M, Burkhardt H (2009) Image analysis of arabidopsis trichome patterning in 4D confocal datasets. In: IEEE international symposium on biomedical imaging: from nano to macro, 2009. ISBI’09, pp 742–745
Goldberg DE, Holland JH (1988) Genetic algorithms and machine learning. Mach Learn 3(2):95–99
Hua S, Sun Z (2001) Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17(8):721–728
Collinet C, Stöter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, Nagel WE, Fava E, Kalaidzidis Y, Zerial M (2010) Systems survey of endocytosis by multiparametric image analysis. Nature 464(7286):243–249
Liu K, Schmidt T, Blein T, Durr J, Palme K, Ronneberger O (2013) Joint 3d cell segmentation and classification in the arabidopsis root using energy minimization and shape priors. In: IEEE 10th international symposium on biomedical imaging (ISBI), pp 422–425
Sankar M, Nieminen K, Ragni L, Xenarios I, Hardtke CS (2014) Automated quantitative histology reveals vascular morphodynamics during Arabidopsis hypocotyl secondary growth. eLife 3
Carlsson K, Danielsson P-E, Liljeborg A, Majlöf L, Lenz R, Åslund N (1985) Three-dimensional microscopy using a confocal laser scanning microscope. Opt Lett 10(2):53–55
Racine V, Sachse M, Salamero J, Fraisier V, Trubuil A, Sibarita JB (2007) Visualization and quantification of vesicle trafficking on a three-dimensional cytoskeleton network in living cells. J Microsc 225(Pt 3):214–228
Saxton MJ, Jacobson K (1997) Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct 26(1):373–399
Swedlow JR, Eliceiri KW (2009) Open source bioimage informatics for cell biology. Trends Cell Biol 19(11):656–660
Phillips R, Milo R (2009) A feeling for the numbers in biology. Proc Natl Acad Sci USA 106(51):21465–21471
Acknowledgment
This work was supported in part by the National Science Foundation (NSF) Integrative Graduate Education and Research Traineeship (IGERT) in Video Bioinformatics (DGE-0903667) and NSF Graduate Research Fellowship Program (GRFP). Nolan Ung is an IGERT and GRFP Fellow.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Ung, N., Raikhel, N.V. (2015). Video Bioinformatics: A New Dimension in Quantifying Plant Cell Dynamics. In: Bhanu, B., Talbot, P. (eds) Video Bioinformatics. Computational Biology, vol 22. Springer, Cham. https://doi.org/10.1007/978-3-319-23724-4_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-23724-4_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23723-7
Online ISBN: 978-3-319-23724-4
eBook Packages: Computer ScienceComputer Science (R0)